Abstract
Homosexuality is a common occurrence in humans and other species, yet its genetic and evolutionary basis is poorly understood. Here, we formulate and study a series of simple mathematical models for the purpose of predicting empirical patterns that can be used to determine the form of selection that leads to polymorphism of genes influencing homosexuality. Specifically, we develop theory to make contrasting predictions about the genetic characteristics of genes influencing homosexuality including: (i) chromosomal location, (ii) dominance among segregating alleles and (iii) effect sizes that distinguish between the two major models for their polymorphism: the overdominance and sexual antagonism models. We conclude that the measurement of the genetic characteristics of quantitative trait loci (QTLs) found in genomic screens for genes influencing homosexuality can be highly informative in resolving the form of natural selection maintaining their polymorphism.
Keywords: homosexuality, genetics, maintenance, evolution, mathematical models, testable predictions
1. Introduction
There are several reasons for the long-standing interest among evolutionary biologists (e.g. Hutchinson 1959; Wilson 1975; Hammer & Copeland 1994; McKnight 1997; Miller 2000) in explaining persistent, low levels of human homosexuality. First, there is evidence that homosexual males and females have lower lifetime offspring production in some modern Western societies (up to 80% lower; Bell et al. 1981), and that this may also have been true in human ancestors (reviewed in Pillard & Bailey 1998). Second, there are two lines of evidence that homosexuality is influenced by polymorphic genes: (i) twin studies indicate that there are both genetic and environmental factors that contribute to the expression of the homosexual phenotype (Pillard & Bailey 1998; Bailey et al. 1999; Dawood et al. 2000), and (ii) male homosexuality appears to be inherited more frequently from the matriline (Pillard et al. 1981, 1982; Pattatucci 1998; Camperio-Ciani et al. 2004), suggesting the existence of polymorphic, heritable maternal effects and/or polymorphic X-linked genes influencing male homosexuality. Third, even if one assumes only a small fitness cost to the expression of homosexuality, it appears to be more common in both males and females than can be plausibly explained by mutation–selection balance (Kinsey et al. 1948, 1953; Gebhard 1972; Diamond 1993; Sell et al. 1995).
Maternal effects may contribute to the homosexual phenotype. For example, there is a curious relationship between birth order and the incidence of male homosexuality. Among sibs, the occurrence of male homosexuality is positively correlated with the number of older brothers but not the number of older sisters (Blanchard & Bogaert 1996; Blanchard & Klassen 1997; Blanchard 2004). This birth-order effect may be a result of the different social environment experienced by younger brothers, but it may also reflect the progressive immunization of some mothers to unspecified male-specific antigens with each successive male foetus and the increasing effects of such immunization on sexual differentiation of the brain with each successive male foetus (Blanchard & Klassen 1997; Blanchard 2004, but see Bearman 2005).
There have been a few attempts to localize the specific genes that influence male homosexuality. The complex nature of the occurrence of male homosexuality in human pedigrees indicates that its inheritance is not a simple Mendelian trait (Pillard et al. 1981; Camperio-Ciani et al. 2004), making the mapping of individual genes more difficult. A quantitative trait locus (QTL) for homosexuality (Xq28) has been localized to the X chromosome (Hamer et al. 1993; Hu et al. 1995), but the methodology used in these studies was questioned later (McKnight 1997) and the findings have been difficult to replicate (Bailey et al. 1999; Rice et al. 1999). Recently, a genome-wide QTL screen for male homosexuality (Mustanski et al. 2005) found three ‘nominally significant linkage peaks’, indicating three autosomal genes that may influence male sexual orientation, as well as limited support for the previously reported X-linked QTL (Xq28). These initial results are only preliminary and require confirmation from additional genetic studies.
