Abstract
Mutation has traditionally been considered a random process, but this paradigm is challenged by recent evidence of divergence rate heterogeneity in different genomic regions. One facet of mutation rate variation is the propensity for genetic change to correlate with the number of germ cell divisions, reflecting the replication-dependent origin of many mutations. Haldane was the first to connect this association of replication and mutation to the difference in the number of cell divisions in oogenesis (low) and spermatogenesis (usually high), and the resulting sex difference in the rate of mutation. The concept of male-biased mutation has been thoroughly analysed in recent years using an evolutionary approach, in which sequence divergence of autosomes and/or sex chromosomes are compared to allow inference about the relative contribution of mothers and fathers in the accumulation of mutations. For instance, assuming that a neutral sequence is analysed, that rate heterogeneity owing to other factors is cancelled out by the investigation of many loci and that the effect of ancestral polymorphism is properly taken into account, the male-to-female mutation rate ratio, αm, can be solved from the observed difference in rate of X and Y chromosome divergence. The male mutation bias is positively correlated with the relative excess of cell divisions in the male compared to the female germ line, as evidenced by a generation time effect: in mammals, αm is estimated at approximately 4–6 in primates, approximately 3 in carnivores and approximately 2 in small rodents. Another life-history correlate is sexual selection: when there is intense sperm competition among males, increased sperm production will be associated with a larger number of mitotic cell divisions in spermatogenesis and hence an increase in αm. Male-biased mutation has implications for important aspects of evolutionary biology such as mate choice in relation to mutation load, sexual selection and the maintenance of genetic diversity despite strong directional selection, the tendency for a disproportionate large role of the X (Z) chromosome in post-zygotic isolation, and the evolution of sex.
Keywords: point mutation, nucleotide substitution, germ line biology, sex chromosomes
1. Introduction
Mutations constitute the ultimate source of genetic novelty required for evolution by natural selection. Mutational events are likely to arise in all cells of an organism but only those originating in germ cells are transmitted to subsequent generations, and thus are relevant to evolution. However, mutation is not a homogenous process that occurs at a constant rate among lineages or within genomes, and variation in the rate and pattern of germ line mutation are therefore key factors in many aspects of evolutionary and population genetics. For instance, in the absence of selection, the rate of molecular evolution and the level of standing genetic variation are directly governed by the rate of mutation. Moreover, the rate of mutation must be taken into account in order to infer the type and the intensity of selection in DNA sequences in the analyses of divergence data.
If we were able to identify all de novo mutations that our parents provided us with, we would have the possibility to directly measure the rate of mutation per generation. While the necessary technology is not yet available to allow such an endeavour, we can make one particular prediction when it comes to the relative contribution from mothers and fathers: we expect to see most mutations originating from our father's germ cells. This expectation derives from what now seems to be a widespread phenomenon among higher organisms, the rate of germ line mutation is typically higher in spermatogenesis than in oogenesis—in other words, the mutation rate is male biased (Vogel & Motulsky 1997; Hurst & Ellegren 1998; Li et al. 2002). The concept of male-biased mutation builds upon the mechanistic basis of mutation. Since replication error during cell division has been seen as a major source of mutation, the usually higher number of cell division in spermatogenesis than in oogenesis means that, everything else being equal, more mutations will be generated in the male than in the female germ line. In other words, the number of accumulated mutations in a DNA sequence should increase when it is replicated many times. Here, I review how recent work in this area has contributed to the realization of the extent to which sex-specific mutation rates can explain (and cannot explain) the overall pattern of mutation rate heterogeneity across the genome, and to the insight in the evolutionary implications of male-biased mutation.
2. A historical perspective
(a) Indirect studies using human disease and Mendelian genetics
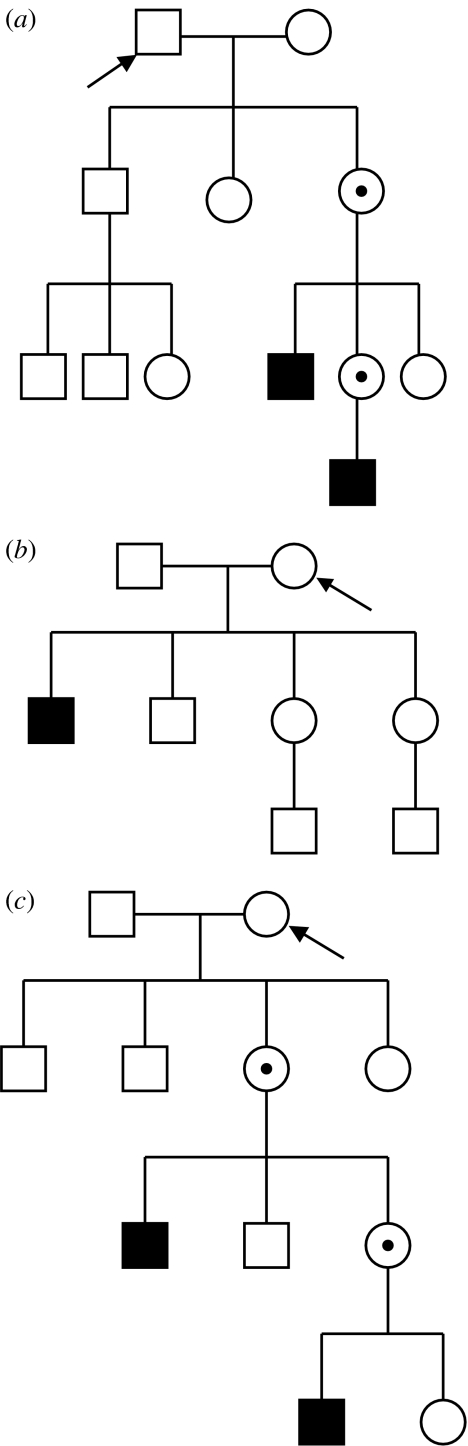
Haldane (1935, 1947, 1948) made the observation that spontaneous cases of X-linked haemophilia B were most often discovered in families with mother carriers. Recessive mutations arising on the X chromosome will have different immediate consequences depending on the sex of parent and offspring (figure 1). When they arise in the male germ line, and given that fathers transmit their single X chromosome to daughters only, paternally generated X-linked mutations will always exist in heterozygous state in the first (female) offspring generation, without expression of the mutant phenotype. Mothers, on the other hand, transmit an X chromosome to both daughters and sons, with equal likelihood. When a new X-linked recessive arises in the female germ line, first generation (hemizygote) sons will be directly affected. Given that X-linked mutations arisen in both parents can thus give female carriers while directly affected sons only follow from maternally derived mutations, Haldane's null hypothesis was that female carriers should be twice as common as directly affected sons. However, the excess of families with mother carriers was observed to be much larger than expected. Based on this observation he suggested that, in humans, fathers contribute more mutations than mothers.
Figure 1.
Pedigrees showing the emergence of a recessive X-linked disease from a de novo mutation. (a) Paternal origin of the mutation. (b) Maternal origin of a mutation first transmitted to a son. (c) Maternal origin of a mutation first transmitted to a daughter. The individual in which the mutation initially arose in one of his/her gametes is arrowed. Squares are males, circles are females, filled squares are affected males and circles with a dot are heterozygote female carriers.
Haldane's idea of male-biased mutation did not spark a great deal of immediate research among contemporary investigators. Indirectly inferring the parental origin of new mutations using X-linked recessives required pedigrees with de novo mutations causing disease, or other distinct phenotypes, and such cases were rare and hindered further research. Thus, the question of a sex difference in the rate of germ line mutation was not studied actively for several decades and, to the extent it was appreciated, it was generally considered an issue mainly for human or medical geneticists (Winter et al. 1983; see also Crow 1993, 1997).
