Abstract
Inter-helix hydrogen bonding involving asparagine (Asn, N), glutamine (Gln, Q), aspartic acid (Asp, D) or glutamic acid (Glu, E) can drive efficient di- or trimerization of transmembrane helices in detergent micelles and lipid bilayers. Likewise, Asn–Asn and Asp–Asp pairs can promote the formation of helical hairpins during translocon-mediated membrane protein assembly in the endoplasmic reticulum. By in vitro translation of model integral membrane protein constructs in the presence of rough microsomes, we show that Asn- or Asp-mediated interactions with a neighbouring transmembrane helix can enhance the membrane insertion efficiency of a marginally hydrophobic transmembrane segment. Our observations suggest that inter-helix hydrogen bonds can form during Sec61 translocon-assisted insertion and thus could be important for membrane protein assembly.
Keywords: endoplasmic reticulum, helix–helix interaction, membrane protein assembly, Sec61, transmembrane helix
Introduction
In eukaryotic cells, most α-helical membrane proteins insert co-translationally, fold and oligomerize in the endoplasmic reticulum membrane (von Heijne, 2003). The initial insertion step is mediated by the Sec61 translocon, a hetero-trimeric complex that forms a protein-conducting channel for secreting water-soluble proteins into the lumen of the endoplasmic reticulum and shunting integral membrane proteins laterally into the endoplasmic reticulum membrane (Alder & Johnson, 2004; White & von Heijne, 2004).
In a recent effort to define the sequence characteristics required for the insertion of transmembrane α-helices into the endoplasmic reticulum membrane (Hessa et al, 2005a), we challenged the Sec61 translocon with a large set of systematically engineered hydrophobic model segments (H segments) and were able to derive a first ‘biological' hydrophobicity scale for membrane proteins on the basis of the measured membrane insertion efficiency of the H segments. The study of Hessa et al (2005a) focused on the insertion of a single transmembrane segment; studies of natural, but multi-spanning membrane proteins suggest that at least some transmembrane helices need to interact with neighbouring helices in the protein to insert properly into the membrane (Skach et al, 1993; Lin & Addison, 1995; Sato et al, 2002; Heinrich & Rapoport, 2003; Sadlish et al, 2005). Conceivably, a more amino-terminally located transmembrane helix might be held in or near the translocon while a more carboxy-terminally located one is entering, making it possible for the two to partition into the surrounding lipid as a pair rather than as two independent helices (Heinrich & Rapoport, 2003; Sadlish et al, 2005).
It is well established that hydrogen bonding between polar residues such as asparagine (Asn, N) or aspartic acid (Asp, D) can drive helix–helix interactions in both detergent micelles and biological membranes (Choma et al, 2000; Zhou et al, 2000, 2001; Gratkowski et al, 2001), and can also facilitate the formation of helical hairpins during translocon-mediated insertion (Hermansson & von Heijne, 2003). What is not clear, however, is whether and to what extent inter-helix hydrogen bonding can drive the process of transmembrane helix insertion itself, and whether the separation between the two helices in the sequence might influence any such interaction. To address these questions in a quantitative way, we have extended the systematic approach established earlier (Hessa et al, 2005a) to study the effects of mutual helix–helix interactions on the efficiency of membrane insertion. We found that the apparent free energy of membrane insertion of a marginally hydrophobic model transmembrane helix can be decreased by up to 1 kcal/mol by position-specific Asn- or Asp-mediated interactions with a neighbouring, stably inserted transmembrane helix.
Results
Model protein and topology assay
As in earlier studies of protein insertion into the endoplasmic reticulum membrane (Hermansson & von Heijne, 2003; Hessa et al, 2005a), we used the well-characterized Escherichia coli inner membrane protein leader peptidase (Lep) as a model protein. Lep consists of two transmembrane segments (H1, H2) connected by a short cytoplasmic loop (P1), and a large C-terminal cytoplasmic domain (P2; Fig 1A). When expressed in vitro in the presence of dog pancreas rough microsomes, Lep adopts the same topology as in E. coli, with both the N terminus and the C-terminal P2 domain located in the lumen of the microsome (Nilsson & von Heijne, 1993).
Figure 1.