Two mechanisms for the maintenance of polymorphism in genes that cause homosexuality have been most frequently mentioned in evolutionary biology literature: overdominance and frequency-dependent selection via kin altruism. The former mechanism assumes that genes inducing homosexuality provide superior fitness in heterozygous conditions, for example, men heterozygous for a homosexual gene may have higher success in attracting women and/or their sperm may have a competitive advantage over that of other men (e.g. Hutchinson 1959; Weinrich 1987; Kirsch & Weinrich 1991; MacIntyre & Estep 1993; Miller 2000). The kin-altruism mechanism assumes that homosexuals assist their close relatives, thereby increasing their own inclusive fitness (Trivers 1974; Pillard & Bailey 1998). A third mechanism, which was briefly mentioned by Hammer & Copeland (1994; see also McKnight 1997; Pillard & Bailey 1998) but that has never been rigorously explored previously, is a sexually antagonistic selection (e.g. Rice 1984; Rice & Holland 1997; Arnqvist & Rowe 2005) under which alleles that decrease fitness of one sex are maintained in the population because they increase the fitness of the other sex. The potential importance of this mechanism is highlighted by recent data which indicate that female maternal relatives of homosexuals (Camperio-Ciani et al. 2004) or relatives of gay men for both maternal and paternal lines (King et al. 2005) have increased fecundity.
The topic of homosexuality has so far received only very limited attention in theoretical evolutionary genetics and we are aware of only two previous papers that have attempted to model it. The first paper was by MacIntyre & Estep (1993), who studied a model of overdominance. The second paper was by Getz (1993), who assumed that reduced mating success of homosexual men was compensated by increased rearing success of females or increased joint fecundity and cooperation of couples. Both these papers studied the case of a single autosomal, diallelic locus, and they concentrated on the conditions for invasion of an allele promoting homosexuality.
Our goal is to formulate a series of simple mathematical models for the purpose of predicting empirical patterns that can be used to guide future genetic analysis of homosexuality. We specifically wanted to generate testable predictions that will provide a foundation for the generation of empirical evidence for or against alternative evolutionary hypotheses for the maintenance of polymorphic genes that influence homosexuality. Accordingly, we develop theory to make predictions about: (i) the chromosomal location, (ii) the dominance among segregating alleles and (iii) their effect sizes that are predicted by two of the major models for the maintenance of polymorphism: the overdominance and sexual antagonism. Because homosexuality has previously received very little attention in the context of sexually antagonistic alleles, our main focus will be on this model, but we will also extend the previous work on overdominance. Lastly, our approach uses as a foundation extant simple models of sexually antagonistic genes (Rice 1984) and of maternal and parental selections (Gavrilets 1998; Spencer 2003; Miller et al. 2006), which we extend to the context of homosexuality. We will assume throughout that males are the heterogametic sex, but all our results can be applied reciprocally to the case of female heterogamety.
We do not attempt to analyse the altruism towards kin model. Biological intuition suggests that for this mechanism to work, the number of ‘extra’ children raised by heterosexual kin with the help of a homosexual relative has to be larger than the number of children the extended family ‘lost’ owing to the homosexuality of the relative. Because neither any existing data nor any mathematical models known to us support its plausibility, we consider it premature to include the kin-altruism mechanism in our analysis.
2. Results
We consider a one-locus, two-allele diploid population with genotypes AA, Aa and aa. Generations are discrete and non-overlapping. The population size is effectively infinite. Fitness is understood as viability (i.e. the probability of surviving to reproduction), mating success (i.e. the probability of entering the mating pool) or fertility (i.e. the contribution to the number of offspring assuming that maternal and paternal contributions interact multiplicatively). Mating is random among the individuals who enter the mating pool.
Throughout the manuscript, A is an allele that has little or no influence on sexual orientation, and allele a masculinizes or feminizes both sexes, and thereby increases the probability of homosexuality in the discordant sex. For example, it is well known that environmental chemicals can produce intersexual individuals in fishes and other vertebrates (Devlin & Nagahama 2002). A feminizing allele a would be one that canalized development towards the female sex-determination pathway. Such an allele would be favoured in females because it protects them when exposed to masculinizing environmental conditions, but this same allele, when expressed in males, would feminize them and could thereby lead to homosexuality. Below, our main focus is on the conditions required for the maintenance of genetic variation. The dynamic equations describing specific models are given in appendix A.