(b) The evolutionary approach
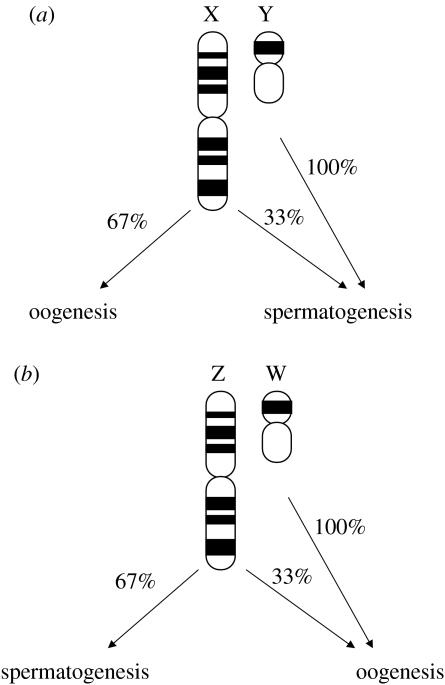
Miyata et al. (1987) offered a new way to attack the problem. Given the mode of inheritance of autosomes and sex chromosomes, they showed that it is possible to infer the relative contribution of males and females to evolutionarily accumulated mutations by studying sequence divergence in different chromosomal classes. Specifically, in the XY system, the Y chromosome evolves under the influence of male-originating mutations only, while the X chromosome and the autosomes are hit by mutations of both male and female origin. Knowing the rate of male mutation from the rate of Y chromosome divergence, the female rate can be solved from X-chromosomal data taking into account that X is two-thirds of the time in the female germ line and one-third in the male germ line (figure 2). Alternatively, relative sex-specific rates can be solved by comparisons of autosomes and either of the sex chromosomes. The male-to-female mutation rate ratio (αm) is then estimated from either of the following equations:
| (2.1) |
Figure 2.
Schematic view of the pattern of sex chromosomal inheritance in organisms with (a) male heterogamety (XY sex determination) and (b) female heterogamety (ZW sex determination).
| (2.2) |
Correspondingly, in a ZW system of female heterogamety, the female mutation rate is given by W chromosome divergence (W is only transmitted through the female germ line) and the male rate can be indirectly obtained through data from the Z chromosome (Miyata et al. 1987).
3. Genomic determinants of the male mutation bias
(a) Inferring mutation rates from neutral substitution rates
A starting point for the evolutionary approach to αm estimation is that the rate of mutation can be inferred from the neutral rate of nucleotide substitution or sequence divergence. While not controversial in itself, a more difficult issue is how to define truly neutral sequences. Synonymous sites (particularly fourfold degenerate sites) and non-coding DNA such as introns, intergenic sequence and pseudogenes have traditionally been seen as selectively neutral, but this may not necessarily be true (Smith & Hurst 1999). For instance, recent analysis suggests that selectively driven codon usage may occur in organisms previously thought to be free of a codon bias, including mammals (Hellman et al. 2003; Urrutia & Hurst 2003; Chamary & Hurst 2004). Moreover, there is an overall tendency for sequence conservation in first introns (Chamary & Hurst 2004) and, more generally, in the ends of introns (Hare & Palumbi 2003; Sorek & Ast 2003; Chamary & Hurst 2004). Furthermore, a bulk of evidence now exists to demonstrate both negative and positive (Andolfatto 2005) selection on non-coding DNA, including the widespread presence of conserved sequence blocks in intergenic and intronic DNA (Majewski & Ott 2002).
In practice, one may have to accept that a fraction of the presumably neutral sequence used for αm estimation is not entirely free of constraint but that the frequency of such constraint is similar for sequences from the chromosomal classes being compared. Alternatively, one may try to exclude potentially conserved sites or blocks of non-coding DNA from the analysis. Using a distant out-group to identify conserved elements can be a means for such masking of the sequence dataset.
(b) To what extent does chromosomal divergence reflect sex-specific mutation rates?
An underlying argument in the evolutionary approach is that the time the sequences spend in the male and female is the main factor governing chromosome-specific mutation rates. However, potential violations of this assumption occur at different levels. Based on the theory of adaptive mutation rate variation, one potential problem is that the X chromosome may have evolved a lower mutation rate to reduce the exposure of deleterious recessive mutations when in hemizygous state (McVean & Hurst 1997). While there was some support from initial mouse–rat comparisons (McVean & Hurst 1997; Lercher et al. 2001), more recent analysis based on larger datasets of human–mouse and mouse–rat coding sequence comparisons do not support this hypothesis (Malcom et al. 2003). Moreover, in ZW systems, where the analogous argument would imply that the Z chromosome had evolved a reduced mutation rate, αm estimates using Z–A, Z–W or A–W comparisons of chicken–turkey divergence do not differ significantly (Axelsson et al. 2004); had Z rate been reduced for other reasons than male-biased mutation, αm estimated from Z–A and Z–W data would be lower than when estimated from A–W.
More seriously, an important conclusion from recent large-scale genome analysis is that there is significant local and regional variation in the substitution rate along chromosomes (e.g. MGSC 2002; Smith et al. 2002; Hardison et al. 2003; CSAC 2005). This is likely to reflect neutral processes in which mutation rate variation is associated with factors such as recombination rate variation (Lercher & Hurst 2002; Hellman et al. 2003), nucleotide composition (Hardison et al. 2003) and perhaps also chromatin structure, although it cannot be completely excluded that such heterogeneity will be seen, in a selectionist view, as evidence for adaptive mutation rate variation at the level of individual genes or genomic segments (Smith & Hurst 1999; Chuang & Li 2004). In any case, mutation rate variation within chromosomes is problematic to studies of the male mutation bias since αm estimation will be dependent on which genomic regions are used for the underlying estimates of sequence divergence in different chromosomal classes. Importantly, it clearly shows that the time a particular sequence spends in the male and female germ line is not the sole, and in many cases not even the main, cause of mutation rate variation (Gaffney & Keithley 2005). For instance, human–chimpanzee autosomal divergence varies between 0.8 and 2.0% even when regions as large as 1 Mb are compared; when smaller windows are analysed, the variation is considerable (CSAC 2005). Most of this variation occurs within chromosomes but there is significant support also for rate variation among autosomes in the human–chimpanzee comparison (CSAC 2005). In birds, a further aspect of autosomal rate heterogeneity is that the smaller microchromosomes mutate at a higher rate than the larger macrochromosomes (ICGSC 2004; Axelsson et al. 2005). To conclude, it is quite possible that the variation in αm estimates obtained in studies focusing on one or a few genomic regions only has been due at least in part to within-genome mutation rate heterogeneity.
(c) Taking large-scale mutation rate heterogeneity into account
How should mutation rate heterogeneity be handled when one is interested in estimating the male mutation bias? For a start, whole-genome sequences are now steadily accumulating and offer an outstanding source of information for the study of sex-specific mutation rates using the evolutionary approach (Makova et al. 2004; RGSPC 2004; Lindblad-Toh et al. 2005; CSAC 2005). One possibility when such large-scale data are available is to focus on interspersed repeat elements, either using divergence data between individual copies and an ancestral master repeat sequence (for which data from one genome is sufficient; Erlandsson et al. 2000; IHGSC 2001), or by estimating the divergence of homologous repeats in two or more species (Makova et al. 2004; RGSPC 2004). Using interspersed repeats has the advantage of facilitating the identification of orthologous sequence in the non-coding parts of relatively divergent genomes. It might also be argued that focusing on the same sequence inserted at many different locations in the genome reduces the confounding effects of mutation rate heterogeneity related to inherent features of the sequence context, like nucleotide composition. However, interspersed elements tend to evolve towards the GC content of surrounding regions (Filipski et al. 1989; Casane et al. 1997; IHGSC 2001). Moreover, a potential pitfall in the analysis of repeat elements is that they may be subject to events of gene conversion (Roy et al. 2000). If such events are biased with respect to chromosomal classes involved (Jurka 2004; Webster et al. 2005), estimates of sex-specific mutation rates would also be biased.