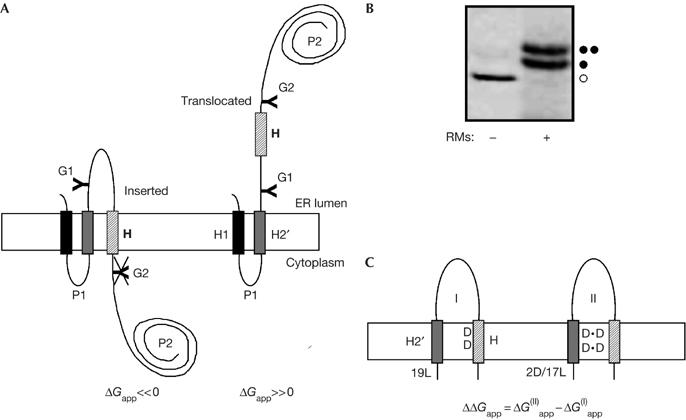
The leader peptidase model protein. (A) Wild-type leader peptidase (Lep) has two transmembrane helices (H1, H2) and a large lumenal domain (P2). It inserts into rough microsomes in an Nlum–Clum orientation. In the studies reported here, the H2 segment was replaced by an H2′ segment of the general composition L19−nNn or L19−nDn (n=0, 1 or 2), and a 19-residue-long H segment (hatched), containing one or two Asn or Asp residues, was inserted into the P2 domain. Asn-X-Thr glycosylation acceptor sites (G1, G2) were introduced on both sides of the H segment, as shown. Constructs in which the H segment was integrated into the endoplasmic reticulum (ER) membrane as a transmembrane helix were glycosylated only on the G1 site (left), whereas those in which the H segment was translocated across the ER membrane were glycosylated on both the G1 and G2 sites (right). (B) In vitro translation in the absence (−) and presence (+) of dog pancreas rough microsomes (RMs) of a construct containing the 2N/17L H2′ segment and the 2N/9L/8A (N3N) H segment (see Table 1). Unglycosylated, single glycosylated and double glycosylated forms of the protein are indicated by an open circle, one filled circle and two filled circles, respectively. For this construct, ΔGapp=0.1 kcal/mol. (C) To measure the effect of Asp- or Asn-mediated interactions between the H2′ and H segments, two constructs were compared for each H segment: one with a uniformly hydrophobic 19-Leu H2′ segment (I) and one with an H2′ segment in which one or two of the Leu residues have been replaced by Asp or Asn residues (II). The interaction free energy is expressed as the difference (ΔΔGapp) in the apparent free energy of insertion of the H segment between the two constructs. In the example shown, the H segment contains two Asp residues and the appropriate number of Leu and Ala residues to make ΔG(I)app≈0 kcal/mol. A, Ala, alanine; D, Asp, aspartic acid; L, Leu, leucine; N, Asn, asparagine; T, Thr, threonine.
In our recent study (Hessa et al, 2005a), we introduced an engineered version of Lep that allowed quantitative measurements of the efficiency of membrane insertion of short polypeptide segments (H segments). Briefly, an H segment was placed in the P2 domain, 150 residues downstream of the H2 transmembrane segment (Fig 1A). In addition, two Asn-X-Thr (threonine, T) acceptor sites for N-linked glycosylation were introduced into the P2 domain: one (G1) between the H2 and H segments, and the other (G2) just downstream of the H segment. After in vitro translation in the presence of dog pancreas rough microsomes (Fig 1B), the degree of membrane integration of a given H segment was quantified by comparing the fractions of single glycosylated (that is, integrated) and double glycosylated (that is, non-integrated) molecules, which we expressed as an apparent free energy of membrane insertion (ΔGapp; see Methods).
Helix–helix interaction assay
To examine possible Asn- or Asp-mediated interactions between the H2′ and H segments, we replaced the endogenous H2 transmembrane segment with simplified 19-residue segments (H2′ segments) and measured changes in the insertion efficiency of Asp- or Asn-containing H segments caused by the presence or absence of Asp or Asn residues in the H2′ segment (Fig 1C). The H2′ segments were designed to insert stably (∼100% insertion) regardless of their Asp or Asn content, whereas the H segments were designed to have about 50% insertion efficiency (ΔGapp≈0 kcal/mol), because the membrane insertion assay is sensitive for ΔGapp measurements in the interval of [−1, +1] kcal/mol. To avoid data interpretation on the basis of absolute ΔGapp values, we calculated ΔΔGapp values, that is, the change in ΔGapp for a given H segment caused by replacing an Asp- or Asn-containing H2′ segment with the 19-Leu (leucine, L) H2′ segment (Fig 1C). A negative ΔΔGapp value means that the H segment inserts more efficiently into the membrane when there are Asp or Asn residues in the H2′ segment as compared with the situation with the uniformly hydrophobic 19-Leu H2′ segment.