3. Direct selection
First, we analyse the case when fitness and sexual orientation in both sexes are influenced solely by the direct genetic effects of genes residing in a zygote. In later sections, we will consider the cases, where maternal effects influence these characters.
(a) Autosomal gene
Assume that the locus under consideration is autosomal. Let female fitnesses be f1, f2 and f3 and male fitnesses be m1, m2 and m3 in genotypes AA, Aa and aa, respectively. In this system, the polymorphism is protected (i.e. both alleles increase in frequency when rare) if
| 3.1a |
| 3.1b |
(e.g. eqn 2.7 in Karlin 1972). Note that inequality (3.1a) is the condition for a successful invasion of allele a in a population initially monomorphic for allele A and inequality (3.1b) is the condition for a successful invasion of allele A in a population initially monomorphic for allele a. We consider three different cases.
(i) Overdominance in both sexes
If heterozygotes have the highest fitness in both sexes (i.e. m2>m1, m3 and f2>f1, f3), then conditions (3.1) are satisfied and the population evolves to a polymorphic state (Karlin 1972, p. 706). Although overdominance in both sexes is possible, the more plausible scenario for overdominance of a feminizing or masculinizing allele would be overdominance in one sex and directional selection in the other sex, as described subsequently.
(ii) Overdominance in one sex and directional selection in the other sex
For illustration, we assume that allele a is a feminizing allele that increases female fitness so that f1≤f2≤f3, where at least one inequality is strict. However, the same conclusions are reached if the roles of the two sexes are reversed. Also assume that one copy of allele a increases male fitness but two copies of a reduce male fitness by causing homosexual behaviour so that m2>m1>m3 (see Hutchinson 1959; Weinrich 1987 for the rationale for this form of overdominance; Kirsch &Weinrich 1991; MacIntyre & Estep 1993; Miller 2000). Without any loss of generality, we can set f1=m1=1. Because f2>f1 and m2>m1, condition (3.1a) is satisfied, i.e. if the feminizing allele is favoured in both females and heterozygous males, it will always increase when rare. Let us write f2=1+hG, f3=1+G, , m3=1−C′, where G characterizes the strength of directional selection in females (and is equal to the maximum fitness gain for females), 0≤h≤1 is a measure of dominance of the allele a in females, is the overdominant fitness gain to heterozygous males and 0≤C′≤1 is the cost to homozygous males. Here and throughout, primed and unprimed symbols denote fitness effects in opposite sexes. Condition (3.2) for the maintenance of genetic variation can be represented as
| 3.2 |
The left-hand side of inequality (3.2) increases with both and C′, whereas the right-hand side of this inequality increases with G and decreases with h. Inequality (3.2) implies that simple overdominance in males (i.e. ) is not sufficient in general, but the combination of the fitness gain () to heterozygous males and the cost to aa male homozygotes (C′) has to be large enough relative to the fitness gain (G) and level of dominance (h) in females, i.e. the constraint for polymorphism is that the cost to homozygous males (C′) or advantage to heterozygous males () has to be sufficiently large to prevent fixation of this allele. Interestingly, it implies that increasing the fitness cost to homozygous males () promotes the maintenance of genetic variation; if homozygous males have zero fitness (i.e. C′=1), the variation is always maintained. Overall, overdominance need not be strong to maintain polymorphism. Increasing the degree of dominance h of allele a in females promotes the maintenance of genetic variation; if allele a is dominant in females (i.e. h=1), the variation is always maintained.