For broad analyses of many taxa and, in particular, for studies of genetically less well-characterized organisms, whole-genome approaches are not yet feasible. However, owing to within-chromosomal mutation rate heterogeneity, it is still advisable to estimate αm using sequence data from several genomic regions, rather than sampling the same total amount of sequence from a single locus. It has been argued that the analysis of homologous sequences in different chromosomal classes, like genes (Shimmin et al. 1993b) or other genomic segments (Bohossian et al. 2000) shared between the X and the Y chromosomes, would have some merit. However, Berlin et al. (2006) found no evidence for a correlation in the substitution rate of introns of genes shared between the sex chromosomes.
(d) Site heterogeneity in the male mutation bias
Mutation rate variation among and within chromosomes clearly represents a confounding factor in studies of sex-specific mutation rates. Now, if such heterogeneity can be dealt with by sampling many loci, can we then safely estimate the male mutations bias? Miyata et al. (1987) originally made the assumption that the majority of mutations are replication-dependent, which laid the ground for derivation of the expected rates of sequence evolution of chromosomal classes. In retrospect, it would seem that this assumption was an oversimplification since it has been known for long that damages and lesions unrelated to replication can introduce mutations in DNA (Macphee 1995). Recently, a clear-cut illustration of how this affects αm estimation was provided by Taylor et al. (2006) who used a whole-genome human–chimpanzee comparison to show that while CpG sites have only a modest male mutation bias (αm≈2–3), non-CpG sites have αm≈6–7, which is close to what might be expected if the majority of mutation is owing to replication errors. The weak bias seen at CpG sites is consistent with the nature of most CpG mutations, which are the result of methylated cytosine converting into thymine upon spontaneous, replication-independent deamination. Taylor et al. make the important note that the proportion of CpG sites in sequences used for estimating chromosomal divergence will therefore affect the magnitude of the male mutation bias.
Generally, we should distinguish between replication-dependent and replication-independent mutations when studying sex-specific mutation rates, with the former expected to show a more pronounced sex-bias than the latter. This applies as well to mutations other than nucleotide substitution. For example, length changes in microsatellite sequences are generally thought to arise from replication slippage (Ellegren 2004). Consistent with this, the excess of paternally derived mutations in simple repeats is of similar magnitude as point mutations, at least in humans (Ellegren 2000). Moreover, small insertion and deletion polymorphisms in unique sequence also show an excess of male mutations, suggesting that they primarily occur during DNA replication (Sundström et al. 2003; Makova et al. 2004). A practical consequence of high male mutation rates at hypervariable repeat loci used in DNA profiling is that paternity tests would need to take into account the higher chance of male-derived mutations as an alternative to non-paternity (Ibarguchi et al. 2004).
(e) The rate of mutation per cell generation
The assumption that αm should be equal to the male-to-female ratio in the number of germ line cell divisions, c (Miyata et al. 1987), builds on the premise that the rate of mutation per cell division is equal in spermatogenesis and oogenesis. In essence, we still do not know if this is the case. In humans, and probably so also in other organisms, the two sexes differ in germ line methylation levels (Driscoll & Migeon 1990; Bestor 1998), so CpG mutability may certainly be expected to vary in a sex-specific manner (Huttley et al. 2000). However, also when replication-dependent mutations are considered, there might be variation in the per cell generation mutation rate. The testis and ovary clearly represent two different mutational environments, which show distinct differences in physiology and morphology, and in the rate and timing of mitotic division of germ cells. For instance, while cell division in oogenesis is generally believed to be prenatal for species with arrested meiosis in females (but see Johnson et al. 2004), most cell divisions in spermatogenesis occur after sexual maturity. Moreover, genome-wide microarray hybridization experiments demonstrate that many genes are differentially expressed in the male and female germ line (e.g. Ranz et al. 2003; Parisi et al. 2003), including genes involved in DNA repair (Allen et al. 1995; Blackshear et al. 1998). Furthermore, the continuous production of sperm during adulthood may imply that male germ cells are more sensitive to the effects of ageing and senescence, e.g. from the accumulation of somatic mutations in genes involved in the replication machinery. Such effects could be reinforced by the fact that males and females often reproduce at different ages. The trade-off between the benefit of avoidance of transferring deleterious mutations to offspring and the cost incurred from efficient replication and repair (Leigh 1970; Kondrashov 1995) may find different optima in the two sexes.
4. Population genetic aspects of estimating αm
In the absence of horizontal transfer of genetic information, orthologous sequences of two species coalesce in a sequence of a common ancestor. For any pair of species, the coalescence time will vary from sequence to sequence given that their most recent common ancestor is likely to have contained ancestral polymorphism. We shall thus expect to obtain slightly different divergence estimates for different genomic regions, even if the rate of mutation is uniform. This is in itself not a problem when estimating average mutation rates from divergence data, as long as it is kept in mind that the number of mutations observed is the sum of the number that arose after the split of the two species plus the number that gave rise to the ancestral polymorphism in the most recent common ancestor. However, for estimates of the male mutation bias based on observed sequence divergence in autosomes and/or sex chromosomes, there is a problem if different chromosomal classes were associated with different levels of sequence diversity in a common ancestor. This is likely to be the case for comparisons involving the Y chromosome in organisms with male heterogamety and the W chromosome in organisms with female heterogamety. The Y (W) is mostly non-recombining and has a smaller effective population size (Ne) than the X (Z) chromosome and the autosomes. Population genetics theory predicts that these factors should lead to reduced levels of nucleotide diversity (Charlesworth & Charlesworth 2000), something which is supported by empirical data from several organisms (Filatov et al. 2000; ISMWG 2001; Bachtrog & Charlesworth 2002; Berlin & Ellegren 2004). Sequences on the Y (W) chromosome are therefore likely to coalesce more quickly than sequences on the X (Z) chromosome and the autosomes. As a consequence, estimates of αm based on comparisons of sequence divergence in, for example, X and Y chromosomes will be underestimates (The other way around, male-biased mutation has implications for deriving neutral expectations about the amount of genetic variability to be seen in a particular chromosome. Sequence diversity (θ) is usually modelled by the expression θ=4Neμ. Clearly, differences in the mutation rate (μ) among chromosomal classes will affect the expected level of diversity (ISMWP 2001).
The effect of ancestral polymorphism will only matter at relatively recent divergences; for species that diverged long ago, the vast majority of mutations contributing to the observed divergence will have occurred after the split of species. Makova & Li (2002) estimated the magnitude of male-biased mutation among primate species and found a much lower αm estimate in the human–chimpanzee comparison (approx. 2) than in the comparisons of human versus more distantly related primates (approx. 5), which is not expected on biological grounds (see Ellegren 2002). Moreover, both Bartosch-Härlid et al. (2003) and Sandstedt & Tucker (2005) obtained lower αm estimates in terminal than in internal branches of large avian and rodent phylogenies, respectively. This hints that the most unbiased estimates would be obtained using divergence data mapped down to internal branches only (cf. Makova & Li 2002). Alternatively, the contribution of ancestral polymorphism to divergence can be corrected for by using the current level of nucleotide diversity as an approximation for ancestral levels of polymorphism, and then subtract this from observed pairwise divergences.
Species divergence is a process that may be associated with a long period of interbreeding and introgression between the two diverging lineages. There is as yet only limited information on the genomic consequences of this process, although introgression will generally increase the heterogeneity in regional sequence divergence. It was recently shown that sequence divergence in a whole-genome sequence comparison of human and chimpanzee is unexpectedly low for the X chromosome (Patterson et al. 2006). One possible explanation for this observation is that X-linked loci of human and chimpanzee more often than autosomal loci share ancestry at the time of hybridization between the diverging lineages. Such discrepancy between autosomes and X chromosome divergence would be compatible with the tendency for the heterogametic sex to be more sensitive to hybrid sterility and inviability than the homogametic sex (Haldane 1922). Important to this discussion, differences between autosomal and X chromosome divergence for reasons related to the process of speciation will bias estimates of sex-specific mutation rates using the evolutionary approach.
A summary of issues of concern when estimating the male mutation bias using the evolutionary approach is provided in table 1.
Table 1.
