Abstract
Genetic diversity can benefit social insects by providing variability in immune defences against parasites and pathogens. However, social parasites of ants infest colonies and not individuals, and for them a different relationship between genetic diversity and resistance may exist. Here, we investigate the genetic variation, assessed using up to 12 microsatellite loci, of workers in 91 Formica lemani colonies in relation to their infestation by the specialist social parasite Microdon mutabilis. At the main study site, workers in infested colonies exhibited lower relatedness and higher estimated queen numbers, on average, than uninfested ones. Additionally, estimated queen numbers were negatively correlated with estimated average numbers of mates per queen within infested colonies. At another site, infested colonies also exhibited significantly lower worker relatedness, and estimated queen numbers were comparable in trend. In contrast, in two populations of F. lemani where M. mutabilis was absent, relatedness within colonies was high (40 and 90% with R>0.6). While high genetic variation can benefit social insects by increasing their resistance to pathogens, there may be a cost in the increased likelihood of infiltration by social parasites owing to greater variation in nestmate recognition cues. This study provides the first empirical test of this hypothesis.
Keywords: myrmecophily, microsatellites, polygyny, monogyny, social parasites, colony gestalt
1. Introduction
It is widely accepted that high levels of genetic variation maintain flexibility in a species' response to a changing environment, including disease, parasite and predator pressures. It is also recognized that animals living at high densities and in social groups are especially vulnerable to parasites and disease (Alexander 1974), as the aggregation of highly related individuals increases the chance of transmission and infection (e.g. Brown & Brown 1986). In contrast to this, social animals achieve advantages over non-social species through the co-operation of closely related individuals within family groups, leading to a potential conflict between the benefits of social living and vulnerability to disease.
Mechanisms that increase genetic variation within the colonies of most ants (and other social hymenoptera) include the occurrence of multiple functional queens (polygyny) and the multiple mating of queens (polyandry). Both can lead to another set of within-colony conflicts over resource allocation or reproduction, with a trade-off between producing more sexual siblings and lower relatedness between these siblings. In some species, polyandry and polygyny may be negatively correlated, suggesting that an optimum level of genetic diversity may be selected for (Keller & Reeve 1994). However, Schmid-Hempel & Crozier (1999) found no overall relationship between the number of queens and mates in ants once phylogeny was accounted for, and the pattern may vary within species and between populations (Keller & Reeve 1994).
Thus, individuals within colonies of social insects are often less related than was once predicted. Moreover, several studies found that colonies containing close relatives are less fit and less productive than those that contain distantly related individuals (for ants: Sundström 1995; Cole & Wiernasz 1999; Hughes & Boomsma 2004; but see Fjerdingstad et al. 2003). Hypotheses to explain why hymenoptera are polygynous or polyandrous (Page 1986; Sherman et al. 1988; Boomsma & Ratnieks 1996) suggest that genetic diversity allows the expression of a more complete genetically based caste system, as well as expanding the environmental tolerances of a species, reducing the proportion of diploid males and allowing the enhancement of queens' inclusive fitness to induce workers to produce males. In addition, increased genetic diversity is thought to be adaptive (Page 1986; Crozier & Fjerdingstad 2001), where productivity is reduced due to susceptibility to parasites (Hamilton 1987; Sherman et al. 1988; Schmid-Hempel 1998). Evidence supporting this idea comes from bumble- and honeybees (Liersch & Schmid-Hempel 1998; Baer & Schmid-Hempel 1999; Tarpy 2003; Tarpy & Seeley 2006), from comparative studies involving large numbers of social insect host species and their associated parasite species (Schmid-Hempel & Crozier 1999), and from variation in colonies of the ant Acromyrmex octospinosus, both to resist a virulent fungal parasite (Hughes & Boomsma 2004) and to hinder the ability of the parasites to adapt while cycling within genetically diverse colonies (Hughes & Boomsma 2006).
Most evidence to date for the genetic variation versus parasitism hypothesis comes from studies of parasites and pathogens that attack individuals within colonies. However, generalizations have also been made for parasites that possess very different life histories. For example, the viruses, bacteria, microsporidea, macro-parasites, parasitoids and social parasites assayed by Schmid-Hempel (1994) employ a diversity of mechanisms for infesting insect societies—ranging from the antigenic surface protein recognition modes of pathogens to the chemical mimicry used by some myrmecophilous arthropods (Akino et al. 1999; Lenoir et al. 2001)—and it is not self-evident that a simple relationship between genetic variability and resistance applies across all types of interactions (Akino et al. 1999; Lenoir et al. 2001; van Baalen & Beekman 2006). Interactions between social parasites and their hosts differ fundamentally from those of pathogens, since the former infiltrate insect societies to exploit the resources of the colony as a whole, whereas the latter parasitize individual ants (Thomas et al. 2005). In the few social parasites studied, successful infiltration of the host society involves chemical camouflage or mimicry of host-recognition signals (Hölldobler & Wilson 1990; Akino et al. 1999; Lenoir et al. 2001; Schönrogge et al. 2004). The increase of genetic diversity within highly polygynous–polydomous social insect species may increase resistance to colony pathogens, but it might simultaneously make these societies penetrable by social parasites. Here, we test this latter hypothesis through studies of the ant Formica lemani and its syrphid social parasite, Microdon mutabilis. We compare the genetic variation between infested and uninfested colonies in two F. lemani populations where M. mutabilis occurs, and contrast these with two other F. lemani populations with no record of M. mutabilis parasitization.