(iii) Sexually antagonistic selection
For illustration, we assume that a feminizing allele a increases the fitness of females but decreases the fitness of males. However, the conclusions are not changed, if the roles of the two sexes are reversed. Let female fitnesses be f1=1, f2=1+hG, f3=1+G and male fitnesses be m1=1, m2=1−hC′, m3=1−C′, where G>0 is the maximum (i.e. homozygous) gain in fitness of females, 0≤C′≤1 is the maximum fitness loss of males and 0≤h≤1 is a measure of dominance. Then, allele a increases in frequency when rare if
| 3.3a |
i.e. if the fitness gain to females is larger than the fitness loss to males (Rice 1984, p. 736). Allele a does not go to fixation if
| 3.3b |
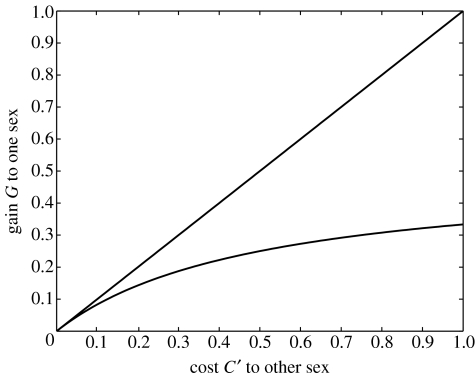
i.e. if the fitness loss of males is sufficiently large. These conditions are illustrated in figure 1. Note that if G is small, the conditions for the maintenance of genetic variation are very strict. For example, if G=0.1, then C′ must be between 0.083 and 0.10. Also note that conditions (3.3) do not depend on the degree of dominance h.
Figure 1.
Conditions for the maintenance of genetic variation by sexually antagonistic selection in an autosomal locus. Variation is maintained for parameter values between the two lines.
In the above example, we assumed that the degree of dominance is equal in both sexes. If this is not so, then the most favourable scenario for the maintenance of variation is when allele a is dominant in the sex, where it is advantageous and is recessive in the sex where it is deleterious. For example, if the feminizing allele a is fully dominant in females and fully recessive in males, then polymorphism is maintained, no matter what the gain to females (G>0) and cost to males (C′>0). In sum, when dominance is similar between the sexes, polymorphism on the autosomes is unlikely, owing to the narrow parameter space supporting polymorphism (for feasible combinations of G and C′), but strong counterbalancing dominance relationships reverses this outcome.
(b) X-linked gene
Assume that the locus under consideration is X-linked. Let f1, f2 and f3 be fitnesses of females AA, Aa and aa, respectively, and m1 and m2 be fitnesses of males A and a. In this model, polymorphism is protected if
| 3.4a |
| 3.4b |
where is the average of male fitnesses (e.g. p. 80 in Edwards 1977). Note that inequality (3.4a) is the condition for a successful invasion of allele a in a population initially monomorphic for allele A and inequality (3.4b) is the condition for a successful invasion of allele A in a population initially monomorphic for allele a.
We consider three different cases.
(i) Overdominance in the homogametic sex
Let f2>f1, f3 and, for definiteness, m1>m2. Without any loss of generality, we can set f1=m1=1, f2=1+God, m2=1−C′, where God>0 is the fitness gain of heterozygous females and 0<C′≤1 is the fitness loss in males. Then, the condition (3.4b) is always satisfied and polymorphism is maintained, if
| 3.5 |
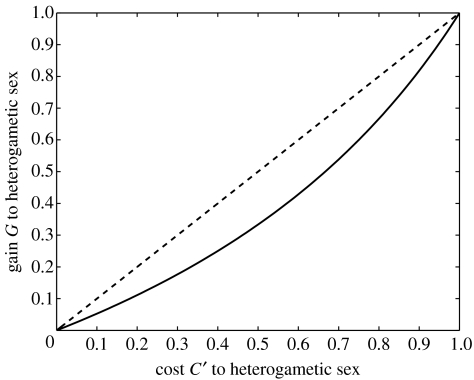
The right-hand side of the last inequality is positive and increases with C′. Thus, simple overdominance in females (i.e. God>0) is not enough but the gain in heterozygous female fitness God has to be sufficiently large relative to cost to male hemizygotes C′ (see figure 2).
Figure 2.
Conditions for the maintenance of genetic variation in an X-linked gene when there is overdominance in one sex and directional selection in the other sex. Variation is maintained for parameter values above the solid line (see inequality (3.5)). The dashed line shows the diagonal G=C′ for comparison.
