Abstract
Cdt1 is an essential protein required for licensing of replication origins. Here, we show that in Schizosaccharomyces pombe, Cdt1 is proteolysed in M and G1 phases in response to DNA damage and that this mechanism seems to be conserved from yeast to Metazoa. This degradation does not require Rad3 and Cds1, indicating that it is independent of classic DNA damage and replication checkpoint pathways. Damage-induced degradation of Cdt1 is dependent on Cdt2 and Ddb1, which are components of a Cul4 ubiquitin ligase. We also show that Cdt2 and Ddb1 are needed for cell-cycle changes in Cdt1 levels in the absence of DNA damage. Cdt2 and Ddb1 have been shown to be involved in the degradation of the Spd1 inhibitor of ribonucleotide reductase after DNA damage, and we speculate that Cdt1 downregulation might contribute to genome stability by reducing demand on dNTP pools during DNA repair.
Keywords: licensing, Mcm2–7, pre-RC formation, replication origin, Schizosaccharomyces pombe
Introduction
DNA replication is a strictly regulated process which ensures that the genome is faithfully duplicated in each cell cycle. Replication origins are marked by the presence of the origin recognition complex (ORC) on DNA. An important regulatory step in replication is the assembly of Mcm2–7 (mini-chromosome maintenance) proteins at the ORC (reviewed by Nishitani & Lygerou, 2002; Blow & Dutta, 2005). This process—termed licensing or pre-replicative complex (pre-RC) formation—requires Cdt1 (Cdc10-dependent transcript 1) and Cdc6/18, and allows initiation to occur at the origin in S phase. The transition to S phase requires activation of cyclin-dependent kinase and Cdc7/Hsk1. These trigger the binding of proteins such as GINS (composed of Sld5-Psf1-Psf2-Psf3) and Cdc45 to the origin, allowing replication forks to be established. The Mcm2–7 complex probably provides helicase activity during DNA synthesis and is displaced from chromatin on completion of replication.
Pre-RC assembly is confined to M and G1 phases by several mechanisms which ensure that origins are activated only once per cell cycle, and these include regulation of Cdt1 (reviewed by Kearsey & Cotterill, 2003). In Schizosaccharomyces pombe, Cdt1 is expressed during M and G1 phases and is degraded in S phase (Hofmann & Beach, 1994; Nishitani et al, 2000), thus ensuring that no pre-RCs are assembled after initiation of replication. S-phase degradation of Cdt1 also occurs in other eukaryotes and two E3 ubiquitin ligases, Cul4–Ddb1 and SCFSkp2 (Skp1-Cullin 1-F-box protein), have been implicated in this process (Zhong et al, 2003; Liu et al, 2004; Sugimoto et al, 2004; Nishitani et al, 2006; Senga et al, 2006). Cdt1 regulation is important in mammalian cells, as overexpression can lead to genome instability (Vaziri et al, 2003) and has oncogenic potential (Seo et al, 2005).
In addition to the cell-cycle-dependent degradation of Cdt1, proteolysis of this factor has been reported after DNA damage. In human and Drosophila cells, DNA damage causes proteolysis of Cdt1 by means of a pathway that is dependent on the Cul4–Ddb1 ubiquitin ligase (Higa et al, 2003; Hu et al, 2004; Senga et al, 2006). Ultraviolet (UV) damage has also been reported to activate an SCFSkp2-dependent degradation pathway (Kondo et al, 2004).
Here, we have studied the effects of DNA damage on Cdt1 in S. pombe. DNA damage effects Cdt1 proteolysis by a mechanism that is independent of the DNA damage checkpoint pathway, but dependent on Ddb1 (homologous to the DDB1 subunit of the mammalian damaged DNA-binding protein DDB). We show that the WD40 repeat-containing factor Cdt2 (Cdc10-dependent transcript 2) is also involved in this proteolysis, a protein previously implicated in the turnover of the Spd1 inhibitor of ribonucleotide reductase (Liu et al, 2005). In addition, we show that Ddb1 and Cdt2, presumably as subunits of a Cul4 ubiquitin ligase, are relevant to the regulation of Cdt1 in a normal cell cycle.
Results
Cdt1 levels fall after UV irradiation
In an analysis of the effects of DNA damage on gene expression in meiosis, we observed that Cdt1 protein levels were depressed after UV irradiation (E.R. & S.E.K., unpublished data). Cells were arrested in G1 phase by nitrogen starvation and induced to enter meiosis by re-feeding and inactivating the Pat1 meiotic repressor. Cdt1 was present in G1-arrested cells, but after UV exposure Cdt1 levels decreased in a dose-dependent manner (Fig 1A).