2. Material and methods
(a) Study system
Little is known of the genetics or demography of F. lemani except that it is at least facultatively polygynous and polydomous, and is more apt to form colonies by budding than the closely related Formica fusca (Collingwood 1979). Microdon mutabilis lives almost its entire life cycle within F. lemani nests, where the larvae are predators of ant brood (Elmes et al. 1999; Schönrogge et al. 2002). Like many social parasite species, M. mutabilis is rare in comparison with its host (Schönrogge et al. 2002), and shows extreme host specificity not simply to one species of ant but to individual host populations and possibly to particular family groups within a F. lemani population (Elmes et al. 1999). In this sense, the F. lemani–M. mutabilis host–parasite interaction is analogous to some highly host-adapted systems of true parasites (e.g. Dybdahl & Lively 1996).
(b) Field collections
Our main infested study site was on the Isle of Mull in Scotland (NM 518 463 UK grid), where, in May 2001, 267 F. lemani colonies were identified under stones on either side of a 4 km track within a forest. Ant colonies were found continuously in the cleared area about 2 m on either side of the road but not within the forest. From 41 of these colonies (ranging from 1.8 to 3982 m apart), we collected 1952 workers (mean 47.6 workers per colony ±s.e. 2.6) and 26 queens (0–8 per colony). A second infested population was studied on the Burren, an area of limestone pavement in Ireland (M2708 Irish grid). Here, we collected 1394 workers (45.07±3.89) and 32 queens (0–8 per colony) from 30 colonies (2–517 m apart) on a 500 m×500 m grid in 2002, representing all the colonies within the grid. In 2002, we collected F. lemani from two well-known sites where M. mutabilis has never been recorded: 377 workers (37.7±2.07) were taken from 10 colonies (4.0–188.7 m apart) from coastal heath in St. Agnes (StAg), Cornwall (SW 699 502 UK grid); and 358 workers (35.8±2.16) from 10 colonies (3.1–74.8 m apart) in a field with limestone outcrops near Limerick (Lim), Ireland (R2951 Irish grid). Nests located under stones within 1 m of each other, and with interconnecting tunnels, were considered to be from the same colony. Ants were stored in 96% ethanol until genetic analysis.
(c) Evidence for parasitism
Nests were examined in May when larvae of M. mutabilis move to the top of the nests to pupate. About 60% of nests were excavated to detect immature larvae.
(d) Microsatellite analyses
DNA was extracted from whole individual workers using a modified Chelex 100 method (Walsh et al. 1991). Individual ants were screened for variation at 12 microsatellite loci (FL12 from Chapuisat (1996); 11 FE loci from Gyllenstrand et al. (2002), see Table 1 in the electronic supplementary material). Seven of these loci were separately amplified (see Table 1 in electronic supplementary material) using standard Bioline Taq polymerase, while the remaining five co-amplified using the Qiagen Multiplex kit (Qiagen). Samples from the Burren, StAg and Lim were amplified with a subset of the most variable nine loci (see Table 1 in the electronic supplementary material). The 5′ end of each forward primer was fluorescently labelled with 6-FAM, HEX or TET (see Table 1 in the electronic supplementary material) to enable detection by a MJ Research Basestation automatic genotyping machine. Amplification conditions (see Table 1 in the electronic supplementary material) for the singly amplified loci were: initial denaturation at 95°C for 5 min, then cycles of denaturation at 94°C for 45 s, annealing for 45 s, extension for 1 min at 72°C followed by a final extension for 10 min at 72°C. Conditions for the multiplex amplifications were: initial denaturation for 15 min at 95°C followed by 32 cycles of 94°C for 30 s, 57°C for 90 s, 72°C for 1 min with a final extension of 60°C for 30 min. Products from all singularly amplified reactions were mixed equally (2.5 μl each) and then diluted 1/3 with sigma water. One microlitre of each dilution was then mixed with 4 μl formamide and 0.17 μl ROX 400HD (Applied Biosystems) size standard per well for electrophoresis. Multiplex products were treated similarly, except that they were diluted 1/30. A sample of 1.5 μl from each mixed loci sets was electrophoresed for 90 min at 40°C using the supplied KBB buffer system (MJ Research). The gel results were analysed with Cartographer software (MJ Research).
(e) Marker variation
The Mull population was tested for deviations from Hardy–Weinberg proportions, using the program Genepop v. 3.1 (Raymond & Rousset 1995), by examining the genotype frequency distributions and using one individual from each colony. Exact p-values were estimated by the Markov chain method implementing 1000 dememorization steps, 1000 batches and 1000 iterations per batch for all tests. Global tests across loci were constructed using Fisher's combined probability method as implemented in Genepop. Genotypic linkage disequilibrium among alleles was also tested using Genepop and significance levels adjusted using sequential Bonferroni corrections (Hochberg 1988). Null allele frequencies were estimated using Micro-Checker (Van Oosterhout et al. 2004) on a subset of workers that amplified for all loci in order to disregard possible errors due to degraded DNA. Observed heterozygosity per locus was determined using the Microsatellite Toolkit (Park 2001).
(f) Population substructuring
The F statistic for the effect of subpopulations compared to the total population (Fst) was calculated, and its significance tested, to assist in the evaluation of loci by testing the hypothesis that there was population substructuring on Mull. These data were arranged into individuals within colonies within the total population for the Mull site only and the tests were implemented in FSTAT v.2.9.3.2 (Goudet 2001).
(g) Relatedness estimations
Relatedness estimates (R) were calculated from worker and queen genotypes using the program Relatedness v.5.08 (Goodnight & Queller 1998), which implements the methods described by Queller & Goodnight (1989). Colonies were weighted equally and standard errors were estimated by jackknifing over loci. Background allele frequencies for the relatedness estimates for each site were calculated separately using data from all genotyped workers at each site. Relatedness values between infested and uninfested colonies were compared using R−R′ unpaired difference, with p-values estimated from jackknifing over loci, in Relatedness.