The 19-residue H segments, which were designed with the aid of the biological hydrophobicity scale (Hessa et al, 2005a), were composed of Leu, Ala (alanine, A) and one or two Asn or Asp residues such that ΔGapp≈0 kcal/mol (Table 1). Single Asn or Asp residues were placed in position 10 of the H segment, and pairs of Asn or Asp residues were placed in positions 8 and 12, 7 and 13, or 9 and 11. The H2′ segments were designed with the overall composition L19−nNn or L19−nDn (with n=0, 1 or 2) to ensure stable (100%) insertion. The Asn and Asp residues in H2′ were placed either in position 10 or in positions 8 and 12 (that is, one helical turn apart) in H2′. We used the same GGPG…GPGG flanking segments as used previously for both the H2′ and H segments (Hessa et al, 2005a).
Table 1.
Constructs analysed in this study
| H2′ name | H2′ sequence | H name | H sequence | ΔGapp (kcal/mol) |
|---|---|---|---|---|
|
H2′ control experiments | ||||
| Wild type |
…TGASVFPVLAIVLIV… |
3L/16A |
AAAALAAAALAAAALAAA |
0.1; ND |
| CCH2 |
…LAIVLIVSPSAQA+AY… |
3L/16A |
AAAALAAAALAAAALAAA |
−0.2; ND |
| pPL |
pPL signal peptide |
3L/16A |
AAAALAAAALAAAALAAA |
0.1; ND |
| 19L |
LLLLLLLLLLLLLLLLLLL |
3L/16A |
AAAALAAAALAAAALAAA |
0.1; ND |
| 1D/18L |
LLLLLLLLLDLLLLLLLLL |
3L/16A |
AAAALAAAALAAAALAAA |
−0.3; ND |
| 1N/18L |
LLLLLLLLLNLLLLLLLLL |
3L/16A |
AAAALAAAALAAAALAAA |
−0.2; ND |
| 1D/18L |
LLLLLLLDLLLLLLLLLLL |
3L/16A |
AAAALAAAALAAAALAAA |
−0.2; ND |
| 1N/18L |
LLLLLLLNLLLLLLLLLLL |
3L/16A |
AAAALAAAALAAAALAAA |
−0.0; ND |
| 2D/17L |
LLLLLLLDLLLDLLLLLLL |
3L/16A |
AAAALAAAALAAAALAAA |
−0.0; ND |
| 2N/17L |
LLLLLLLNLLLNLLLLLLL |
3L/16A |
AAAALAAAALAAAALAAA |
−0.0; ND |
|
Single Asp and Asn experiments | ||||
| 19L |
LLLLLLLLLLLLLLLLLLL |
1N/5L/13A (N) |
AAAAAAALLNLLLAAAAA |
0.3; 0.8 |
| 1N/18L |
LLLLLLLLLNLLLLLLLLL |
1N/5L/13A (N) |
AAAAAAALLNLLLAAAAA |
0.2; 0.8 |
| CCH2 |
…LAIVLIVSPSAQA+AY… |
1N/5L/13A (N) |
AAAAAAALLNLLLAAAAA |
0.0, ND |
| 19L |
LLLLLLLLLLLLLLLLLLL |
1D/7L/11A (D) |
AAAAAALLLDLLLLAAAA |
0.1; 0.4 |
| 1D/18L |
LLLLLLLLLDLLLLLLLLL |
1D/7L/11A (D) |
AAAAAALLLDLLLLAAAA |
−0.1; 0.3 |
|
Double Asp and Asn experiments | ||||
| 19L |
LLLLLLLLLLLLLLLLLLL |
2N/8L/9A (N5Nl) |
AAAALLNLLLLLNLAAAA |
−0.1; ND |
| 2N/17L |
LLLLLLLNLLLNLLLLLLL |
2N/8L/9A (N5Nl) |
AAAALLNLLLLLNLAAAA |
−0.3; ND |
| 19L |
LLLLLLLLLLLLLLLLLLL |
2N/9L/8A (N5Ns) |
AAAALLNLLLLLNLLAAAA |
ND; −0.5 |
| 2N/17L |
LLLLLLLNLLLNLLLLLLL |
2N/9L/8A (N5Ns) |
AAAALLNLLLLLNLLAAAA |
ND; −0.7 |
| 19L |
LLLLLLLLLLLLLLLLLLL |
2N/9L/8A (N3N) |
AAAALLLNLLLNLLLAAAA |
0.3; 0.6 |
| 2N/17L |
LLLLLLLNLLLNLLLLLLL |
2N/9L/8A (N3N) |
AAAALLLNLLLNLLLAAAA |
0.1; 0.0 |
| 19L |
LLLLLLLLLLLLLLLLLLL |
2N/9L/8A (N1N) |
AAAALLLLNLNLLLLAAAA |
−0.3; −0.2 |
| 2N/17L |
LLLLLLLNLLLNLLLLLLL |
2N/9L/8A (N1N) |
AAAALLLLNLNLLLLAAAA |
−0.5; −0.6 |
| 19L |
LLLLLLLLLLLLLLLLLLL |
2D/11L/6A (D5D) |
AAALLLDLLLLLDLLLAAA |
−0.3; −0.1 |
| 2D/17L |
LLLLLLLDLLLDLLLLLLL |
2D/11L/6A (D5D) |
AAALLLDLLLLLDLLLAAA |