Figure 1.
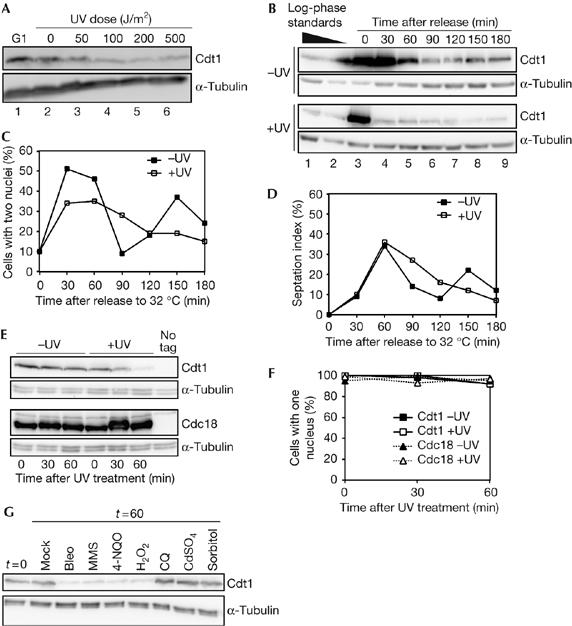
DNA damage reduces Cdt1 levels in M- and G1-phase cells. (A) Western blot analysis of Cdt1–Myc levels after ultraviolet (UV) irradiation of nitrogen-starved G1-arrested cells (P1424). Cells used for lanes 2–6 were released from the block for 30 min by re-feeding at 34°C. (B) Western blot of Cdt1–Myc levels in the mitotic cell cycle after UV irradiation. nda3 cells (P1451) were arrested in mitosis, UV irradiated (100 J/m2) and then released from the block by shifting to 32°C. Log-phase extracts are shown for comparison. (C) Nuclear counts for cells in experiment shown in (B) showing the timing of anaphase. (D) Septation index of cells for experiment shown in (B). (E) Western blot analysis of Cdt1–Myc and Cdc18–TAP levels in mitotically arrested cells after UV irradiation (100 J/m2). Strains P1451 and P1452 were treated as in (B), except that cells were kept at 20°C after irradiation to maintain the mitotic arrest. (F) Nuclear counts of the experiment shown in (E), showing that UV irradiation does not lead to the release of cells from the nda3 mitotic block. (G) Cdt1–Myc levels after treatment with DNA damage and stress agents. Cells (P1451) were arrested at the nda3 block, treated with 10 μg/ml bleomycin (bleo), 0.2% MMS, 0.6 μg/ml 4-NQO, 0.5 mM hydrogen peroxide (H2O2), 32 μg/ml chloroquin (CQ), 0.5 mM CdSO4 or 1 M sorbitol, and extracts were prepared 60 min later. Cells were kept at 20°C to maintain the mitotic arrest. MMS, methyl methane sulphonate; 4-NQO, 4-nitroquinoline 1-oxide; TAP, tandem affinity purification tag.
To determine whether this effect could be seen in the mitotic cell cycle, M-phase-arrested cells were subjected to UV irradiation. Mitotic arrest was achieved using the nda3-311 mutation, which arrests cells in prometaphase as the spindle checkpoint is activated (He et al, 1997). Arrested cells had high levels of Cdt1 (Fig 1B, compare lanes 1–3). On releasing mock-irradiated cells from the block, Cdt1 levels remained high for 30 min and then decreased over 60–90 min (Fig 1B; −UV), which corresponds to the timing of S phase as judged by cytokinesis and Mcm4 chromatin binding (Fig 1C,D; supplementary Fig S1 online). After UV irradiation (100 J/m2, ∼70% survival), there was a significant reduction in the level of Cdt1 by 30 min (Fig 1B; +UV). UV irradiation does not block nuclear division, cytokinesis or Mcm4 chromatin binding, although these events seem to be slightly delayed compared with control cells (Fig 1C,D; supplementary Fig S1 online).