(h) Estimates of queen and mate numbers
Individual worker genotypes were used as input for the maximum likelihood program Colony (Wang 2004) to estimate the number of queens and their mates contributing to the workers, per colony, at each site. Allelic dropout and other errors were assumed to be 5%. As only one sex is able to be designated as multiply mating in the program, we expected queens to be the polygamous sex with males expected to be monogamous. We compared the number of queens, estimated for uninfested and infested colonies, using a generalized linear model with Poisson errors and a logit link implemented in SPlus v.7.0. Since the variance is overdispersed, significance was assessed on the deletion of terms using an F-test. Similarly, we compared the average number of mates per queen per colony (see §3). For simplicity, standard errors in text and error bars in the figure are calculated on untransformed data, which makes little practical difference. We did not estimate the genetically effective queen number (Pedersen & Boomsma 1999) as the number of mates per queen was estimated to be high (see §3) and thus the assumption of singly mated queens was invalidated. In any case, comparisons of the R estimates above give a comparable test of genetic diversity. A bivariate Pearson's correlation was used to test whether a relationship existed between the estimates of queen numbers and the average number of mates per queen at each site. Additionally, infestation levels of infested colonies only were compared to R estimates using a bivariate Pearson's correlation in SPSS.
3. Results
(a) Infestation
Of the 267 F. lemani colonies on Mull, 42 (18.7%) were parasitized by M. mutabilis. Of the 41 colonies sampled for genetic analysis, 22 (54%) were infested (mean number±s.e.: larvae 0.40±0.22, fresh pupae 3.70±1.22, old pupal cases 5.01±2.80) and 19 (46%) were uninfested. The Burren site was significantly (Fisher's exact two-tailed p<0.0001) more heavily infested as compared with the surveyed Mull colonies, with 20 (66%) colonies containing M. mutabilis (mean number±s.e.: larvae 1.70±0.57, fresh pupae 2.05±0.76, old pupal cases 2.85±0.66) and 10 (33%) not. No colony from StAg or Lim was infested.
(b) Relatedness
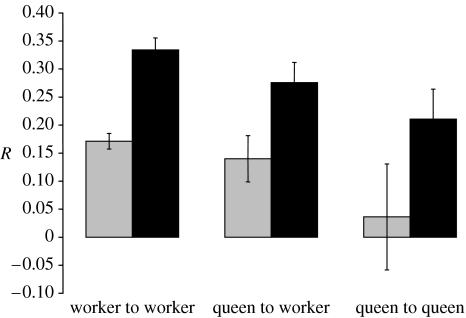
Information on variation of the loci used can be found in the electronic supplementary material to this paper. For all Mull colonies, the average value of within-colony relatedness, R, between workers was 0.244±0.015 s.e. The average relatedness of workers within infested colonies on Mull was significantly lower than in uninfested colonies (jackknife over 12 loci: R−R′ difference=−0.163; s.e.=0.021; p<0.0001; figure 1). Similarly, the average R value of queens to workers was significantly lower (jackknife over 12 loci: R−R′ difference=−0.128; s.e.=0.046; p<0.0001) within infested Mull colonies than within the uninfested ones (figure 1). Finally, the average queen to queen R value on Mull was also significantly lower (jackknife over 12 loci: R−R′ difference=−0.175; s.e.=0.098; p<0.0001) in infested colonies than in uninfested ones (figure 1), although this comparison is based on a very small sample (multiple queens were collected from two uninfested colonies, containing three or eight queens, and two infested colonies each containing three queens).
Figure 1.
Average relatedness (R) of Formica lemani ants on Mull (with standard errors) determined using 12 microsatellite loci. Comparisons are within or between workers and queens within Microdon mutabilis infested (grey) and uninfested (black) colonies. All differences were highly significant (see text).
The same strong pattern was found at our second infested site, the Burren. The average R value among workers within all colonies was 0.492±0.017 s.e. Significant differences existed between the relatedness of workers within infested (mean R=0.458±0.019 s.e.) and uninfested (0.565±0.024) Burren colonies (jackknife over nine loci: R−R′ difference=−0.108; s.e.=0.026; p<0.0001). Similarly, the average R value of queens to workers was significantly lower (jackknife over nine loci: R−R′ difference=−0.367; s.e.=0.124; p<0.0001) within infested Burren colonies than within uninfested ones. However, this comparison is based on a small sample from uninfested colonies (two colonies with one sampled queen each), and no queen to queen comparison could therefore be made. The average within-colony worker relatedness in the M. mutabilis uninfested populations was 0.49±0.029 and 0.77±0.034 for Lim and StAg, respectively.
(c) Queen and mate numbers
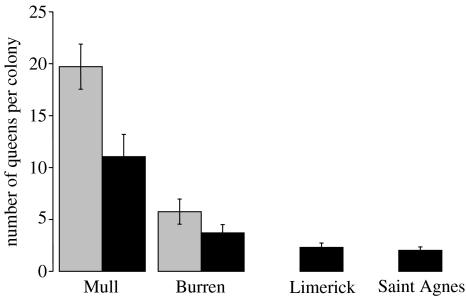
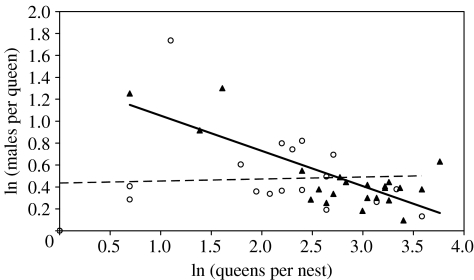
On Mull, the estimated average number of queens per infested colony was significantly greater (F1,40=7.73, p=0.008) than that of uninfested colonies (figure 2). There was no difference (F1,40=0.057, p=0.81) in the average number of mates per queen (infested, 1.69±0.14 s.e.; uninfested, 1.76±0.23). Interestingly, there was a significantly negative correlation (Pearson correlation, r22=−0.556, two-tailed p=0.007) between the estimated queen number and the average number of mates per queen in infested colonies. The same comparison among uninfested colonies from Mull was not significant (r19=−0.207, p=0.396). The linear regression with log-transformed estimates for queens and mates clearly demonstrates this difference (figure 3).