To compare the conditions for the maintenance of genetic variation by overdominance in the cases of autosomal and X-linked genes, let us first rewrite inequality (3.5) as
| 3.6a |
Let us also assume that in the autosomal case, the corresponding gene is recessive (i.e. h=0) and that the gain to heterozygous males is small (i.e. ). These assumptions are conservative in the sense that they narrow the conditions for the maintenance of polymorphism as specified by inequality (3.2). Then, inequality (3.2) can be rewritten as
| 3.6b |
Comparing inequalities (3.6a) and (3.6b) shows that with overdominance when the cost of homosexuality C′ is large, it is substantially easier to maintain genetic variation in an autosomal gene than in a X-linked gene.
(ii) Sexually antagonistic selection: feminization allele
Assume that allele a increases female fitness but decreases male fitness. Let female fitnesses be f1=1, f2=1+hG, f3=1+G and male fitnesses be m1=1, m2=1−C′, where G is the maximum gain in fitness of females, C′ is the loss of males and h is a measure of dominance. Then, allele a increases in frequency when rare if
| 3.7a |
i.e. if the fitness gain to females is sufficiently large (Rice 1984, eqn 6), and it does not go to fixation if
| 3.7b |
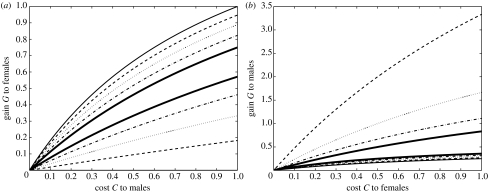
i.e. if the fitness loss to males is large enough. Combining inequalities (3.7) and solving for h demonstrate that h>0.5 is required to support polymorphism, which means that allele a has to be sufficiently dominant. Increasing h broadens the conditions for the maintenance of variation (see figure 3a). Assuming that G and C′ are small, a dominant (h=1) feminization allele will invade, if G>C′/2 (which happens under less strict conditions than in the autosomal case) but will never go to fixation, i.e. the maintenance of a feminization allele requires its fitness advantage to females to be sufficiently large.
Figure 3.
Conditions for the maintenance of genetic variation by sexually antagonistic selection in an X-linked locus. Variation is maintained for parameter values between the two lines of the same color. (a) Feminization allele (solid thin lines, h=0.6; dashed lines, h=0.7; dotted lines, h=0.8; dash–dot lines, h=0.9; solid thick lines, h=1.0). Note that the second solid thick line coincides with the x-axis. (b) Masculinization allele (solid thin lines, h=0.0; dashed lines, h=0.1; dotted lines, h=0.2; dash–dot lines, h=0.3; solid thick lines, h=0.4). Note that the second solid thin line coincides with the y-axis.
(iii) Sexually antagonistic selection: masculinization allele
Assume that allele a increases male fitness but decreases female fitness. Let f1=1, f2=1−hC′, f3=1−C′, m1=1, m2=1+G, where G is the gain in fitness of males, C′ is the maximum fitness loss of females and h is a measure of dominance. Then, allele a increases in frequency when rare if
| 3.8a |
i.e. if the fitness gain to males is sufficiently large (Rice 1984, eqn 3), and it does not go to fixation if
| 3.8b |
i.e. if the fitness loss to females is large enough. Combining inequalities (3.8) and solving for h demonstrate that h<(1+G)/(2+G) is required to support polymorphism, which means that allele a has to be sufficiently recessive. Decreasing h broadens the conditions for the maintenance of variation (see figure 3a). Assuming that G and C′ are small, a recessive (h=0) masculinization allele will always invade and will not go to fixation if G<2C′, i.e. the maintenance of a masculinization allele requires its fitness advantage to males to be sufficiently small.
4. Maternal selection
Next, we consider the case when fitnesses in both sexes are determined by maternal effects (i.e. a maternal effect that partially masculinizes or feminizes all zygotes irrespective of their chromosomal sex determination) and that masculinization of females and feminization of males reduces fitness. Let f1, f2, f3 and m1, m2, m3 be fitnesses (owing to maternal effect influences) of daughters and sons of females that are AA, Aa and aa, respectively. In this model, the polymorphism is protected if conditions (3.1) are satisfied, i.e. these conditions are exactly the same as when direct selection acts on autosomal locus. Note that because the gene under consideration is expressed in females only, whether it is autosomal or X-linked is irrelevant.