Issues of concern when estimating the male mutation bias using the evolutionary approach.
| aspect | comment |
|---|---|
| sequence neutrality | can selective neutrality of synonymous sites be assumed? Is non-coding DNA free of selective constraint? |
| adaptive mutation rate variation | theoretical arguments would suggest that the X (Z) chromosome has evolved a lower mutation rate, for adaptive reasons. However, there is at present limited support for this phenomenon |
| regional mutation rate variation | owing to regional mutation rate variation, it is advisable to estimate the male mutation bias by sampling sequence data from several genomic regions, rather retrieving the same amount of sequence from a single region only |
| site heterogeneity | non-replication-dependent mutations, such as deamination of cytosine at CpG dinucleotides, are not expected to show the same sex-specific rates of mutation as mutation induced by replication errors |
| mutation rate per cell division | it is still unclear if the mutation rate per cell division is the same in the male and the female germ line. This is also the case for the mutation rate in relation to parental age |
| ancestral polymorphism | for closely related taxa, polymorphism in a common ancestor may make a significant contribution to the observed pairwise divergence. If polymorphism levels differ between, for example, autosomes and sex chromosomes, estimates of the male mutation bias may thus be flawed. This can be handled by using more divergent comparisons, or by trying to correct divergence estimates for ancestral polymorphism |
5. Life history and the male mutation bias
In the discussion above, I have focused on the patterns and causes of variation in sex-specific mutation rates across the genome, and the estimation of the male mutation bias. What about variation among genomes (lineages)? Two life-history parameters that potentially could correlate with αm are generation time and intensity of sexual selection. Each of these parameters can be expected to covary with the ratio of the number of cell divisions in the male and female germ line; indeed, if c varies among species, so should αm. Specifically, while the number of mitotic cell divisions in oogenesis may be relatively similar across at least many vertebrates, the number of cell divisions in spermatogenesis can be expected to be higher in species that (i) live longer, (ii) reproduce at an older age (have long pre-reproductive time), and/or (iii) reproduce several times in life. Similarly, sperm competition among males can lead to increased sperm production and hence a larger number of cell divisions in spermatogenesis.
It is not straightforward to test the predictions of generation time and sexual selection effects because the evolutionary approach for αm estimation measures the number of mutations accumulated over long periods of time, during which the life history of ancestral species along lineages may have varied. However, whole-genome data from different mammalian lineages are supportive of a generation time effect: αm is of the order of 4–6 in primates (CSAC 2005; Taylor et al. 2006), around 3 in the dog lineage (Lindblad-Toh et al. 2005) and around 2 in mice and rats (Makova et al. 2004; RGSPC 2004); these values are in qualitative agreement with (i) the long generation time typically seen in primates, (ii) the short generation time of small rodents, and (iii) carnivores being intermediate in this respect. Clades of long-lived birds were found to have a higher mean αm than clades with shorter life span. Moreover, clades of birds with high intensity of sexual selection had higher mean αm than clades with more monogamous species (Bartosch-Harlid et al. 2003).
Most studies of male-biased mutation so far reported using the evolutionary approach have analysed either mammals (Shimmin et al. 1993a, 1994; Chang et al. 1994; Chang & Li 1995; Agulnik et al. 1997; Huang et al. 1997; McVean & Hurst 1997; Bohossian et al. 2000; Lawson & Hewitt 2002; Makova & Li 2002; Malcom et al. 2003; Tucker et al. 2003; Sandstedt & Tucker 2005) or birds (Ellegren & Fridolfsson 1997; Kahn & Quinn 1999; Carmichael et al. 2000; Fridolfsson & Ellegren 2000; Garcia-Moreno & Mindell 2000; Bartosch-Härlid et al. 2003; Axelsson et al. 2004; Berlin et al. 2006), so the correlates with life history are mainly restricted to these groups of species. A study of salmonid fishes revealed an αm estimate of 6 (Ellegren & Fridolfsson 2003) and in dioecious plants there are also evidence for higher rate of mutation on the Y than the X chromosome (Filatov & Charlesworth 2002; Whittle & Johnston 2002); indeed, there are more cell divisions in pollen than in ovule production. In Drosophila, the observation of similar rates of sequence divergence in the different chromosomal classes (Bauer & Aquadro 1997) ties in nicely with the number of cell divisions in the male and female germ line also being similar.
6. The evolutionary biology and genetics of the male mutation bias
(a) Female preference for old males
Mutations that effect fitness are generally negative, i.e. they introduce a genetic or mutation load when segregating in the population (Haldane 1937). Although the precise rate of deleterious mutation can be difficult to estimate (Eyre-Walker & Keightley 1999), the continuous sperm production throughout life and the concomitant accumulation of new mutations in the germ line means that the mutation load (Haldane 1937; Muller 1950) might increase as males age (Hansen & Price 1995). While intuitively making sense, Hansen & Price (1999) showed formally that this holds true at mutation–selection balance under various age-specific fitness component scenarios and when mutations have unconditionally deleterious effects. Mutation load is an important factor in the evolution of mate choice (Heisler 1984; Rice 1988; Pomiankowski et al. 1991), a common assumption being that old males are genetically superior owing to viability selection (Manning 1985; Kokko & Lindström 1996; Kokko 1998). Accordingly, female preference should be biased towards older males. However, if mutation load tends to increase in all males as they age, the trade-off in female choice between obtaining either direct or indirect (the ‘good genes hypothesis’) benefits and avoiding paternally derived deleterious mutation may select for preference for subadult males. This is an important topic worthy of further research.
(b) Male-biased mutation and the enigma of maintenance of genetic variance in viability genes
Another evolutionary aspect of male-biased mutation is the link between the intensity of sexual selection and the elevation of male mutation rates. The maintenance of additive genetic variance for fitness traits despite strong directional selection by choosy females represents a classical problem of sexual selection theory (Andersson 1994). As discussed earlier, there is some evidence for an increase in the male mutation bias as a consequence of increased sperm production resulting from sperm competition (Bartosch-Härlid et al. 2003). Since the former increase should primarily be owing to an elevated rate of male mutation, not a slowdown in the female mutation rate, the overall effect on the total (sex-average) mutation rate is that of an increase. Based on these lines of thinking, it has been suggested that there is a positive correlation between the intensity of sexual selection and the genetic variability of viability genes (Bartosch-Härlid et al. 2003, Møller & Cuervo 2003). In other words, at the same time as depletion of fitness-related genetic variability accelerates through an increased variance in male reproductive success, the generation of new variability also accelerates. This is also an aspect that should be subject to further research, through modelling.
(c) The rate of adaptive evolution
The rate of adaptive evolution could be limited by the supply of mutations favoured by positive selection. In itself, the efficacy of selection will differ between autosomal and sex chromosomal genes as the latter are not always in diploid state. Additionally, sex-biased gene expression and sexual antagonism have different consequences to selection whether or not a gene is sex-linked (Rice 1984; Charlesworth et al. 1987). However, male-biased mutation causing mutation rate heterogeneity among chromosomal classes will also affect the rate of adaptive evolution in different classes. Kirkpatrick & Hall (2004) showed that for additive mutations (dominance coefficient h=0.5) subject to equal selection in the two sexes, the relative substitution rate of such X-linked mutations will be lower, and that of Z-linked higher, than for autosomal alleles. Moreover, when h≠0.5, X-linked alleles will only evolve faster than autosomal alleles when mutations are quite recessive. On the other hand, Z-linked alleles will evolve faster than autosomal alleles over a broad range of dominance values.