Figure 2.
Estimated queen numbers of Formica lemani (with standard errors) from each site within Microdon mutabilis infested (grey) and uninfested (black) colonies. The comparison between Mull infested and uninfested colonies was highly significant; the uninfested populations at St Agnes and Limerick did not differ significantly, and both contained significantly fewer estimated queens per colony than the infested populations (Mull and Burren) (see text).
Figure 3.
A linear regression of log-transformed [ln(x)] estimated number of queens (polygyny) and estimated number of mates (polyandry) in F. lemani Mull colonies infested by M. mutabilis (solid triangles, solid line) and without M. mutabilis (open circles, dashed line). For infested nests the correlation is highly significant (F1,20=29.6, p<0.001), while in non-infested nests it is not (F1,17=0.04, p>0.8).
Infested colonies on the Burren also contained a greater estimated average number of queens than uninfested colonies (figure 2), although the difference is not significant (F1,29=1.45, p=0.24), perhaps due to the small number of uninfested colonies located there. The estimated average number of mates per queen per colony did not differ significantly (F1,29=0.74, p=0. 4) between the infested (1.96±0.227) and uninfested (1.68±0.138) Burren colonies. No significant correlations between queen and mate estimates were found for either infested (r20=−0.073, p=0.761) or uninfested (r10=0.088, p=0.809) Burren colonies. Correlations between queen and mate numbers at Lim (r10=0.189, p=0.601) and StAg (r10=−0.341, p=0.335) were also not significant.
There was no significant difference between Lim and StAg for either estimated queen numbers (figure 2, F1,19=0.38, p>0.05) or average estimated mate numbers (F1,19=1.39, p>0.05; Lim mean=2.03±0.321; StAg mean=1.52±0.294) per colony. When the Lim and StAg data were combined, all queen number comparisons with either Mull infested (F1,41=111.02, p<0.0001), Mull uninfested (F1,38=31.02, p<0.0001), Burren infested (F1,39=12.0, p<0.01) or Burren uninfested (F1,29=6.11, p<0.05) colonies were significantly lower. Similar comparisons with average mate number per queen per colony were not significant (all p>0.55: Lim/StAg versus Mull infested, F1,41=0.094; Mull uninfested, F1,38=0.002; Burren infested, F1,39=0.357; Burren uninfested, F1,29=0.084).
(d) Correlations with infestation levels
No significant correlations were found between R and the number of old pupae, current pupae or combined infestation levels (Pearson's correlation r22=1.09, p=0.315; r22=−0.08, p=0.362; r22=0.06, p=0.396, respectively) in Mull colonies. Similarly, for the Burren samples, we found no significant correlations of R with old pupae (r20=0.111, p=0.64), present pupae (r20=−0.172, p=0.467) or combined pupae count (r20=−0.071, p=0.765). As we did not estimate the size of colonies, we were unable to factor this into our comparisons of R and infestation levels, limiting our ability to detect existing relationships.
4. Discussion
Our results clearly indicate that populations and individual colonies of F. lemani that are genetically diverse are more likely to be infested with M. mutabilis than the homogeneous ones. Although our findings are correlational, they are in stark contrast to the negative relationship between relatedness and susceptibility reported for somatic parasites and pathogens (e.g. Baer & Schmid-Hempel 1999; Tarpy 2003; Hughes & Boomsma 2004; Tarpy & Seeley 2006). In the case of pathogens, resistance can be chemical or involve behaviour that suppresses transmission, such as allogrooming, ‘dead worker removal’, and general nest hygiene; and it is increasingly evident that a lack of genetic diversity in the innate immune response promotes pathogen/parasite success in highly related ant colonies (Rolff & Siva-Jothy 2003). In contrast, most known examples of ca 10 000 species of ant social parasite employ subterfuge (mimicry, camouflage and stealth) to penetrate host societies (Hölldobler & Wilson 1990; Thomas et al. 2005), and while resistance via the immune response against pathogens relies on a lock-and-key recognition, resistance to myrmecophile attack depends on the ability of worker ants to detect differences between an intruder's chemical signature and the mean chemical template of the colony to which the workers belong (Vander Meer & Morel 1998).
In most insect societies, species-, nestmate- and in some cases kin-recognition are primarily based on their members' ability to detect characteristic cocktails of surface lipids, often referred to as cuticular hydrocarbons (CHCs), on fellow members (Crozier 1987; Hölldobler & Wilson 1990; Hannonen & Sundström 2003). The origin of CHCs is still under debate but has been demonstrated to have a genetic component in some insect species (Drosophila melanogaster: Coyne et al. 1999; Takahashi et al. 2001). In ants, CHCs have been shown to be largely inherited, for example, in Formica (Beye et al. 1998), and to be under selection, as shown by certain invasive species (Nowbahari et al. 1990; Tsutsui et al. 2000; Boomsma et al. 2003). At the same time, environmental factors such as diet may modify CHC signatures (e.g. Provost 1989; Elmes et al. 2004). The cue-template model (Vander Meer & Morel 1998) hypothesises that each worker in a colony produces a distinctive chemical signature, providing a baseline against which the odours of other individuals are assessed. In polygynous ants (like F. lemani), the less closely related colony members exchange CHCs by allogrooming, and the template that workers accept is a mean signature for the whole nest, known as the colony gestalt odour (Crozier & Dix 1979; Crozier 1987; Vander Meer 1998). As a result, the colony gestalt odour in monogynous species or colonies tends to be tighter and better defined than that in polygynous species or colonies.