To confirm that this result was not due to an indirect effect on cell-cycle progression in itself, we examined cells in which the mitotic arrest was maintained after UV irradiation. Cdt1 levels were constant in arrested cells but decreased in 60 min after UV irradiation (Fig 1E, top). To determine whether this effect was specific to Cdt1, we examined Cdc18, which shows a cell-cycle pattern of expression similar to that of Cdt1. In contrast to Cdt1, Cdc18 was not affected by UV irradiation (Fig 1E, bottom). The prometaphase arrest was maintained after UV irradiation (Fig 1F), showing that the reduction in the Cdt1 level was not due to progression into S phase, when Cdt1 is normally degraded. Overall, these results indicate that Cdt1 levels decrease as a direct consequence of UV irradiation.
Other forms of DNA damage affect Cdt1 levels
To determine whether the effect of UV irradiation on Cdt1 is caused by other types of DNA damage, similar experiments were carried out using mutagens and stress agents. 4-Nitroquinoline 1-oxide (4-NQO), the γ-irradiation mimetic bleomycin (bleo) and the alkylating agent methyl methane sulphonate (MMS) all caused a reduction in Cdt1 levels (Fig 1G). Hydrogen peroxide (H2O2), which causes oxidative DNA damage, also reduced Cdt1 levels. Chloroquine (CQ: a DNA intercalating agent), heavy metal (cadmium) and osmotic stress did not affect Cdt1. These results indicate that different types of DNA damage lead to a reduction in Cdt1 levels, but other forms of cellular stress have no effect.
Cdt1 turnover is due to induction of proteolysis
The reduction in Cdt1 levels induced by DNA damage could result from either induction of proteolysis or inhibition of Cdt1 synthesis, if proteolysis is constant. To distinguish between these possibilities, we studied the effects of cycloheximide (CHX) on Cdt1 levels in nda3-arrested cells. Without UV irradiation, Cdt1 levels did not decrease after 60 min in CHX, indicating that Cdt1 is stable during the nda3 block (Fig 2; −CHX). However, UV irradiation still led to a decrease in Cdt1 levels when CHX was present (Fig 2, +CHX). This suggests that Cdt1 proteolysis is induced by UV treatment and is independent of protein synthesis.
Figure 2.
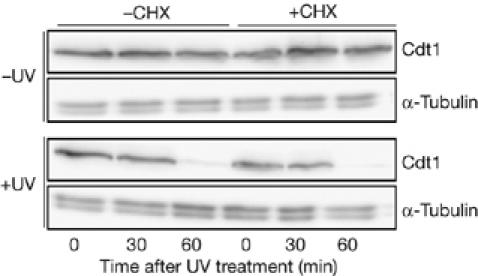
Reduction in Cdt1 levels after ultraviolet irradiation is proteolysis dependent. Western blot of Cdt1–Myc in nda3 (P1451) cells that were arrested in mitosis at 20°C, UV irradiated (100 J/m2) and then incubated with or without cycloheximide (CHX; 100 μg/ml). Cells were kept at 20°C to maintain the mitotic arrest.
Cdt1 proteolysis is checkpoint Rad independent
In mammalian cells, Cdt1 proteolysis induced by ionizing radiation is independent of ATM (ataxia telangiectasia mutated) and CHK2 (Higa et al, 2003). By contrast, Kondo et al (2004) have reported that UV-induced Cdt1 proteolysis is caffeine sensitive, suggesting that ATR (ataxia telangiectasia and Rad3-related kinase)/ATM might be required in human cells. To clarify the situation in fission yeast, we examined how Cdt1 proteolysis was affected by the rad3Δ and cds1Δ mutations, in which the DNA damage and replication checkpoints are inactivated, respectively. UV treatment still led to Cdt1 proteolysis after deletion of either gene (Fig 3A,B), indicating that the activation of degradation is independent of a classical checkpoint pathway.
Figure 3.