5. Combined direct and maternal selection
Finally, we consider the case when female fitness is determined exclusively by her genotype while male fitness is determined by a maternal effect. Let f1, f2 and f3 be the fitnesses of females AA, Aa and aa. Let m1, m2 and m3 be the maternally determined fitnesses of their sons. In this model, the polymorphism is protected
| 5.9a |
| 5.9b |
Note that inequality (5.9a) is the condition for a successful invasion of allele a in a population initially monomorphic for allele A and inequality (5.9b) is the condition for a successful invasion of allele A in a population initially monomorphic for allele a. As in the previous case, because the gene under consideration is expressed in females only, whether it is autosomal or X-linked is irrelevant.
To illustrate these conditions, assume that allele a increases female fitness but decreases her sons' fitness so that f1=1, f2=1+hG, f3=1+G but m1=1, m2=1−hC′, m3=1−C′, where G and C′ are the maximum fitness gain and maximum fitness loss for females and males, respectively. Then, allele a will invade if
| 5.10a |
i.e. if the fitness gain to females is sufficiently large, and it does not go to fixation if
| 5.10b |
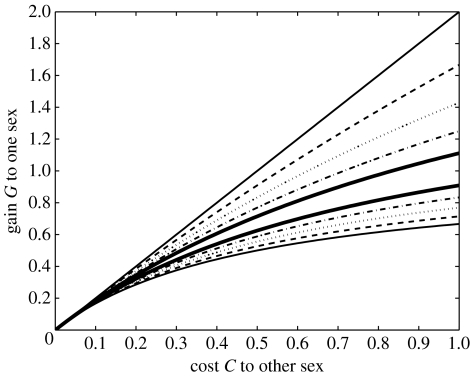
i.e. if the fitness loss to males is large enough. In this case, the condition for the invasion of allele a is less strict that that when both fitnesses are determined genetically, but the condition for allele a not getting fixed is more strict. Figure 4 illustrates the conditions for the maintenance of genetic variation in this model. Note that decreasing h (i.e. the degree of dominance of allele a) increases the range of parameters values resulting in polymorphism. Variation is maintained the easiest if allele a is recessive (i.e. h=0) but even in this case, the conditions for the maintenance of variation are strict (G/(1+2G)<C′/2<G) especially if selection is weak. Comparing these conditions with those in the case of direct selection on an autosomal gene (which can be written as G/(1+2G)<C′<G by combining inequalities (3.3)) shows that mixed selection can maintain polymorphism at higher costs (i.e. higher values of C′) than direct selection.
Figure 4.
Conditions for the maintenance of genetic variation by sexually antagonistic maternal selection. Variation is maintained for parameter values between the two lines of the same type (solid thin lines, h=0.0; dashed lines, h=0.2; dotted lines, h=0.4; dash–dot lines, h=0.6; solid thick lines, h=0.8).
6. Discussion
Our population genetic models indicate that genes influencing homosexuality can readily spread and become polymorphic under a wide range of conditions. One of the main goals in our population genetic analysis of homosexuality was to use theory as a guide to focus future research on the genetic basis of homosexuality. We were especially interested in determining whether explicit predictions concerning the genetic attributes (genomic locations, dominance and effect size) of homosexuality genes could distinguish between two of the major competing evolutionary explanations for the maintenance of polymorphic homosexuality genes: overdominance versus sexual antagonism. Because there is an empirically established maternal effect influencing the expression of homosexuality, we also sought to examine how heritable maternal effects might influence these genetic predictions.