The more diffuse the colony gestalt odour (owing to genetically diverse workers), the greater the variance around the mean template that workers have to tolerate if they are not to exclude nest mates (Starks et al. 1998). In theory, this makes a cosmopolitan ant society more susceptible to intruders, even if they are imperfect mimics of the colony's broad gestalt odour. Although this hypothesis is only intended for F. lemani, it may also apply to other Serviformica species and indeed, social insects in general.
In practice, it has long been recognized that social parasite species are usually associated with polygynous and polydomous ant species (Buschinger 1970; Alloway et al. 1982; Thomas et al. 2005). Among the few social parasites whose secretions have been analysed, Maculinea butterflies do indeed synthesize CHCs that mimic those of their host's societies so closely that the worker ants rescue and feed the parasitic caterpillars in preference to their own larvae (Akino et al. 1999; Nash et al. 2002; Schönrogge et al. 2004; Thomas et al. 2005).
The exact mechanism used by M. mutabilis to infiltrate F. lemani colonies is unknown, but it undoubtedly involves camouflage and probably mimicry (Elmes et al. 1999; Napper 2004). The four populations we studied represent a range of biotopes: coastal heath in StAg; a grassy roadside on Mull; limestone outcrops in Lim; and the largest limestone pavement in Europe (Burren). Using the populations where M. mutabilis is absent (StAg and Lim) as a baseline, we suggest that F. lemani populations generally consist of colonies that have low levels of polygyny, with a substantial proportion being monogynous. Thus, not only do the nests on the Burren and Mull contain significantly higher levels of polygyny than at Lim and StAg, but within the Mull and Burren populations, colonies attacked by M. mutabilis were more polygynous than unparasitized ones. Although other explanations may explain the lower genetic diversity and lack of parasites at the two uninfested sites, our intrapopulation comparison on both Mull and the Burren provides compelling evidence for a relationship between M. mutabilis infestation and genetic diversity.
Slave-making ants provide the only comparable well-studied system in which the degree and distribution of polygyny in a host ant may be affected at the population level by an intruder. However, this involves a different interaction in which slave-making workers release powerful alarm pheromones during raids that cause the host ants to attack each other. This diminishes the latter's ability to protect their brood, which is captured and reared as slaves in the slave-makers' colonies. In at least one system (raider, Harpagoxenus sublaevis; host, Leptothorax acervorum), monogynous host colonies are better able to resist slave-making raids (Foitzik et al. 2003). Furthermore, Leptothorax longispinosus (host) colonies in the vicinity of Protomognathus americanus (slave-maker) colonies are less polygynous, have higher relatedness and also fewer workers compared with host colonies away from slave-makers. The fact that slave colonies contain host workers which are genetically more diverse than workers in the neighbouring colonies may simply result from raids on multiple-host colonies.
The similarities between these studies and ours are intriguing, although in both systems the cause and the effect are unclear. Slave-makers might raid genetically diverse colonies more easily, leaving monogynous or weakly polygynous colonies as survivors in areas where the slave-makers occur. Alternatively, the slave-makers may preferentially settle in areas that have small monogynous colonies of their hosts (Foitzik et al. 2003).
The hypothesis that only genetically diverse colonies and populations of F. lemani can be infested by M. mutabilis—because it is easier for the parasite to enter colonies whose odour diversity is great—is not the only explanation of our results. Another non-exclusive possibility is that the lower relatedness of infested nests is a consequence of being parasitized. In this scenario, colonies become infested with M. mutabilis and subsequently acquire additional queens and/or recruit workers from neighbouring nests to minimize the impact of the parasite. We have often found that the same host nest is attacked in three or more consecutive years, as indicated by the simultaneous presence of old pupal cases, fresh pupae and young biennial larvae of M. mutabilis in the same nest (Schönrogge et al. 2006). In these nests, worker numbers must inevitably decline or be compensated, owing to the large amounts of ant brood eaten by M. mutabilis (Schönrogge et al. 2000, 2006). Extra-nest queens can be adopted to increase a colony's ergonomic efficiency (Oster & Wilson 1978) by either increasing its work-force size or increasing the genetic diversity of the colony as a defence against strong parasitism (Rosengren et al. 1993).
Our finding of a negative correlation between estimated queen numbers and average numbers of mates per queen suggests that, at least on Mull, these two pathways to increased genetic diversity, polygyny and polyandry do not operate independently, suggesting that there may some other benefit to polyandry/polygyny distinct from its implications for infection by Microdon. There would be no relationship between polygyny and polyandry if colonies adopt queens in order to combat the presence of the parasite, as colonies would not have the ability to increase the numbers of mates of their queen(s). Thus, the association between genetic diversity and Microdon infestation is more likely to be due to genetically diverse nests being more susceptible to invasion rather than to infested colonies adopting additional queens.
We suggest that a negative correlation between estimated queen numbers and average numbers of mates per queen could arise in two ways: (i) for the Mull population at least, there may a trade-off between susceptibility to Microdon and resistance to pathogens. To date there is no information about pathogen pressures on F. lemani colonies, but if colonies with low queen numbers tend to be more polyandrous, and selection is strong throughout the population, pathogen exposure could determine which F. lemani populations are susceptible to Microdon attacks; (ii) we recently found that F. lemani colonies infested by Microdon produce about twice as many queens per worker compared with uninfested ones (Schönrogge et al. 2006). The fate of new queens produced in infested nests is currently unknown. However, the decrease in mate numbers with increasing numbers of queens in infested colonies could arise through a higher inclination to swarm by these new queens, leading to multiple mating and subsequent colony founding, either independently or in small groups. This would also be consistent with the lack of such a correlation among non-infested colonies. Clearly, the population level impact by M. mutabilis on its host F. lemani warrants further investigation.