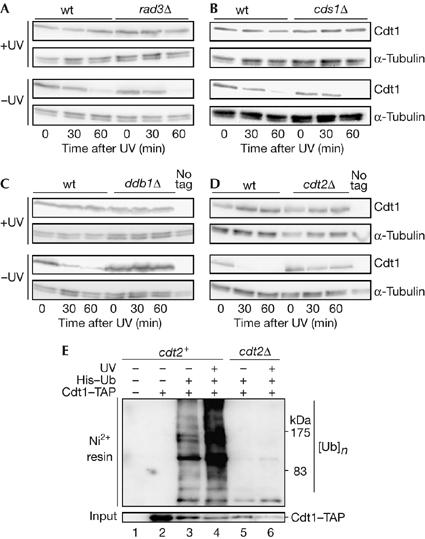
Ultraviolet-induced proteolysis of Cdt1 is not dependent on Rad3 and Cds1, but requires Ddb1 and Cdt2. (A) Cdt1–Myc levels after UV treatment in nda3 (wild type (wt), P1451) and nda3 rad3Δ cells (P1630) arrested at 20°C. (B) Cdt1–Myc levels after UV treatment in nda3 (wt, P1451) and nda3 cds1Δ (P1623) cells arrested at 20°C. (C) Cdt1–Myc levels after UV treatment in nda3 (wt, P1451) and nda3 ddb1Δ (P1615) cells arrested at 20°C. (D) Cdt1–Myc levels after UV treatment in nda3 (wt, P1451) and nda3 cdt2Δ (P1706) cells arrested at 20°C. In (A–D), the mitotic arrest was maintained throughout the time course and all UV treatments were carried out 100 J/m2. (E) High-molecular-weight ubiquitylated Cdt1 levels are increased after UV irradiation in cdt2+ strains. Ubiquitylated proteins were purified under denaturing conditions from strains expressing His6–ubiquitin (His-Ub) by Ni2+-NTA agarose affinity chromatography and probed with peroxidase–anti-peroxidase soluble complex to detect Cdt1–TAP. Irradiated strains were exposed to UV (100 J/m2) and grown for 20 min before preparing extracts. Strains used were P137 (lane 1), P1517 (lane 2), P1783 (lanes 3,4) and P1785 (lanes 5,6). NTA, nitrilo-triacetic acid; TAP, tandem affinity purification tag.
Cdt1 turnover is dependent on Ddb1 and Cdt2
In Metazoa, one pathway of Cdt1 proteolysis requires ubiquitylation by the Cul4–Ddb1 ubiquitin ligase, both in S phase and after DNA damage (Hu et al, 2004; Arias & Walter, 2006; Nishitani et al, 2006; Senga et al, 2006). We therefore examined whether Ddb1 is involved in this process in S. pombe. Deletion of the ddb1 gene blocks Cdt1 degradation after UV irradiation of nda3-arrested cells (Fig 3C). Thus, a need for Ddb1 in Cdt1 proteolysis after DNA damage seems to be conserved between S. pombe and Metazoa. Overexposure of western blots showed a ladder of Cdt1 bands in the wild-type strain, consistent with the role of Ddb1 in polyubiquitylation of Spd1 (Liu et al, 2005), but this was not observed in the ddb1 mutant (supplementary Fig S2 online).
We also examined the role of Cdt2 (Yoshida et al, 2003) in Cdt1 turnover after UV irradiation. Cdt2 has been recently identified as a part of the Cul4–Ddb1 complex required for Spd1 ubiquitylation in fission yeast (Liu et al, 2005). Spd1 is an inhibitor of ribonucleotide reductase that is degraded during S phase and after DNA damage (Holmberg et al, 2005; Liu et al, 2005), similar to Cdt1. In a cdt2Δ mutant, Cdt1 is stable after UV irradiation (Fig 3D), suggesting that the Cul4–Ddb1–Cdt2 complex is also responsible for Cdt1 turnover. We also observed that formation of high-molecular-weight conjugates of His6–ubiquitin with Cdt1 was stimulated by UV irradiation, but not in a cdt2Δ strain (Fig 3E). These results predict that Pcu4 (cullin 4) is also involved in the degradation of Cdt1 in S. pombe, as in mammalian cells (Higa et al, 2003; Liu et al, 2005; Nishitani et al, 2006). Interestingly, Spd1 degradation after DNA damage in G2 or replication arrest requires accumulation of the Cdt2 protein by a mechanism that is dependent on Rad checkpoint pathways (Liu et al, 2005). This regulation is clearly different from that of Cdt1 proteolysis, which does not require synthesis of Cdt2, or Rad3- and Cds1-dependent checkpoint pathways.
As the purpose of Cdt1 degradation after DNA damage might be to delay licensing and perhaps subsequent DNA replication, we investigated whether Ddb1 inactivation and consequent stabilization of Cdt1 affected the timing of Mcm4 chromatin binding after UV irradiation. Although UV irradiation delays anaphase and subsequent Mcm4 chromatin binding, deletion of the ddb1 gene did not abolish this UV-induced delay (supplementary Fig S1 online).