−0.4; −0.2 |
| 19L |
LLLLLLLLLLLLLLLLLLL |
2D/12L/5A (D3D) |
AAALLLLDLLLDLLLLLAA |
0.0; 0.0 |
| 2D/17L |
LLLLLLLDLLLDLLLLLLL |
2D/12L/5A (D3D) |
AAALLLLDLLLDLLLLLAA |
−0.5; −0.9 |
| 19L |
LLLLLLLLLLLLLLLLLLL |
2D/12L/5A (D1Dl) |
AAALLLLLDLDLLLLLLAA |
−1.0; ND |
| 2D/17L |
LLLLLLLDLLLDLLLLLLL |
2D/12L/5A (D1Dl) |
AAALLLLLDLDLLLLLLAA |
−1.1; ND |
| 19L |
LLLLLLLLLLLLLLLLLLL |
2D/11L/6A (D1Ds) |
AAALLLLLDLDLLLLLAAA |
ND; 0.2 |
| 2D/17L | LLLLLLLDLLLDLLLLLLL | 2D/11L/6A (D1Ds) | AAALLLLLDLDLLLLLAAA | ND; −0.1 |
The H2′ and H segments are listed for each construct (abbreviated H segment names in parentheses refer to Figure 2). All H2′ segments (except wild type, CCH2 and pPL) and all H segments have GGPG…GPGG flanks. Two ΔGapp values are given: the first refers to constructs with a 150-residue-long loop between H2′ and H segments and the second to constructs with a 55-residue loop (ND, not done). Pairs of constructs used to calculate the ΔΔGapp values shown in Figure 2 are indicated. Note that the composition of the H segments are in some cases different for the long- and short-loop constructs (indicated by subscripts l and s in the abbreviated H segment names, respectively), necessitated by the requirement that ΔGapp≈0 kcal/mol. The standard deviation in repeat ΔGapp measurements is σ≈±0.1 kcal/mol (Hessa et al, 2005a). A, Ala, alanine; D, Asp, aspartic acid; L, Leu, leucine; N, Asn, asparagine.
To assess the influence of the length of the loop connecting the H2′ and H segments on the insertion of the H segments, we also made constructs with the loop shortened from 150 to 55 residues. This allowed investigation of the hypothesis that H segments with short loops have a higher probability of encountering H2′ segments during insertion than those with long loops. The H2′ and H segment sequences of all constructs are listed in Table 1.
Effects of H2′ on insertion of a non-polar H segment
Before studying the Asn–Asn and Asp–Asp pairs, it was necessary to ascertain whether different Asn- or Asp-containing H2′ sequences could affect the insertion of a purely hydrophobic H segment. To this end, several different H2′ segments were combined with an H segment of the overall composition 3L/16A, as detailed in Table 1 (‘H2′ control experiments'). In the context of the original Lep construct, which had the wild-type H2 sequence, ΔGapp=−0.1 kcal/mol for the 3L/16A H segment (Hessa et al, 2005a). As seen in Table 1, ΔGapp for the 3L/16A H segment varied by only ±0.2 kcal/mol when different H2′ sequences were used, when a signal peptidase cleavage site was introduced in H2′ (CCH2) or even when the entire H1–H2 region was replaced by the signal peptide from preprolactin. This variation is within the estimated standard deviation of the ΔGapp measurement (2σ=±0.2 kcal/mol; Hessa et al, 2005a). Therefore, we conclude that the H2′ sequence has little influence on ΔGapp when the H segment is composed of only hydrophobic residues.