Kirkpatrick & Hall (2004) highlight two important consequences of the contrasting predictions made for XY and ZW systems. Firstly, as there is a propensity for genes encoding sexually selected traits to map to the X and the Z chromosome (Reinhold 1998), the ease by which adaptive evolution can occur on the Z chromosome would favour the rapid evolution of male sexual display in female heterogametic organisms. Indeed, birds and butterflies (ZW sex determination) are often recognized as having pronounced sexual dimorphism and exaggerated display traits. Secondly, a high rate of accumulation of non-synonymous substitution on the Z chromosome could potentially contribute to a strong role of the Z chromosome in post-zygotic isolation, analogous to the ‘large-X effect’ (Turelli & Orr 2000) described for male heterogametic species. It also has a bearing on the ‘faster-X hypothesis’ (Charlesworth et al. 1987), which suggests that X-linked genes would evolve faster than autosomal genes when the two sexes mutate at similar rates. However, the empirical support for faster-X evolution is elusive (Counterman et al. 2004; Thornton et al. 2006). In light of this, it would be valuable to obtain comparative data on the ratio of non-synonymous (dN) to synonymous (dS) substitution in autosomes and the Z chromosome of ZW species such as birds or butterflies.
7. Conclusions and further research
There is now firm support that the overall rate of mutation differs between the sexes for taxa in which the number of germ line cell divisions differs between males and females. However, early work in this area probably had an overly simplistic view on the male mutation bias, as it was then assumed that a sex difference in the mutation rate was the main denominator of mutation rate heterogeneity among genomic regions (with the exception of non-replication-dependent CpG mutations). We know now that this is not the case. On the other hand, while other factors are of predominant importance at any individual locus, the genome-wide mean rate difference between sexes effects the total number of mutations contributed by fathers and mothers. On this basis, the term ‘male-driven evolution’ was coined (Shimmin et al. 1993a).
Given the importance of mutations to central aspects of evolutionary biology such as sexual selection and speciation, and that mutation rate represents a key parameter in population genetics, there is a need for increased understanding of the character and magnitude of the male mutation bias across phylogenetically divergent lineages. Moreover, additional work is required to model ramifications of mutation rate on topics such as mate choice and post-zygotic isolation. Furthermore, to the evolutionary aspects discussed earlier should be added the call for further studies of the interesting link between male mutation bias and evolution of sex. One common explanation for the twofold cost of sexual reproduction for females is that meiosis reduces the influence of deleterious mutation. However, in a seminal study by Redfield (1994), it was shown that when sex-specific mutation rates are highly skewed, female benefit incurred from sex can be outweighed by the negative effects of new deleterious mutations given to progeny by male gametes. Could it be that the cost–benefit balance of sexual reproduction introduces a limit for how high male mutation rates can be?
It is also clear that studies of sex-specific mutation rates can offer insight into germ line biology and provide knowledge of the nature of mutations at individual loci, notably such loci that have a strong effect of fitness. Analyses of the parental origin of spontaneous mutation causing human disease (Hurst & Ellegren 1998) have mostly found an excess of paternally derived mutations, in some cases almost skewed towards male-only origin. Sex differences in germ line methylation levels can contribute to such extreme biases if disease-causing mutations are confined to CpG sites (El-Maarri et al. 1998). However, this may represent exceptions rather than a role as Taylor et al. (2006) found an overall tendency for more sex-equal mutation rates at CpG than non-CpG sites. A better knowledge of the pattern at individual loci, and even at individual sites, is needed and this applies particularly to studies of organisms other than humans.
Finally, another interesting dimension of male-biased mutation is the relationship between the continuous generation of new mutations as males age and the actual accumulation of mutations in male gametes (Jung et al. 2003). Selection for or against non-neutral mutations in primordial germ cells and/or sperm can affect both the mutation load carried by male gametes and the contribution of advantageous mutation to progeny. This should be taken into account when modelling the evolutionary consequences of male-biased mutation. That the issue might be more complicated than perhaps intuitively thought is illustrated by the observation of selection for a pathogenic mutation in the male germ line at a disease-causing locus (Goriely et al. 2003).
Acknowledgments
I thank the Swedish Research Council for financial support and Judith Mank for discussion.
References
- Agulnik A.I, Bishop C.E, Lerner J.L, Agulnik S.I, Solovyev V.V. Analysis of mutation rates in the SMCY/SMCX genes shows that mammalian evolution is male-driven. Mamm. Genome. 1997;8:134–138. doi: 10.1007/s003359900372. doi:10.1007/s003359900372 [DOI] [PubMed] [Google Scholar]
- Allen J.W, Ehling U.H, Moore M.M, Lewis S.E. Germ line specific factors in chemical mutagenesis. Mutat. Res. 1995;330:219–231. doi: 10.1016/0027-5107(95)00042-h. [DOI] [PubMed] [Google Scholar]
- Andersson M. Princeton University Press; Princeton, NJ: 1994. Sexual selection. [Google Scholar]
- Andolfatto P. Adaptive evolution of non-coding DNA in Drosophila. Nature. 2005;437:1149–1152. doi: 10.1038/nature04107. doi:10.1038/nature04107 [DOI] [PubMed] [Google Scholar]
- Axelsson E, Smith N.G, Sundström H, Berlin S, Ellegren H. Male-biased mutation rate and divergence in autosomal, Z-linked and W-linked introns of chicken and turkey. Mol. Biol. Evol. 2004;21:1538–1547. doi: 10.1093/molbev/msh157. doi:10.1093/molbev/msh157 [DOI] [PubMed] [Google Scholar]
- Axelsson E, Webster M.T, Smith N.G, Burt D.W, Ellegren H. Comparison of the chicken and turkey genomes reveals a higher rate of nucleotide divergence on microchromosomes than macrochromosomes. Genome Res. 2005;15:120–125. doi: 10.1101/gr.3021305. doi:10.1101/gr.3021305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D, Charlesworth B. Reduced adaptation of a non-recombining neo-Y chromosome. Nature. 2002;416:323–326. doi: 10.1038/416323a. doi:10.1038/416323a [DOI] [PubMed] [Google Scholar]
- Bartosch-Härlid A, Berlin S, Smith N.G, Møller A.P, Ellegren H. Life history and the male mutation bias. Evolution. 2003;57:2398–2406. doi: 10.1554/03-036. doi:10.1554/03-036 [DOI] [PubMed] [Google Scholar]
- Bauer V.L, Aquadro C.F. Rates of DNA sequence evolution are not sex-biased in Drosophila melanogaster and D. simulans. Mol. Biol. Evol. 1997;14:1252–1257. doi: 10.1093/oxfordjournals.molbev.a025734. [DOI] [PubMed] [Google Scholar]