Acknowledgments
We thank Kerry Cutter for assistance with microsatellite typing, Boyd Barr for discussions on life history of Microdon mutabilis and the Mull site, and Judith Wardlaw for maintenance of Formica colonies. We are indebted to anonymous reviewers whose comments greatly improved the manuscript.
Supplementary Material
Variation of markers used in the study and a table summarising characteristics and amplification conditions of loci examined.
References
- Akino T, Knapp J.J, Thomas J.A, Elmes G.W. Chemical mimicry and host specificity in the butterfly Maculinea rebeli, a social parasite of Myrmica ant colonies. Proc. R. Soc. B. 1999;266:1419–1426. doi:10.1098/rspb.1999.0796 [Google Scholar]
- Alexander R.D. The evolution of social behaviour. Annu. Rev. Ecol. Syst. 1974;5:325–383. doi:10.1146/annurev.es.05.110174.001545 [Google Scholar]
- Alloway T.M, Buschinger M, Stuart R, Thomas C. Polygyny and polodomy in North American species of the three ant genus Leptothorax Mayr (Hymenoptera: Formicidae) Psyche. 1982;89:249–274. [Google Scholar]
- Baer B, Schmid-Hempel P. Experimental variation in polyandry affects parasite loads and fitness in a bumble-bee. Nature. 1999;397:151–154. doi:10.1038/16451 [Google Scholar]
- Beye M, Neumann P, Chapuisat M, Pamilo P, Moritz R.F.A. Nestmate recognition and the genetic relatedness of nests in the ant Formica pratensis. Behav. Ecol. Sociobiol. 1998;43:67–72. doi:10.1007/s002650050467 [Google Scholar]
- Boomsma J.J, Ratnieks F.L.W. Paternity in eusocial Hymenoptera. Phil. Trans. R. Soc. B. 1996;351:947–975. [Google Scholar]
- Boomsma J.J, Nielsen J, Sundstrom L, Oldham N.J, Tentschert J, Petersen H.C, Morgan E.D. Informational constraints on optimal sex allocation in ants. Proc. Natl Acad. Sci. USA. 2003;100:8799–8804. doi: 10.1073/pnas.1430283100. doi:10.1073/pnas.1430283100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C, Brown M.B. Ectoparasitism as a cost of coloniality in cliff swallows (Hirundo pyrrhonota) Ecology. 1986;67:1206–1218. doi:10.2307/1938676 [Google Scholar]
- Buschinger A. Neue Vorstellungen zur Evolution des Sozialparasitismus und der Dulosis bei Ameisen (Hym, Formicidae) Biologisches Zentralblatt. 1970;88:273–299. [Google Scholar]
- Chapuisat M. Characterization of microsatellite loci in Formica lugubris B and their variability in other ant species. Mol. Ecol. 1996;5:599–601. doi: 10.1111/j.1365-294x.1996.tb00354.x. doi:10.1046/j.1365-294X.1996.00124.x [DOI] [PubMed] [Google Scholar]
- Cole B.J, Wiernasz D.C. The selective advantage of low relatedness. Science. 1999;285:891–893. doi: 10.1126/science.285.5429.891. doi:10.1126/science.285.5429.891 [DOI] [PubMed] [Google Scholar]
- Collingwood C.A. Scandinavian Science Press Ltd; Klampenborg, Denmark: 1979. The Formicidae (Hymenoptera) of Fennoscandia and Denmark. [Google Scholar]
- Coyne J.A, Wicker-Thomas C, Jallon J.M. A gene responsible for a cuticular hydrocarbon polymorphism in Drosophila melanogaster. Genet. Res. 1999;73:189–203. doi: 10.1017/s0016672398003723. doi:10.1017/S0016672398003723 [DOI] [PubMed] [Google Scholar]
- Crozier R.H. Genetic aspects of kin recognition: concepts, models, and synthesis. In: Fletcher D.J.C, Michener C.D, editors. Kin recognition in animals. Wiley; New York, NY: 1987. pp. 55–73. [Google Scholar]
- Crozier R.H, Dix M.W. Analysis of two genetic models for the innate components of colony odor in social Hymenoptera. Behav. Ecol. Sociobiol. 1979;4:217–224. doi:10.1007/BF00297645 [Google Scholar]
- Crozier R.H, Fjerdingstad E.J. Polyandry in social Hymenoptera—disunity in diversity? Ann. Zool. Fenn. 2001;38:267–285. [Google Scholar]
- Dybdahl M.F, Lively C.M. The geography of coevolution: comparative population structures for a snail and its trematode parasite. Evolution. 1996;50:2264–2275. doi: 10.1111/j.1558-5646.1996.tb03615.x. doi:10.2307/2410696 [DOI] [PubMed] [Google Scholar]
- Elmes G.W, Barr B, Thomas J.A, Clarke R.T. Extreme host specificity by Microdon mutabilis (Diptera: Syrphidae), a social parasite of ants. Proc. R. Soc. B. 1999;266:447–453. doi:10.1098/rspb.1999.0658 [Google Scholar]
- Elmes G.W, Wardlaw J.C, Schonrogge K, Thomas J.A, Clarke R.T. Food stress causes differential survival of socially parasitic caterpillars of Maculinea rebeli integrated in colonies of host and non-host Myrmica ant species. Entomol. Exp. Appl. 2004;110:53–63. doi:10.1111/j.0013-8703.2004.00121.x [Google Scholar]
- Fjerdingstad E.J, Gertsch P.J, Keller L. The relationship between multiple mating by queens, within- colony genetic variability and fitness in the ant Lasius niger. J. Evol. Biol. 2003;16:844–853. doi: 10.1046/j.1420-9101.2003.00589.x. doi:10.1046/j.1420-9101.2003.00589.x [DOI] [PubMed] [Google Scholar]
- Foitzik S, Fischer B, Heinze J. Arms races between social parasites and their hosts: geographic patterns of manipulation and resistance. Behav. Ecol. 2003;14:80–88. doi:10.1093/beheco/14.1.80 [Google Scholar]
- Goodnight, K. F. & Queller, D. C. 1998 Relatedness 5.08.: Available at http://www.gsoftnet.us/GSoft.html
- Goudet, J. 2001 FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). Available from http://www.unil.ch/izea/softwares/fstat.html Lausanne.