Single Asn–Asn and Asp–Asp pairs
To test the effect of Asn–Asn or Asp–Asp interactions on the membrane integration efficiency of the H segment, we compared pairs of constructs with one Asn or Asp in the middle of the H segment and either zero or one Asn or Asp in the middle of the H2′ segment. As explained above, interactions between the H2′ and H segments mediated by Asn–Asn or Asp–Asp pairing should be reflected in a change in ΔGapp (that is, ΔΔGapp should be significantly different from zero) for the H segment depending on the presence or absence of an Asn or Asp residue in the H2′ segment.
For the constructs with a uniformly hydrophobic 19-Leu H2′ segment, the membrane insertion efficiency of the designed H segments 1N/5L/13A and 1D/7L/11A was correctly estimated by the biological hydrophobicity scale to be about 50%, corresponding to ΔGapp≈0 kcal/mol (Table 1). The introduction of one Asn or Asp into H2′ did not lead to a significant decrease in ΔGapp for the 1N/5L/13A and 1D/7L/11A H segments (ΔΔGapp≈−0.1 kcal/mol; Table 1; Fig 2; constructs N and D; black bars). Similar results were obtained with the shorter 55-residue loop (white bars). We also tested whether the introduction of a signal peptidase cleavage site in H2 (CCH2) would affect the insertion efficiency of the 1N/5L/13A H segment, but found only a marginal difference compared with the 19L and 1N/18L H2′ segments (Table 1).
Figure 2.
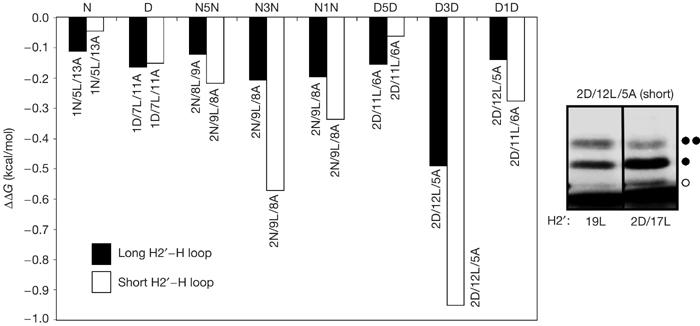
Effects on membrane insertion of single or pairs of Asn or Asp residues in the H2′ and H segments. The difference in the apparent free energy of insertion for constructs with and without Asn (N) or Asp (D) residues in H2′ (ΔΔGapp) for the different H segments is shown (see Table 1 for sequences). Black bars: 150-residue-long loop between the H2′ and H segments; white bars: 55-residue-long loop between the H2′ and H segments. The gel shows the two D3D constructs (H2′: 19L; H: 2D/12L/5A) and (H2′: 2D/17L; H: 2D/12L/5A), both with a short loop, translated in the presence of rough microsomes (unglycosylated, single glycosylated and double glycosylated forms of the protein are indicated by an open circle, one filled circle and two filled circles, respectively; the dye front is visible at the bottom of the gel). The corresponding ΔGapp values are 0.0 and −0.9 kcal/mol (Table 1), and ΔΔGapp=−0.9 kcal/mol (as shown by the white bar for the D3D pair in the bar graph). A, Ala, alanine; D, Asp, aspartic acid; L, Leu, leucine; N, Asn, asparagine.
Double Asn–Asn and Asp–Asp pairs
We next examined how the presence of two Asn–Asn or Asp–Asp pairs, one in H2′ and the other in the H segment, influenced the membrane insertion efficiency of the latter. We placed the Asn and Asp residues in the H segment with three different spacings: in positions 8 and 12, 7 and 13, and 9 and 11. The Asn and Asp residues in H2′ were placed in positions 8 and 12, that is, one helical turn apart. To counterbalance the effect of the Asn and Asp residues on the insertion efficiency of the H segment such that again ΔGapp≈0 kcal/mol, the overall composition of the H segments was chosen as between 8 and 12 Leu residues.
As seen in Table 1 and Fig 2, the introduction of two Asn–Asn pairs in H2′ decreased ΔGapp for the 2N/9L/8A H segments by ⩽0.2 kcal/mol for all constructs with a 150-residue-long loop between the H2′ and H segments (constructs N5N, N3N and N1N; black bars). For the corresponding constructs with two Asp–Asp pairs in the H segment, ΔGapp decreased by ⩽0.2 kcal/mol for the D1D and D5D segments, whereas the decrease was significantly larger (ΔΔGapp=−0.5 kcal/mol) for the D3D construct, in which the two Asp residues in the H segment are one helical turn apart. The results were similar for the constructs with the shorter, 55-residue-long loop, except that the decrease in ΔGapp was significant for both the N3N and D3D constructs, with ΔΔGapp≈−1 kcal/mol for the latter. We conclude that pairs of appropriately spaced Asn or Asp residues can mediate helix–helix interactions during membrane protein assembly in the endoplasmic reticulum and help control membrane protein topology.