- Berlin S, Ellegren H. Chicken W: a genetically uniform chromosome in a highly variable genome. Proc. Natl Acad. Sci. USA. 2004;101:15 967–15 969. doi: 10.1073/pnas.0405126101. doi:10.1073/pnas.0405126101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin S, Brandström M, Backström N, Axelsson E, Smith N.G.C, Ellegren H. Substitution rate heterogeneity and the male mutation bias. J. Mol. Evol. 2006;62:226–233. doi: 10.1007/s00239-005-0103-6. doi:10.1007/s00239-005-0103-6 [DOI] [PubMed] [Google Scholar]
- Bestor T.H. Cytosine methylation and the unequal developmental potentials of the oocyte and sperm genomes. Am. J. Hum. Genet. 1998;62:1269–1273. doi: 10.1086/301891. doi:10.1086/301891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackshear P.E, et al. Brca1 and Brca2 expression patterns in mitotic and meiotic cells of mice. Oncogene. 1998;16:61–68. doi: 10.1038/sj.onc.1201506. doi:10.1038/sj.onc.1201506 [DOI] [PubMed] [Google Scholar]
- Bohossian H.B, Skaletsky H, Page D.C. Unexpectedly similar rates of nucleotide substitution found in male and female hominids. Nature. 2000;406:622–625. doi: 10.1038/35020557. doi:10.1038/35020557 [DOI] [PubMed] [Google Scholar]
- Carmichael A.N, Fridolfsson A.K, Halverson J, Ellegren H. Male-biased mutation rates revealed from Z and W chromosome-linked ATP synthase alpha-subunit (ATP5A1) sequences in birds. J. Mol. Evol. 2000;50:443–447. doi: 10.1007/s002390010046. [DOI] [PubMed] [Google Scholar]
- Casane D, Boissinot S, Chang B.H, Shimmin L.C, Li W.H. Mutation pattern variation among regions of the primate genome. J. Mol. Evol. 1997;45:216–226. doi: 10.1007/pl00006223. doi:10.1007/PL00006223 [DOI] [PubMed] [Google Scholar]
- Chamary J.V, Hurst L.D. Similar rates but different modes of sequence evolution in introns and at exonic silent sites in rodents: evidence for selectively driven codon usage. Mol. Biol. Evol. 2004;21:1014–1021. doi: 10.1093/molbev/msh087. doi:10.1093/molbev/msh087 [DOI] [PubMed] [Google Scholar]
- Chang B.H, Li W.H. Estimating the intensity of male-driven evolution in rodents by using X-linked and Y-linked Ube 1 genes and pseudogenes. J. Mol. Evol. 1995;40:70–77. doi: 10.1007/BF00166597. doi:10.1007/BF00166597 [DOI] [PubMed] [Google Scholar]
- Chang B.H, Shimmin L.C, Shyue S.K, Hewett-Emmett D, Li W.H. Weak male-driven molecular evolution in rodents. Proc. Natl Acad. Sci. USA. 1994;91:827–831. doi: 10.1073/pnas.91.2.827. doi:10.1073/pnas.91.2.827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. The degeneration of Y chromosomes. Phil. Trans. R. Soc. B. 2000;355:1563–1572. doi: 10.1098/rstb.2000.0717. doi:10.1098/rstb.2000.0717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Coyne J.A, Barton N.H. The relative rates of evolution of sex-chromosomes and autosomes. Am. Nat. 1987;130:113–146. doi:10.1086/284701 [Google Scholar]
- Chimpanzee Sequencing and Analysis Consortium (CSAC) Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. doi:10.1038/nature04072 [DOI] [PubMed] [Google Scholar]
- Chuang J.H, Li H. Functional bias and spatial organization of genes in mutational hot and cold regions in the human genome. PLoS Biol. 2004;2:e29. doi: 10.1371/journal.pbio.0020029. doi:10.1371/journal.pbio.0020029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counterman B.A, Ortiz-Barrientos D, Noor M.A. Using comparative genomic data to test for fast-X evolution. Evolution. 2004;58:656–660. doi:10.1554/03-413 [PubMed] [Google Scholar]
- Crow J.F. How much do we know about spontaneous human mutation rates? Env. Mol. Mutagen. 1993;21:122–129. doi: 10.1002/em.2850210205. [DOI] [PubMed] [Google Scholar]
- Crow J.F. The high spontaneous mutation rate: is it a health risk? Proc. Natl Acad. Sci. USA. 1997;94:8380–8386. doi: 10.1073/pnas.94.16.8380. doi:10.1073/pnas.94.16.8380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll D.J, Migeon B.R. Sex difference in methylation of single-copy genes in human meiotic germ cells: implications for X chromosome inactivation, parental imprinting, and origin of CpG mutations. Somat. Cell Mol. Genet. 1990;16:267–282. doi: 10.1007/BF01233363. doi:10.1007/BF01233363 [DOI] [PubMed] [Google Scholar]
- Ellegren H. Heterogeneous mutation processes in human microsatellite DNA sequences. Nat. Genet. 2000;24:400–402. doi: 10.1038/74249. doi:10.1038/74249 [DOI] [PubMed] [Google Scholar]
- Ellegren H. Human mutation—blame (mostly) men. Nat. Genet. 2002;31:9–10. doi: 10.1038/ng0502-9. doi:10.1038/ng0502-9 [DOI] [PubMed] [Google Scholar]
- Ellegren H. Microsatellites: simple sequences with complex evolution. Nat. Rev. Genet. 2004;5:435–445. doi: 10.1038/nrg1348. doi:10.1038/nrg1348 [DOI] [PubMed] [Google Scholar]
- Ellegren H, Fridolfsson A.K. Male-driven evolution of DNA sequences in birds. Nat. Genet. 1997;17:182–184. doi: 10.1038/ng1097-182. doi:10.1038/ng1097-182 [DOI] [PubMed] [Google Scholar]
- Ellegren H, Fridolfsson A.K. Sex-specific mutation rates in salmonid fish. J. Mol. Evol. 2003;56:458–463. doi: 10.1007/s00239-002-2416-z. doi:10.1007/s00239-002-2416-z [DOI] [PubMed] [Google Scholar]
- El-Maarri O, et al. Methylation levels at selected CpG sites in the factor VIII and FGFR3 genes, in mature female and male germ cells: implications for male-driven evolution. Am. J. Hum. Genet. 1998;63:1001–1008. doi: 10.1086/302065. doi:10.1086/302065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlandsson R, Wilson J.F, Pääbo S. Sex chromosomal transposable element accumulation and male-driven substitutional evolution in humans. Mol. Biol. Evol. 2000;17:804–812. doi: 10.1093/oxfordjournals.molbev.a026359. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A, Keightley P.D. High genomic deleterious mutation rates in hominids. Nature. 1999;397:344–347. doi: 10.1038/16915. doi:10.1038/16915 [DOI] [PubMed] [Google Scholar]
- Filatov D.A, Charlesworth D. Substitution rates in the X-linked and Y-linked genes of the plants, Silene latifolia and S. dioica. Mol. Biol. Evol. 2002;19:898–907. doi: 10.1093/oxfordjournals.molbev.a004147. [DOI] [PubMed] [Google Scholar]
- Filatov D.A, Moneger F, Negrutiu I, Charlesworth D. Low variability in a Y-linked plant gene and its implications for Y-chromosome evolution. Nature. 2000;404:388–390. doi: 10.1038/35006057. doi:10.1038/35006057 [DOI] [PubMed] [Google Scholar]
- Filipski J, Salinas J, Rodier F. Chromosome localization-dependent compositional bias of point mutations in Alu repetitive sequences. J. Mol. Biol. 1989;206:563–566. doi: 10.1016/0022-2836(89)90501-9. doi:10.1016/0022-2836(89)90501-9 [DOI] [PubMed] [Google Scholar]
- Fridolfsson A.K, Ellegren H. Molecular evolution of the avian CHD1 genes on the Z and W sex chromosomes. Genetics. 2000;155:1903–1912. doi: 10.1093/genetics/155.4.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney D.J, Keithley P.D. The scale of mutational variation in the murid genome. Genome Res. 2005;15:1086–1094. doi: 10.1101/gr.3895005. doi:10.1101/gr.3895005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Moreno J, Mindell D.P. Rooting a phylogeny with homologous genes on opposite sex chromosomes (gametologs): a case study using avian CHD. Mol. Biol. Evol. 2000;17:1826–1832. doi: 10.1093/oxfordjournals.molbev.a026283. [DOI] [PubMed] [Google Scholar]
- Goriely A, McVean G.A, Rojmyr M, Ingemarsson B, Wilkie A.O. Evidence for selective advantage of pathogenic FGFR2 mutations in the male germ line. Science. 2003;301:643–646. doi: 10.1126/science.1085710. doi:10.1126/science.1085710 [DOI] [PubMed] [Google Scholar]