Cell cycle Cdt1 regulation requires Ddb1 and Cdt2
In mammalian cells, inactivation of the Cul4–Ddb1 pathway does not result in constitutive cell-cycle levels of Cdt1 owing to the existence of a second pathway for proteolysis which is dependent on Skp2 (Nishitani et al, 2006; Senga et al, 2006). In S. pombe, Cdt1 proteolysis in S phase has not been well characterized, and we investigated whether Ddb1 and Cdt2 are relevant. In wild-type cells expressing yellow fluorescent protein-tagged Cdt1, the protein is detectable only in the nuclei of some binucleate (G1 and M) cells (Fig 4A, arrows). However, if ddb1 or cdt2 is inactivated, then Cdt1 levels are elevated and the protein is seen in cells at all stages of the cell cycle (Fig 4A,B,D). The Cdt1 level is also elevated in a proteasome (mts3-1) mutant, as expected (Fig 4D). Cdt1 is degraded during a hydroxyurea (HU)-induced S-phase arrest of wild-type cells (Nishitani et al, 2000), but accumulates in cdt2Δ and ddb1Δ cells (Fig 4B). This suggests that a single Cul4–Ddb1–Cdt2 E3 pathway is required for regulation of Cdt1 levels during the cell cycle, unlike the situation in mammalian cells. De-regulation of Cdt1, on cdt2 or ddb1 deletion, does not lead to re-replication of DNA, even when spd1 is deleted to remove inhibition of DNA synthesis (Fig 4C; data not shown). This is consistent with earlier reports that Cdt1 overexpression does not affect DNA replication control (Gopalakrishnan et al, 2001; Yanow et al, 2001) and presumably reflects redundancy in mechanisms preventing re-replication.
Figure 4.
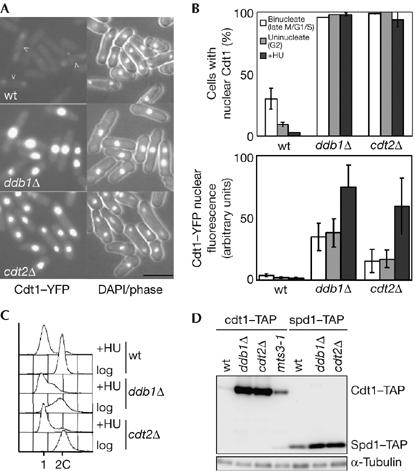
Cdt2 and Ddb1 regulate Cdt1 levels during the mitotic cell cycle. (A) Cdt1-yellow fluorescent protein (YFP) imaged in asynchronous wild-type (wt; P1262), ddb1Δ (P1769) and cdt2Δ (P1674) cells. Scale bar, 10 μm. (B) Strains shown in (A) were analysed to determine the percentage of cells showing nuclear Cdt1–YFP (top) and the fluorescence levels of nuclear Cdt1–YFP (bottom). Cells were analysed in asynchronous culture (data are given for binucleate (late M/G1/S) and uninucleate (G2) cells) and after S-phase arrest (HU for 3 h). (C) Flow-cytometric analysis of cells analysed in (B). (D) Comparison of Cdt1 and Spd1 levels in wt (P1517, P1766), ddb1Δ(P1673, P1767), cdt2Δ (P1731, P1768) and mts3-1 (P1611) cell extracts made from asynchronous cultures. The mts3-1 strain was grown at a semi-permissive temperature. DAPI, 4,6-diamidino-2-phenylindole; HU, hydroxyurea; TAP, tandem affinity purification tag.
Discussion
This study shows that Ddb1 and Cdt2 are needed for Cdt1 proteolysis after DNA damage and are also relevant to the regulation of Cdt1 levels in a normal cell cycle. Cdt2 contains several WD40 repeats and might function to recruit Cdt1 to the Cul4–Ddb1 complex, as suggested for its role in Spd1 recognition (Liu et al, 2005). While this paper was under review, involvement of Cdt2 in Cdt1 proteolysis was also reported in Drosophila, Xenopus and mammalian cells (Higa et al, 2006; Jin et al, 2006).