A final observation relates to the effect of amphiphilicity on the efficiency of membrane insertion. We previously found that transmembrane helices with a higher hydrophobic moment inserted less efficiently than less amphiphilic helices with the same overall amino-acid composition (Hessa et al, 2005a). We observed a similar trend in the present data, with the more amphiphilic N3N and D3D H segments inserting less efficiently by 0.6–1.0 kcal/mol than the N5N, D5D, N1N and D1D H segments (Table 1).
Discussion
Although helix–helix association in the membrane environment in most cases seems to be governed by electrostatic and van der Waals interactions between closely packed interfaces (Curran & Engelman, 2003), polar residues such as Asn, Asp, Gln (glutamine, Q) and Glu (glutamic acid, E) have been shown to drive helix–helix association through the formation of inter-helical hydrogen bonds (Choma et al, 2000; Zhou et al, 2000, 2001; Gratkowski et al, 2001) and to promote the formation of helical hairpins during translocon-mediated membrane insertion (Hermansson & von Heijne, 2003).
By analysing model protein constructs in which zero, one or two Asn or Asp residues have been placed in two neighbouring hydrophobic segments (H2′ and H), we now find that the free energy of insertion (ΔGapp) of a marginally hydrophobic H segment is significantly reduced only if both the H2′ and H segments contain two Asn or two Asp residues with a spacing of three, but not one or five, residues (that is, when they are spaced one helical turn apart in both the H2′ and H segments; Fig 2). These results indicate that inter-helix hydrogen bonds can form during Sec61 translocon-assisted insertion and that H2′ remains in close enough proximity to the translocon to offer its hydrogen bond donor and acceptor sites to the incoming H segment even when the intervening loop is 150-residue long (Heinrich & Rapoport, 2003; Mitra et al, 2005; Sadlish et al, 2005). Although we cannot completely rule out other explanations for our observations, such as Asp and Asn residues in H2′ specifically altering the behaviour of the Sec61 translocon and only indirectly affecting the H segment insertion efficiency, we consider direct H2′–H segment interactions more likely for two reasons: (i) H2′ segment composition does not influence the insertion efficiency of the purely hydrophobic 3L/16A H segment and (ii) a strong H2′–H segment interaction effect is seen only when pairs of Asp and Asn residues are next to each other on one side of the H2′ and H segments (i, i+4 spacings).
The maximal reduction in ΔGapp seen with Asn–Asn or Asp–Asp pairs in both the H2′ and H segments is 0.5–1.0 kcal/mol, which is less than the 1–2 kcal/mol measured for hydrogen-bond-driven dimerization of certain transmembrane peptides in detergent micelles (Gratkowski et al, 2001; Zhou et al, 2001; Lomize et al, 2004). The values measured here are for Asn and Asp residues surrounded by rather bulky Leu residues, and it might be that the sequence context is suboptimal for efficient hydrogen-bond formation between the side chains (Dawson et al, 2003; Doura et al, 2004). Alternatively, the H2′ and H segment helices might be still partly water exposed in the translocon channel when the critical interaction develops.
An implicit assumption behind the original derivation of the biological hydrophobicity scale was that interactions between the H1–H2 transmembrane helices in Lep and the H segment have no, or only minor, effects on the membrane insertion efficiency of the latter (Hessa et al, 2005a). We can now examine this issue explicitly. First, for the apolar 3L/16A H segment, ΔGapp is largely independent of the sequence of H2′ (Table 1). Second, the strongest hydrogen-bond interactions between transmembrane helices are known to be mediated by Asn, Asp, Gln and Glu (Zhou et al, 2001); yet, we see at most a moderate effect on ΔGapp (ΔΔGapp≈−1.0 kcal/mol) even in constructs in which there is potential for two inter-helix Asn–Asn or Asp–Asp interactions. Third, the only potentially hydrogen-bonding residues in the wild-type H1 and H2 transmembrane helices are two Thr residues in H1 and a Thr and a Ser (serine, S) in H2. Both Thr and Ser are poor mediators of helix–helix interactions in membranes compared with Asn and Asp (Zhou et al, 2001; Dawson et al, 2002). We conclude that inter-helix interactions, although clearly not unimportant in natural proteins (Skach et al, 1993; Lin & Addison, 1995; Sato et al, 2002; Heinrich & Rapoport, 2003; Sadlish et al, 2005), can be treated as a second-order correction in the calculation of ΔGapp values based on the biological hydrophobicity scale (Hessa et al, 2005a, 2005b).