- Gyllenstrand N, Gertsch P.J, Pamilo P. Polymorphic microsatellite DNA markers in the ant Formica exsecta. Mol. Ecol. Notes. 2002;2:67–69. [Google Scholar]
- Hamilton W.D. Kinship, recognition, disease, and intelligence: constraints of social evolution. In: Ito Y, Brown J.L, Kikkawa J, editors. Animal societies: theory and facts. Japenese Scientific Society Press; Tokyo, Japan: 1987. pp. 81–102. [Google Scholar]
- Hannonen M, Sundström L. Sociobiology—worker nepotism among polygynous ants. Nature. 2003;421:910. doi: 10.1038/421910a. doi:10.1038/421910a [DOI] [PubMed] [Google Scholar]
- Hochberg Y. A sharper Bonferroni procedure for multiple tests of significance. Biometrika. 1988;75:800–803. doi:10.2307/2336325 [Google Scholar]
- Hölldobler B, Wilson E.O. Springer; Berlin, Germany: 1990. The ants. [Google Scholar]
- Hughes W.O.H, Boomsma J.J. Genetic diversity and disease resistance in leaf-cutting ant societies. Evolution. 2004;58:1251–1260. doi: 10.1554/03-546. doi:10.1554/03-546 [DOI] [PubMed] [Google Scholar]
- Hughes W.O.H, Boomsma J.J. Does genetic diversity hinder parasite evolution in social insect colonies? J. Evol. Biol. 2006;19:132–143. doi: 10.1111/j.1420-9101.2005.00979.x. doi:10.1111/j.1420-9101.2005.00979.x [DOI] [PubMed] [Google Scholar]
- Keller L, Reeve H.K. Genetic-variability, queen number, and polyandry in social Hymenoptera. Evolution. 1994;48:694–704. doi: 10.1111/j.1558-5646.1994.tb01354.x. doi:10.2307/2410479 [DOI] [PubMed] [Google Scholar]
- Lenoir A, D'Ettorre P, Errard C, Hefetz A. Chemical ecology and social parasitism in ants. Annu. Rev. Entomol. 2001;46:573–599. doi: 10.1146/annurev.ento.46.1.573. doi:10.1146/annurev.ento.46.1.573 [DOI] [PubMed] [Google Scholar]
- Liersch S, Schmid-Hempel P. Genetic variation within social insect colonies reduces parasite load. Proc. R. Soc. B. 1998;265:221–225. doi:10.1098/rspb.1998.0285 [Google Scholar]
- Napper, E. 2004 Chemical and behavioural interactions between ants and Microdon hoverflies. Ph.D. thesis. Southampton University, Southampton, UK.
- Nash, D. R., Als, T. D., Tentschert, J., Maile, R., Jungnickel, H. & Boomsma, J. J. 2002 Local adaptation and coevolution of chemical mimicry in the butterfly Maculinea alcon, a social parasite of Myrmica ants. In Proc. XIV Int. Meeting of the IUSSI, Sapporo, Japan, p. 138. Sapporo, Japan.
- Nowbahari E, Lenoir A, Clement J.L, Lange C, Bagneres A.G, Joulie C. Individual, geographical and experimental variation of cuticular hydrocarbons of the ant Cataglyphis cursor (Hymenoptera, Formicidae)—their use in nest and subspecies recognition. Biochem. Syst. Ecol. 1990;18:63–73. doi:10.1016/0305-1978(90)90036-F [Google Scholar]
- Oster G.F, Wilson E.O. Princeton University Press; Princeton, NJ: 1978. Caste and ecology in the social insects. [PubMed] [Google Scholar]
- Page R.E. Sperm utilization in social insects. Annu. Rev. Entomol. 1986;31:297–320. doi:10.1146/annurev.en.31.010186.001501 [Google Scholar]
- Park, S. D. E. 2001 Microsatellite toolkit. Available from http://acer.gen.tcd.ie/~sdepark/ms-toolkit/
- Pedersen J.S, Boomsma J.J. Effect of habitat saturation on the number and turnover of queens in the polygynous ant, Myrmica sulcinodis. J. Evol. Biol. 1999;12:903–917. doi:10.1046/j.1420-9101.1999.00109.x [Google Scholar]
- Provost E. Social environmental factors influencing mutual recognition of individuals in the ant Leptothorax lichtensteini Bondr (Hymenoptera, Formicidae) Behav. Process. 1989;18:35–59. doi: 10.1016/S0376-6357(89)80004-X. doi:10.1016/0376-6357(89)90004-1 [DOI] [PubMed] [Google Scholar]
- Queller D.C, Goodnight K.F. Estimating relatedness using genetic markers. Evolution. 1989;43:258–275. doi: 10.1111/j.1558-5646.1989.tb04226.x. doi:10.2307/2409206 [DOI] [PubMed] [Google Scholar]
- Raymond M, Rousset F. Genepop (version 1.2): population genetics software for exact tests and ecumenicism. J. Hered. 1995;86:248–249. [Google Scholar]
- Rolff J, Siva-Jothy M.T. Invertebrate ecological immunology. Science. 2003;301:472–475. doi: 10.1126/science.1080623. doi:10.1126/science.1080623 [DOI] [PubMed] [Google Scholar]
- Rosengren R, Sundström L, Fortelius W. Monogyny and polygyny in Formica ants: the result of alternative dispersal tactics. In: Keller L, editor. Queen number and sociality in insects. Oxford University Press; Oxford, UK: 1993. pp. 308–333. [Google Scholar]
- Schmid-Hempel P. Infection and colony variability in social insects. Phil. Trans. R. Soc. B. 1994;346:313–321. [Google Scholar]
- Schmid-Hempel P. Princeton University Press; Princeton, NJ: 1998. Parasites in social insects. [Google Scholar]