Given that Cdt1 proteolysis after DNA damage is independent of the DNA damage checkpoint, an important question is how turnover is activated. Recent studies in vertebrate cells suggest that Cdt1 interacts with chromatin-bound proliferating cell nuclear antigen (PCNA) and subsequently the Cul4–Ddb1–Cdt2 E3 complex is recruited, allowing Cdt1 ubiquitylation (Arias & Walter, 2006; Hu & Xiong, 2006; Jin et al, 2006; Senga et al, 2006). As only chromatin-associated PCNA can interact with Cdt1, the chromatin binding of PCNA, which occurs during DNA repair and S phase, is likely to be the regulating step leading to Cdt1 recognition by the E3 complex. A recent report suggested that a reduction in fission yeast Cdt1 levels after DNA damage is also PCNA dependent (Hu & Xiong, 2006), indicating that this mechanism is widely conserved.
Activation of Cdt1 degradation during S phase blocks further pre-RC formation and therefore helps to ensure a single round of DNA replication per cell cycle. However, the function of Cdt1 turnover after DNA damage is less clear. It has been suggested that this represents a checkpoint mechanism that delays S-phase entry (Higa et al, 2003). In S. pombe, UV irradiation has been shown to delay the entry of cdc10-arrested (G1) cells into S phase (Nilssen et al, 2003). However, Cdt1 does not seem to be relevant to this delay, as the protein is not detectable in cdc10-arrested cells and UV irradiation does not affect Cdt1 induction after release (T. Tvegård, B. Grallert, E.A. Nilssen, H. Soltani, S.K. & E.B., unpublished data). In any case, downregulation of Cdt1 alone might be problematic as a mechanism to delay S phase, as studies in Saccharomyces cerevisiae have suggested that a reduction in the efficiency of pre-RC formation leads to chromosome instability, presumably as a result of increased spacing between initiation events (Lengronne & Schwob, 2002; Tanaka & Diffley, 2002). Cdt1 downregulation could be more effective when coupled to a mechanism delaying S-phase initiation. In mammalian cells, a defect in ORC function leads to a delay of cyclin E expression and S-phase entry, which could represent a checkpoint monitoring the efficiency of pre-RC formation (Teer et al, 2006). Furthermore, blocking licensing in primary fibroblasts by geminin overexpression delays S-phase entry in a manner suggestive of a licensing checkpoint (Shreeram et al, 2002).
A second explanation is that Cdt1 proteolysis contributes to efficient DNA repair and error-free DNA replication following DNA damage. Ongoing DNA repair might reduce the dNTP pool, and downregulation of Cdt1 might reduce the number of replication forks in S phase, thus moderating S-phase demand on dNTPs. This would help to maintain dNTP levels in a manner analogous to the Rad53-dependent suppression of late-replication origins during inhibition of DNA synthesis by HU treatment (Santocanale & Diffley, 1998; Shirahige et al, 1998). Coordinated proteolysis of Spd1, relieving inhibition of ribonucleotide reductase, and Cdt1, reducing DNA synthesis, could synergistically help to maintain dNTP levels and provide optimum conditions for maintaining genome stability.
Methods
Fission yeast methods. Strains used are shown in supplementary Table I online. Strain construction, Cdt1 tagging, Mcm4 chromatin binding assay and flow cytometry were as described previously (Gregan et al, 2003; Kearsey et al, 2005).
Cell-cycle synchronization and UV irradiation. Cells were arrested in G1 phase in Edinburgh minimal medium lacking NH4Cl for 16 h at 25°C (Lindner et al, 2002). nda3-311 strains were arrested in M phase by incubation for 4 h at 20°C. For UV exposure, cells were resuspended in water and irradiated with 254 nm UV light in a 6 mm rapidly stirred suspension at 20°C.
Protein methods. For western blot analysis, Cdt1–Myc was detected using antibody M5546 (Sigma, Poole, UK); Tandem affinity purification (TAP)-tagged proteins were detected with peroxidase–anti-peroxidase soluble complex (P1291, Sigma) and α-tubulin was detected with antibody T5168 (Sigma). Analysis of ubiquitin (Ub)-conjugated Cdt1 was carried out using strains expressing His6–ubiquitin (pREP41X-His6Ub from T. Carr) and Cdt1–TAP. Ubiquitylated proteins were purified under denaturing conditions by Ni2+-NTA agarose affinity chromatography (Shiozaki & Russell, 1997). Ubiquitylated Cdt1–TAP was subsequently detected by western blotting.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Figures and Table
Acknowledgments
We thank C. Gordon, D. Hermand, S. Linn, C. Liu, H. Nishitani, T. Toda and H. Yamano for strains. We thank A. Carr, C. Liu and J. Walter for discussions and S.-W. Wang for comments. This work was supported by a Biotechnology and Biological Sciences Research Council studentship, the Norwegian Cancer Society and the Norwegian Research Council.