- Haldane J.B.S. Sex ratio and unidirectional sterility in hybrid animals. J. Genet. 1922;58:237–242. [Google Scholar]
- Haldane J.B.S. The rate of spontaneous mutation of a human gene. J. Genet. 1935;31:317–326-272. doi: 10.1007/BF02717892. [DOI] [PubMed] [Google Scholar]
- Haldane J.B.S. The effect of variation on fitness. Am. Nat. 1937;71:337–349. doi:10.1086/280722 [Google Scholar]
- Haldane J.B.S. The mutation rate of the gene for hemophilia, and its segregation rations in males and females. Ann. Hum. Genet. 1947;13:262–272. doi: 10.1111/j.1469-1809.1946.tb02367.x. [DOI] [PubMed] [Google Scholar]
- Haldane J.B.S. The formal genetics of man. Proc. R. Soc. B. 1948;135:147–170. [Google Scholar]
- Hansen T.F, Price D.K. Good genes and old age: do old males provide superior genes? J. Evol. Biol. 1995;8:759–778. doi:10.1046/j.1420-9101.1995.8060759.x [Google Scholar]
- Hansen T.F, Price D.K. Age- and sex-distribution of the mutation load. Genetica. 1999;106:251–262. doi: 10.1023/a:1003988101586. doi:10.1023/A:1003988101586 [DOI] [PubMed] [Google Scholar]
- Hardison R.C, et al. Covariation in frequencies of substitution, deletion, transposition, and recombination during eutherian evolution. Genome Res. 2003;13:13–26. doi: 10.1101/gr.844103. doi:10.1101/gr.844103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare M.P, Palumbi R.S. High intron sequence conservation across three mammalian orders suggests functional constraints. Mol. Biol. Evol. 2003;20:969–978. doi: 10.1093/molbev/msg111. doi:10.1093/molbev/msg111 [DOI] [PubMed] [Google Scholar]
- Heisler I.L. A quantitative genetic model for the origin of mating preferences. Evolution. 1984;38:1283–1295. doi: 10.1111/j.1558-5646.1984.tb05650.x. doi:10.2307/2408635 [DOI] [PubMed] [Google Scholar]
- Hellman I, Ebersberger I, Ptak S.E, Pääbo S, Przeworski M. A neutral explanation for the correlation of diversity with recombination rates in humans. Am. J. Hum. Genet. 2003;72:1527–1535. doi: 10.1086/375657. doi:10.1086/375657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Chang B.H, Gu X, Hewett-Emmett D, Li W.H. Sex differences in mutation rate in higher primates estimated from AMG intron sequences. J. Mol. Evol. 1997;44:463–465. doi: 10.1007/pl00006166. doi:10.1007/PL00006166 [DOI] [PubMed] [Google Scholar]
- Hurst L.D, Ellegren H. Sex biases in the mutation rate. Trends Genet. 1998;14:446–452. doi: 10.1016/s0168-9525(98)01577-7. doi:10.1016/S0168-9525(98)01577-7 [DOI] [PubMed] [Google Scholar]
- Huttley G.A, Jakobsen I.B, Wilson S.R, Easteal S. How important is DNA replication for mutagenesis? Mol. Biol. Evol. 2000;17:929–937. doi: 10.1093/oxfordjournals.molbev.a026373. [DOI] [PubMed] [Google Scholar]
- Ibarguchi G, Gissing G.J, Gaston A.J, Boag P.T, Friesen V.L. Male-biased mutation rates and the overestimation of extrapair paternity: problem, solution, and illustration using thick-billed murres (Uria lomvia, Alcidae) J. Hered. 2004;95:209–216. doi: 10.1093/jhered/esh029. doi:10.1093/jhered/esh029 [DOI] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consoritum (ICGSC) Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. doi:10.1038/nature03154 [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium (IHGSC) Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. doi:10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- International SNP Map Working Group (ISMWG) A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. doi:10.1038/35057149 [DOI] [PubMed] [Google Scholar]
- Johnson J, Canning J, Kaneko T, Pru J.K, Tilly J.L. Germ line stem cells and follicular renewal in the postnatal mammalian ovary. Nature. 2004;428:145–150. doi: 10.1038/nature02316. doi:10.1038/nature02316 [DOI] [PubMed] [Google Scholar]
- Jung A, Schuppe H.C, Schill W.B. Are children of older fathers at risk for genetic disorders? Andrologia. 2003;35:191–199. doi: 10.1046/j.1439-0272.2003.00579.x. doi:10.1046/j.1439-0272.2003.00579.x [DOI] [PubMed] [Google Scholar]
- Jurka J. Evolutionary impact of human Alu repetitive elements. Curr. Opin. Genet. Dev. 2004;14:603–608. doi: 10.1016/j.gde.2004.08.008. doi:10.1016/j.gde.2004.08.008 [DOI] [PubMed] [Google Scholar]
- Kahn N.W, Quinn T.W. Male-driven evolution among Eoaves? A test of the replicative division hypothesis in a heterogametic female (ZW) system. J. Mol. Evol. 1999;49:750–759. doi: 10.1007/pl00006597. doi:10.1007/PL00006597 [DOI] [PubMed] [Google Scholar]
- Kirkpatrick M, Hall D.W. Male-biased mutation, sex linkage, and the rate of adaptive evolution. Evolution. 2004;58:437–440. doi:10.1554/03-333 [PubMed] [Google Scholar]
- Kokko H. Good genes, old age and life-histroy trade-offs. Evol. Ecol. 1998;12:739–750. doi:10.1023/A:1006541701002 [Google Scholar]
- Kokko H, Lindström J. Evolution of female preference for old mates. Proc. R. Soc. B. 1996;263:1533–1538. [Google Scholar]
- Kondrashov A.S. Modifiers of mutation–selection balance—general-approach and the evolution of mutation rates. Genet. Res. 1995;66:53–69. [Google Scholar]
- Lawson L.J, Hewitt G.M. Comparison of substitution rates in ZFX and ZFY introns of sheep and goat related species supports the hypothesis of male-biased mutation rates. J. Mol. Evol. 2002;54:54–61. doi: 10.1007/s00239-001-0017-x. doi:10.1007/s00239-001-0017-x [DOI] [PubMed] [Google Scholar]
- Leigh E.G. Natural selection and mutability. Am. Nat. 1970;104:301. doi:10.1086/282663 [Google Scholar]
- Lercher M.J, Hurst L.D. Human SNP variability and mutation rate are higher in regions of high recombination. Trends Genet. 2002;18:337–340. doi: 10.1016/s0168-9525(02)02669-0. doi:10.1016/S0168-9525(02)02669-0 [DOI] [PubMed] [Google Scholar]
- Lercher M.J, Williams E.J, Hurst L.D. Local similarity in evolutionary rates extends over whole chromosomes in human–rodent and mouse–rat comparisons: implications for understanding the mechanistic basis of the male mutation bias. Mol. Biol. Evol. 2001;18:2032–2039. doi: 10.1093/oxfordjournals.molbev.a003744. [DOI] [PubMed] [Google Scholar]
- Li W.H, Yi S, Makova K. Male-driven evolution. Curr. Opin. Genet. Dev. 2002;12:650–656. doi: 10.1016/s0959-437x(02)00354-4. doi:10.1016/S0959-437X(02)00354-4 [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. doi:10.1038/nature04338 [DOI] [PubMed] [Google Scholar]
- Macphee D.G. Mismatch repair, somatic mutations, and the origin of cancer. Cancer Res. 1995;55:5489–5492. [PubMed] [Google Scholar]
- Majewski J, Ott J. Distribution and characterization of regulatory elements in the human genome. Genome Res. 2002;12:1827–1836. doi: 10.1101/gr.606402. doi:10.1101/gr.606402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makova K.D, Li W.H. Strong male-driven evolution of DNA sequences in humans and apes. Nature. 2002;416:624–626. doi: 10.1038/416624a. doi:10.1038/416624a [DOI] [PubMed] [Google Scholar]
- Makova K.D, Yang S, Chiaromonte F. Insertions and deletions are male biased too: a whole-genome analysis in rodents. Genome Res. 2004;14:567–573. doi: 10.1101/gr.1971104. doi:10.1101/gr.1971104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcom C.M, Wyckoff G.J, Lahn B.T. Genic mutation rates in mammals: local similarity, chromosomal heterogeneity, and X-versus-autosome disparity. Mol. Biol. Evol. 2003;20:1633–1641. doi: 10.1093/molbev/msg178. doi:10.1093/molbev/msg178 [DOI] [PubMed] [Google Scholar]