- Schmid-Hempel P, Crozier R.H. Polyandry versus polygyny versus parasites. Phil. Trans. R. Soc. B. 1999;354:507–515. doi:10.1098/rstb.1999.0401 [Google Scholar]
- Schönrogge K, Wardlaw J.C, Thomas J.A, Elmes G.W. Polymorphic growth rates in myrmecophilous insects. Proc. R. Soc. B. 2000;267:771–777. doi: 10.1098/rspb.2000.1070. doi:10.1098/rspb.2000.1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönrogge K, Barr B, Wardlaw J.C, Napper E, Gardner M.G, Breen J, Elmes G.W, Thomas J.A. When rare species become endangered: cryptic speciation in myrmecophilous hoverflies. Biol. J. Linn. Soc. 2002;75:291–300. doi:10.1046/j.1095-8312.2002.00019.x [Google Scholar]
- Schönrogge K, Wardlaw J.C, Peters A.J, Everett S, Thomas J.A, Elmes G.W. Changes in chemical signature and host specificity from larval retrieval to full social integration in the myrmecophilous butterfly Maculinea rebeli. J. Chem. Ecol. 2004;30:91–107. doi: 10.1023/b:joec.0000013184.18176.a9. doi:10.1023/B:JOEC.0000013184.18176.a9 [DOI] [PubMed] [Google Scholar]
- Schönrogge K, et al. Host propagation permits extreme local adaptation in a social parasite of ants. Ecol. Lett. 2006;9:1032–1040. doi: 10.1111/j.1461-0248.2006.00957.x. doi:10.1111/j.1461-0248.2006.00957.x [DOI] [PubMed] [Google Scholar]
- Sherman P.W, Seeley T.D, Reeve H.K. Parasites, pathogens, and polyandry in social Hymenoptera. Am. Nat. 1988;131:602–610. doi: 10.1086/286127. doi:10.1086/284809 [DOI] [PubMed] [Google Scholar]
- Starks P.T, Watson R.E, Dipaola M.J, Dipaola C.P. The effect of queen number on nestmate discrimination in the facultatively polygynous ant Pseudomyrmex pallidus (Hymenoptera: Formicidae) Ethology. 1998;104:573–584. [Google Scholar]
- Sundström L. Sex allocation and colony maintenance in monogyne and polygyne colonies of Formica truncorum (Hymenoptera, Formicidae)—the impact of kinship and mating structure. Am. Nat. 1995;146:182–201. doi:10.1086/285794 [Google Scholar]
- Takahashi A, Tsaur S.C, Coyne J.A, Wu C.I. The nucleotide changes governing cuticular hydrocarbon variation and their evolution in Drosophila melanogaster. Proc. Natl Acad. Sci. USA. 2001;98:3920–3925. doi: 10.1073/pnas.061465098. doi:10.1073/pnas.061465098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarpy D.R. Genetic diversity within honeybee colonies prevents severe infections and promotes colony growth. Proc. R. Soc. B. 2003;270:99–103. doi: 10.1098/rspb.2002.2199. doi:10.1098/rspb.2002.2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarpy D.R, Seeley T.D. Lower disease infections in honeybee (Apis mellifera) colonies headed by polyandrous vs monandrous queens. Naturwissenschaften. 2006;93:195–199. doi: 10.1007/s00114-006-0091-4. doi:10.1007/s00114-006-0091-4 [DOI] [PubMed] [Google Scholar]
- Thomas J.A, Schönrogge K, Elmes G.W. Specializations and host associations of social parasites of ants. In: Fellows M.D.E, Holloway G.J, Rohlf J, editors. Insect evolutionary ecology. CABI Publishing; Wallingford, UK: 2005. pp. 479–518. [Google Scholar]
- Tsutsui N.D, Suarez A.V, Holway D.A, Case T.J. Reduced genetic variation and the success of an invasive species. Proc. Natl Acad. Sci. USA. 2000;97:5948–5953. doi: 10.1073/pnas.100110397. doi:10.1073/pnas.100110397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Baalen M, Beekman M. The costs and benefits of genetic heterogeneity in resistance against parasites in social insects. Am. Nat. 2006;167:568–577. doi: 10.1086/501169. doi:10.1086/501169 [DOI] [PubMed] [Google Scholar]
- Van Oosterhout C, Hutchinson W.F, Wills D.P.M, Shipley P. Micro-Checker: software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes. 2004;4:535–538. doi:10.1111/j.1471-8286.2004.00684.x [Google Scholar]
- Vander Meer R.K, editor. Pheromone communication in social insects, ants, wasps, bees, and termites. Westview Press; Oxford, UK: 1998. [Google Scholar]
- Vander Meer R.K, Morel L. Nestmate recognition in ants. In: Vander Meer R.K, Breed M.D, Winston M.L, Espelie K.E, editors. Pheromone communication in social insects. Westview Press; Boulder, CO: 1998. pp. 79–103. [Google Scholar]
- Walsh P.S, Metzger D.A, Higuchi R. Chelex-100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques. 1991;10:506–513. [PubMed] [Google Scholar]
- Wang J.L. Sibship reconstruction from genetic data with typing errors. Genetics. 2004;166:1963–1979. doi: 10.1534/genetics.166.4.1963. doi:10.1534/genetics.166.4.1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Variation of markers used in the study and a table summarising characteristics and amplification conditions of loci examined.