References
- Arias EE, Walter JC (2006) PCNA functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication. Nat Cell Biol 8: 84–90 [DOI] [PubMed] [Google Scholar]
- Blow JJ, Dutta A (2005) Preventing re-replication of chromosomal DNA. Nat Rev Mol Cell Biol 6: 476–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan V, Simancek P, Houchens C, Snaith HA, Frattini MG, Sazer S, Kelly TJ (2001) Redundant control of rereplication in fission yeast. Proc Natl Acad Sci USA 98: 13114–13119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregan J, Lindner K, Brimage L, Franklin R, Namdar M, Hart EA, Aves SJ, Kearsey SE (2003) Fission yeast Cdc23/Mcm10 functions after pre-replicative complex formation to promote Cdc45 chromatin binding. Mol Biol Cell 14: 3876–3887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Patterson TE, Sazer S (1997) The Schizosaccharomyces pombe spindle checkpoint protein mad2p blocks anaphase and genetically interacts with the anaphase-promoting complex. Proc Natl Acad Sci USA 94: 7965–7970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higa LA, Mihaylov IS, Banks DP, Zheng J, Zhang H (2003) Radiation-mediated proteolysis of CDT1 by CUL4–ROC1 and CSN complexes constitutes a new checkpoint. Nat Cell Biol 5: 1008–1015 [DOI] [PubMed] [Google Scholar]
- Higa LA, Banks D, Wu M, Kobayashi R, Sun H, Zhang H (2006) L2DTL/CDT2 interacts with the Cul4/Ddb1 complex and PCNA and regulates CDT1 proteolysis in response to DNA damage. Cell Cycle 5: 1675–1680 [DOI] [PubMed] [Google Scholar]
- Hofmann J, Beach D (1994) cdt1 is an essential target of the cdc10/sct1 transcription factor: requirement for DNA replication and inhibition of mitosis. EMBO J 13: 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg C, Fleck O, Hansen HA, Liu C, Slaaby R, Carr AM, Nielsen O (2005) Ddb1 controls genome stability and meiosis in fission yeast. Genes Dev 19: 853–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Xiong Y (2006) An evolutionarily conserved function of proliferating cell nuclear antigen for cdt1 degradation by the cul4–ddb1 ubiquitin ligase in response to DNA damage. J Biol Chem 281: 3753–3756 [DOI] [PubMed] [Google Scholar]
- Hu J, McCall CM, Ohta T, Xiong Y (2004) Targeted ubiquitination of CDT1 by the DDB1–CUL4A–ROC1 ligase in response to DNA damage. Nat Cell Biol 6: 1003–1009 [DOI] [PubMed] [Google Scholar]
- Jin J, Arias EE, Chen J, Wade Harper J, Walter JC (2006) A family of diverse Cul4–Ddb1-interacting proteins including Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol Cell 23: 709–721 [DOI] [PubMed] [Google Scholar]
- Kearsey S, Cotterill S (2003) Enigmatic variations: divergent modes of regulating eukaryotic DNA replication. Mol Cell 12: 1067–1075 [DOI] [PubMed] [Google Scholar]
- Kearsey SE, Brimage L, Namdar M, Ralph E, Yang X (2005) In situ assay for analyzing the chromatin binding of proteins in fission yeast. Methods Mol Biol 296: 181–188 [DOI] [PubMed] [Google Scholar]
- Kondo T et al. (2004) Rapid degradation of Cdt1 upon UV-induced DNA damage is mediated by SCFSkp2 complex. J Biol Chem 279: 27315–27319 [DOI] [PubMed] [Google Scholar]
- Lengronne A, Schwob E (2002) The yeast CDK inhibitor Sic1 prevents genomic instability by promoting replication origin licensing in late G(1). Mol Cell 9: 1067–1078 [DOI] [PubMed] [Google Scholar]
- Lindner K, Gregan J, Montgomery S, Kearsey S (2002) Essential role of MCM proteins in pre-meiotic DNA replication. Mol Biol Cell 13: 435–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Poitelea M, Watson A, Yoshida SH, Shimoda C, Holmberg C, Nielsen O, Carr AM (2005) Transactivation of Schizosaccharomyces pombe cdt2+ stimulates a Pcu4–Ddb1–CSN ubiquitin ligase. EMBO J 24: 3940–3951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu E, Li X, Yan F, Zhao Q, Wu X (2004) Cyclin-dependent kinases phosphorylate human Cdt1 and induce its degradation. J Biol Chem 279: 17283–17288 [DOI] [PubMed] [Google Scholar]