Methods
Enzymes and chemicals. All enzymes, plasmid pGEM1 and the TNT® Quick transcription/translation system were obtained either from New England Biolabs (Ipswich, MA, USA) or Promega (Madison, WI, USA). 35S-labelled methionine, deoxynucleotides and dideoxyribonucleotides were from GE Healthcare (Uppsala, Sweden). Oligonucleotides were obtained from Cybergene (Stockholm, Sweden) and MWG Biotech (Ebersberg, Germany).
DNA manipulations. All constructs were cloned as described previously (Hermansson & von Heijne, 2003; Hessa et al, 2005a). Site-directed mutagenesis to introduce Asn or Asp residues was performed using the QuikChange® Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA). To shorten the loop between the H2′ and H segments, plasmids were cut with EcoRV and SpeI. After refilling the 5′-end overhang of the SpeI site using DNA polymerase I Klenow fragment, the vector was religated. The resulting frameshift was annulled by re-introducing the missing alanine residue of the SpeI site using the QuikChange® Site-Directed Mutagenesis Kit (Stratagene). This procedure resulted in the deletion of residues 131–226 of Lep. To create model proteins with a cleavable signal sequence in the H1–H2 region, in one set of constructs the C-terminal end of the H2 region was modified to …LIV76SPSAQA+AY81…, where V76 and Y81 denote Lep residues and the + sign indicates the signal peptidase cleavage site (Nilsson et al, 1994), and in a second set the entire H1–H2 region was replaced by the signal peptide of preprolactin (Sasavage et al, 1982), yielding the N-terminal sequence MDSKGSSQKGSRLLLLLVVSNLLLCQGVVS+TPVCPNGPY81…
In vitro expression and quantification of membrane insertion efficiency. Constructs cloned in pGEM1 were transcribed and translated using the TNT® Quick coupled transcription/translation system (Promega). Then, 1 μg DNA template, 1 μl 35S-labelled methionine (5 μCi) and 1 μl dog pancreas rough microsomes (a kind gift from Dr M. Sakaguchi, Hyogo University, Japan) were added at the start of the reaction, and samples were incubated for 90 min at 30°C. Translation products were analysed by using SDS–polyacrylamide gel electrophoresis, and gels were quantified on a Fuji FLA-3000 phosphoimager using the Image Reader 8.1j software. The degree of membrane integration of each H segment was quantified from SDS–polyacrylamide gel electrophoresis gels (Fig 1B) by calculating an apparent equilibrium constant between the membrane-integrated and non-integrated forms: Kapp=f1g/f2g, where f1g is the fraction of single glycosylated and f2g the fraction of double glycosylated Lep molecules (Hessa et al, 2005a). The amount of unglycosylated material (f0g) is <15% in these experiments and was ignored in the calculations. The results were then converted to apparent free energies, ΔGapp=−RT ln Kapp. This approach is justified by the fact that the probability of insertion (=f1g/(f1g+f2g)) of systematically designed H segments follows a Boltzmann distribution (Hessa et al, 2005a), meaning that the data can be treated as simple partitioning between two experimentally identifiable states: inserted and translocated. All data points are mean values from at least two independent experiments.
Acknowledgments
This work was supported by grants from the Swedish Foundation for Strategic Research, the Marianne and Marcus Wallenberg Foundation, the Swedish Cancer Foundation and the Swedish Research Council to G.v.H., the Magnus Bergvall Foundation to I.M.N. and the National Institute of General Medical Sciences to S.H.W.