- Manning J.T. Choosy females and correlates of male age. J. Theor. Biol. 1985;116:349–356. doi:10.1016/S0022-5193(85)80273-3 [Google Scholar]
- McVean G.T, Hurst L.D. Evidence for a selectively favourable reduction in the mutation rate of the X chromosome. Nature. 1997;386:388–392. doi: 10.1038/386388a0. doi:10.1038/386388a0 [DOI] [PubMed] [Google Scholar]
- Miyata T, Hayashida H, Kuma K, Mitsuyasu K, Yasunaga T. Male-driven molecular evolution: a model and nucleotide sequence analysis. Cold Spring Harb. Symp. Quant. Biol. 1987;52:863–867. doi: 10.1101/sqb.1987.052.01.094. [DOI] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium (MGSC) Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. doi:10.1038/nature01262 [DOI] [PubMed] [Google Scholar]
- Møller A.P, Cuervo J.J. Sexual selection, germline mutation rate and sperm competition. BMC Evol. Biol. 2003;3:6. doi: 10.1186/1471-2148-3-6. doi:10.1186/1471-2148-3-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi M, Nuttall R, Naiman D, Bouffard G, Malley J, Andrews J, Eastman S, Oliver B. Paucity of genes on the D. melanogaster X chromosome showing male-biased expression. Science. 2003;299:697–700. doi: 10.1126/science.1079190. doi:10.1126/science.1079190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Richter D.J, Gnerre S, Lander E.S, Reich D. Genetic evidence for complex speciation of humans and chimpanzees. Nature. 2006;441:1103–1108. doi: 10.1038/nature04789. doi:10.1038/nature04789 [DOI] [PubMed] [Google Scholar]
- Pomiankowski A, Iwasa Y, Nee S. The evolution of costly mate preference: I. Fisher and biased mutation. Evolution. 1991;45:1422–1430. doi: 10.1111/j.1558-5646.1991.tb02645.x. doi:10.2307/2409889 [DOI] [PubMed] [Google Scholar]
- Ranz J.M, Castillo-Davis C.I, Meiklejohn C.D, Hartl D.L. Sex-dependent gene expression and evolution of the D. melanogaster transcriptome. Science. 2003;300:1742–1744. doi: 10.1126/science.1085881. doi:10.1126/science.1085881 [DOI] [PubMed] [Google Scholar]
- Rat Genome Sequencing Project Consortium (RGSC) Genome sequence of the brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. doi:10.1038/nature02426 [DOI] [PubMed] [Google Scholar]
- Redfield R.J. Male mutation-rates and the cost of sex for females. Nature. 1994;369:145–147. doi: 10.1038/369145a0. doi:10.1038/369145a0 [DOI] [PubMed] [Google Scholar]
- Reinhold K. Sex linkage among genes controlling sexually selected traits. Behav. Ecol. Sociobiol. 1998;44:1–7. doi:10.1007/s002650050508 [Google Scholar]
- Rice W.R. Sex chromosomes and the evolution of sexual dimorphism. Evolution. 1984;38:735–742. doi: 10.1111/j.1558-5646.1984.tb00346.x. doi:10.2307/2408385 [DOI] [PubMed] [Google Scholar]
- Rice W.R. Heritable variation in fitness as a prerequisite for adaptive female choice: the effect of mutation–selection balance. Evolution. 1988;42:817–820. doi: 10.1111/j.1558-5646.1988.tb02500.x. doi:10.2307/2408873 [DOI] [PubMed] [Google Scholar]
- Roy A.M, Carroll M.L, Nguyen S.N, Salem A.-H, Oldridge M, Wilkie A.O.M, Batzer M, Deininger P.L. Potential gene conversion and source genes for recently integrated Alu elements. Genome Res. 2000;10:1485–1495. doi: 10.1101/gr.152300. doi:10.1101/gr.152300 [DOI] [PubMed] [Google Scholar]
- Sandstedt S.A, Tucker P.K. Male-driven evolution in closely related species of the mouse genus Mus. J. Mol. Evol. 2005;61:138–144. doi: 10.1007/s00239-004-0279-1. doi:10.1007/s00239-004-0279-1 [DOI] [PubMed] [Google Scholar]
- Shimmin L.C, Chang B.H, Li W.H. Male-driven evolution of DNA sequences. Nature. 1993a;362:745–747. doi: 10.1038/362745a0. doi:10.1038/362745a0 [DOI] [PubMed] [Google Scholar]
- Shimmin L.C, Chang B.H, Hewett-Emmett D, Li W.H. Potential problems in estimating the male-to-female mutation rate ratio from DNA sequence data. J. Mol. Evol. 1993b;37:160–166. doi: 10.1007/BF02407351. doi:10.1007/BF02407351 [DOI] [PubMed] [Google Scholar]
- Shimmin L.C, Chang B.H, Li W.H. Contrasting rates of nucleotide substitution in the X-linked and Y-linked zinc finger genes. J. Mol. Evol. 1994;39:569–578. doi: 10.1007/BF00160402. doi:10.1007/BF00160402 [DOI] [PubMed] [Google Scholar]
- Smith N.G, Hurst L.D. The causes of synonymous rate variation in the rodent genome: can substitution rates be used to estimate the sex bias in mutation rate? Genetics. 1999;152:661–673. doi: 10.1093/genetics/152.2.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith N.G.C, Webster M.T, Ellegren H. Deterministic mutation rate variation in the human genome. Genome Res. 2002;12:1350–1356. doi: 10.1101/gr.220502. doi:10.1101/gr.220502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R, Ast G. Intronic sequences flanking alternatively spliced exons are conserved between human and mouse. Genome Res. 2003;13:1631–1637. doi: 10.1101/gr.1208803. doi:10.1101/gr.1208803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundström H, Webster M.T, Ellegren H. Is the rate of insertion and deletion mutation male biased?: molecular evolutionary analysis of avian and primate sex chromosome sequences. Genetics. 2003;164:259–268. doi: 10.1093/genetics/164.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J, Tyekucheva S, Zody M, Chiaromonte F, Makova K.D. Strong and weak male mutation bias at different sites in the primate genomes: insights from the human–chimpanzee comparison. Mol. Biol. Evol. 2006;23:565–573. doi: 10.1093/molbev/msj060. doi:10.1093/molbev/msj060 [DOI] [PubMed] [Google Scholar]
- Thornton K, Bachtrog D, Andolfatto P. X chromosomes and autosomes evolve at similar rates in Drosophila: no evidence for faster-X protein evolution. Genome Res. 2006;16:498–504. doi: 10.1101/gr.4447906. doi:10.1101/gr.4447906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker P.K, Adkins R.M, Rest J.S. Differential rates of evolution for the ZFY-related zinc finger genes, Zfy, Zfx, and Zfa in the mouse genus Mus. Mol. Biol. Evol. 2003;20:999–1005. doi: 10.1093/molbev/msg112. doi:10.1093/molbev/msg112 [DOI] [PubMed] [Google Scholar]
- Turelli M, Orr H.A. Dominance, epistasis and the genetics of postzygotic isolation. Genetics. 2000;154:1663–1679. doi: 10.1093/genetics/154.4.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urrutia A.O, Hurst L.D. The signature of selection mediated by expression on human genes. Genome Res. 2003;13:2260–2264. doi: 10.1101/gr.641103. doi:10.1101/gr.641103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel F, Motulsky A.G. Springer; Berlin, Germany: 1997. Human genetics: problems and approaches. [Google Scholar]
- Webster M.T, Smith N.G.C, Hultin-Rosenberg L, Arndt P.F, Ellegren H. Male-driven biased gene conversion governs the evolution of base composition in human Alu repeats. Mol. Biol. Evol. 2005;22:1468–1474. doi: 10.1093/molbev/msi136. doi:10.1093/molbev/msi136 [DOI] [PubMed] [Google Scholar]
- Whittle C.A, Johnston M.O. Male-driven evolution of mitochondrial and chloroplastidial DNA sequences in plants. Mol. Biol. Evol. 2002;19:938–949. doi: 10.1093/oxfordjournals.molbev.a004151. [DOI] [PubMed] [Google Scholar]
- Winter R.M, Tuddenham E.G.D, Goldman E, Matthews K.B. A maximum likelihood estimate of the sex ratio of mutation rates in haemophilia A. Hum. Genet. 1983;64:156–159. doi: 10.1007/BF00327115. doi:10.1007/BF00327115 [DOI] [PubMed] [Google Scholar]