- Nilssen EA, Synnes M, Kleckner N, Grallert B, Boye E (2003) Intra-G1 arrest in response to UV irradiation in fission yeast. Proc Natl Acad Sci USA 100: 10758–10763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani H, Lygerou Z (2002) Control of DNA replication licensing in a cell cycle. Genes Cells 7: 523–534 [DOI] [PubMed] [Google Scholar]
- Nishitani H, Lygerou Z, Nishimoto T, Nurse P (2000) The Cdt1 protein is required to license DNA for replication in fission yeast. Nature 404: 625–628 [DOI] [PubMed] [Google Scholar]
- Nishitani H et al. (2006) Two E3 ubiquitin ligases, SCF–Skp2 and DDB1–Cul4, target human Cdt1 for proteolysis. EMBO J 25: 1126–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santocanale C, Diffley JF (1998) A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature 395: 615–618 [DOI] [PubMed] [Google Scholar]
- Senga T, Sivaprasad U, Zhu W, Park JH, Arias EE, Walter JC, Dutta A (2006) PCNA is a cofactor for Cdt1 degradation by CUL4/DDB1-mediated N-terminal ubiquitination. J Biol Chem 281: 6246–6252 [DOI] [PubMed] [Google Scholar]
- Seo J, Chung YS, Sharma GG, Moon E, Burack WR, Pandita TK, Choi K (2005) Cdt1 transgenic mice develop lymphoblastic lymphoma in the absence of p53. Oncogene 24: 8176–8186 [DOI] [PubMed] [Google Scholar]
- Shiozaki K, Russell P (1997) Stress-activated protein kinase pathway in cell cycle control of fission yeast. Methods Enzymol 283: 506–520 [DOI] [PubMed] [Google Scholar]
- Shirahige K, Hori Y, Shiraishi K, Yamashita M, Takahashi K, Obuse C, Tsurimoto T, Yoshikawa H (1998) Regulation of DNA-replication origins during cell-cycle progression. Nature 395: 618–621 [DOI] [PubMed] [Google Scholar]
- Shreeram S, Sparks A, Lane DP, Blow JJ (2002) Cell type-specific responses of human cells to inhibition of replication licensing. Oncogene 21: 6624–6632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto N, Tatsumi Y, Tsurumi T, Matsukage A, Kiyono T, Nishitani H, Fujita M (2004) Cdt1 phosphorylation by cyclin A-dependent kinases negatively regulates its function without affecting geminin binding. J Biol Chem 279: 19691–19697 [DOI] [PubMed] [Google Scholar]
- Tanaka S, Diffley JF (2002) Deregulated G1-cyclin expression induces genomic instability by preventing efficient pre-RC formation. Genes Dev 16: 2639–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teer JK, Machida YJ, Labit H, Novac O, Hyrien O, Marheineke K, Zannis Hadjopoulos M, Dutta A (2006) Proliferating human cells hypomorphic for origin recognition complex 2 and pre-replicative complex formation have a defect in p53 activation and Cdk2 kinase activation. J Biol Chem 281: 6253–6260 [DOI] [PubMed] [Google Scholar]
- Vaziri C, Saxena S, Jeon Y, Lee C, Murata K, Machida Y, Wagle N, Hwang DS, Dutta A (2003) A p53-dependent checkpoint pathway prevents rereplication. Mol Cell 11: 997–1008 [DOI] [PubMed] [Google Scholar]
- Yanow SK, Lygerou Z, Nurse P (2001) Expression of Cdc18/Cdc6 and Cdt1 during G2 phase induces initiation of DNA replication. EMBO J 20: 4648–4656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S-h, Al-Amodi H, Nakamura T, McInerny CJ, Shimoda C (2003) The Schizosaccharomyces pombe cdt2+ gene, a target of G1–S phase-specific transcription factor complex DSC1, is required for mitotic and premeiotic DNA replication. Genetics 164: 881–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W, Feng H, Santiago FE, Kipreos ET (2003) CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing. Nature 423: 885–889 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures and Table