References
- Alder NN, Johnson AE (2004) Cotranslational membrane protein biogenesis at the endoplasmic reticulum. J Biol Chem 279: 22787–22790 [DOI] [PubMed] [Google Scholar]
- Choma C, Gratkowski H, Lear JD, DeGrado WF (2000) Asparagine-mediated self-association of a model transmembrane helix. Nat Struct Biol 7: 161–166 [DOI] [PubMed] [Google Scholar]
- Curran AR, Engelman DM (2003) Sequence motifs, polar interactions and conformational changes in helical membrane proteins. Curr Opin Struct Biol 13: 412–417 [DOI] [PubMed] [Google Scholar]
- Dawson JP, Weinger JS, Engelman DM (2002) Motifs of serine and threonine can drive association of transmembrane helices. J Mol Biol 316: 799–805 [DOI] [PubMed] [Google Scholar]
- Dawson JP, Melnyk RA, Deber CM, Engelman DM (2003) Sequence context strongly modulates association of polar residues in transmembrane helices. J Mol Biol 331: 255–262 [DOI] [PubMed] [Google Scholar]
- Doura AK, Kobus FJ, Dubrovsky L, Hibbard E, Fleming KG (2004) Sequence context modulates the stability of a GxxxG-mediated transmembrane helix–helix dimer. J Mol Biol 341: 991–998 [DOI] [PubMed] [Google Scholar]
- Gratkowski H, Lear JD, DeGrado WF (2001) Polar side chains drive the association of model transmembrane peptides. Proc Natl Acad Sci USA 98: 880–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich SU, Rapoport TA (2003) Cooperation of transmembrane segments during the integration of a double-spanning protein into the ER membrane. EMBO J 22: 3654–3663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermansson M, von Heijne G (2003) Inter-helical hydrogen bond formation during membrane protein integration into the ER membrane. J Mol Biol 334: 803–809 [DOI] [PubMed] [Google Scholar]
- Hessa T, Kim H, Bihlmaier K, Lundin C, Boekel J, Andersson H, Nilsson I, White SH, von Heijne G (2005a) Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature 433: 377–381 [DOI] [PubMed] [Google Scholar]
- Hessa T, White SH, von Heijne G (2005b) Membrane insertion of a potassium channel voltage sensor. Science 307: 1427. [DOI] [PubMed] [Google Scholar]
- Lin JL, Addison R (1995) A novel integration signal that is composed of two transmembrane segments is required to integrate the Neurospora plasma membrane H+-ATPase into microsomes. J Biol Chem 270: 6935–6941 [DOI] [PubMed] [Google Scholar]
- Lomize AL, Pogozheva ID, Mosberg HI (2004) Quantification of helix–helix binding affinities in micelles and lipid bilayers. Protein Sci 13: 2600–2612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra K, Schaffitzel C, Shaikh T, Tama F, Jenni S, Brooks CL III, Ban N, Frank J (2005) Structure of the E. coli protein-conducting channel bound to a translating ribosome. Nature 438: 318–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson I, von Heijne G (1993) Determination of the distance between the oligosaccharyltransferase active site and the endoplasmic reticulum membrane. J Biol Chem 268: 5798–5801 [PubMed] [Google Scholar]
- Nilsson I, Whitley P, von Heijne G (1994) The C-terminal ends of internal signal and signal-anchor sequences are positioned differently in the ER translocase. J Cell Biol 126: 1127–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadlish H, Pitonzo D, Johnson AE, Skach WR (2005) Sequential triage of transmembrane segments by Sec61α during biogenesis of a native multispanning membrane protein. Nat Struct Mol Biol 12: 870–878 [DOI] [PubMed] [Google Scholar]
- Sasavage NL, Nilson JH, Horowitz S, Rottman FM (1982) Nucleotide sequence of bovine prolactin messenger RNA. Evidence for sequence polymorphism. J Biol Chem 257: 678–681 [PubMed] [Google Scholar]
- Sato Y, Sakaguchi M, Goshima S, Nakamura T, Uozumi N (2002) Integration of Shaker-type K+ channel, KAT1, into the endoplasmic reticulum membrane: synergistic insertion of voltage-sensing segments, S3–S4, and independent insertion of pore-forming segments, S5–P–S6. Proc Natl Acad Sci USA 99: 60–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skach WR, Calayg MC, Lingappa VR (1993) Evidence for an alternate model of human P-glycoprotein structure and biogenesis. J Biol Chem 268: 6903–6908 [PubMed] [Google Scholar]
- von Heijne G (2003) Membrane protein assembly in vivo. Adv Proteing Chem 63: 1–18 [DOI] [PubMed] [Google Scholar]
- White SH, von Heijne G (2004) The machinery of membrane protein assembly. Curr Opin Struct Biol 14: 397–404 [DOI] [PubMed] [Google Scholar]
- Zhou FX, Cocco MJ, Russ WP, Brunger AT, Engelman DM (2000) Interhelical hydrogen bonding drives strong interactions in membrane proteins. Nat Struct Biol 7: 154–160 [DOI] [PubMed] [Google Scholar]
- Zhou FX, Merianos HJ, Brunger AT, Engelman DM (2001) Polar residues drive association of polyleucine transmembrane helices. Proc Natl Acad Sci USA 98: 2250–2255 [DOI] [PMC free article] [PubMed] [Google Scholar]