(a) Genomic location
The population genetic models that we have developed make it clear that polymorphism for homosexuality genes can be readily achieved at both autosomal and sex-linked loci. The critical factor in maintaining polymorphism is that the homosexual allele has a net selective advantage when rare and disadvantage when common. However, the feasibility of polymorphism on these two types of chromosomes differs substantially. The parameter space permitting polymorphism when overdominance is operating is substantially larger when a gene is autosomal than when it is X-linked (e.g. compare equation 3.6a with equation 3.6b). In contrast, the parameter space permitting polymorphism is far larger on the X compared to the autosomes for sexually antagonistic genes (compare figure 2 with figures 3 and 4) and this effect is exaggerated when the homosexuality genes are masculinizing rather than feminizing (compare figures 3 and 4). Overall, polymorphism for homosexuality via sexually antagonistic alleles is predicted to be strongly over-represented on the X chromosome, whereas polymorphism for homosexuality owing to overdominance will be over-represented on the autosomes, but to a lesser degree. This disparity between the parameter spaces supporting polymorphism on the autosomes and sex chromosomes associated with the overdominant and sexual antagonism models leads to a clear prediction: X-linkage of gene influencing homosexuality is, in and of itself, the evidence supporting sexual antagonism rather than overdominance. Because the X chromosome is a small proportion of the genome of most species, the finding of multiple X-linked gene loci influencing homosexuality (within one species or among different species) would provide strong evidence for sexual antagonism being an important selective force maintaining genetic variation for homosexuality. Similarly, if most genes influencing homosexuality (within a species or among species) are found to be autosomal, then this would provide weaker (because most genes are autosomal), but nonetheless positive, support for overdominance. Additional supporting evidence can be obtained from the dominance and effect size associated with the alleles of these genes.
(b) Dominance
Our population genetic analysis also makes strong predictions about dominance (in the currency of fitness) that can help to discriminate between the overdominance and sexual antagonism models. If a gene influencing homosexuality is X-linked and if the polymorphism is being maintained by sexual antagonism, then a gene causing female homosexuality should show strong dominance and one causing male homosexuality should show a strong level of recessiveness. No such gender by dominance association is predicted on the autosomes, in which case the gene should be dominant in the sex in which it is favoured. Lastly, if an X-linked polymorphism for a gene influencing homosexuality is being maintained by overdominance, then no sex-specific pattern of dominance is predicted.
(c) Effect size
If the effect size (in the currency of fitness) of an autosomal gene influencing homosexuality differs substantially between the sexes (e.g. large influence on fitness in only one sex), then it is unlikely that the gene is maintained by sexual antagonism, unless the gene is strongly dominant in the sex where it is favoured and strongly recessive in the sex where it is disfavoured. This is a strong prediction. If a QTL analysis located an autosomal gene influencing homosexuality, then unless the gene strongly influenced the fitness of both males and females in different directions (under ancestral environmental conditions), sexual antagonism is highly unlikely. In contrast, on the X chromosome, strong asymmetries in effect size (in the currency of fitness) are feasible for sexually antagonistic alleles.
In the case of autosomal overdominance, polymorphism is maintained only when the effect size (i.e. the reduction in fitness, C′, of aa homozygotes) is sufficiently large, whereas in the case of overdominance on the X, the cost to the hemizygous sex (C′ in males) must be sufficiently small. This predicts that if an X-linked homosexuality allele is found that has a large influence on male fitness (under ancestral conditions), then overdominance is unlikely to be the factor responsible for maintaining polymorphism. Similarly, if an autosomal homosexuality allele is found that has little fitness influence when expressed in homozygous homosexuals, then overdominance is unlikely to be the factor maintaining the polymorphism.
(d) Combining location, dominance and effect size
Although none of the genetic characteristics alone is definitive in resolving between overdominance and sexual antagonism, in combination, these characteristics can provide high resolution. For example, if a male homosexuality QTL is X-linked and dominant or a female homosexuality QTL is X-linked and recessive, then there is strong evidence that sexual antagonism is not responsible for maintaining the polymorphism. Similarly, if an autosomal gene influencing homosexuality has a small effect sizes (G and C′) and similar dominance in the two sexes, then sexual antagonism is unlikely to maintain the polymorphism. As a last example, overdominance is unlikely to maintain polymorphism for an X-linked QTL causing male homosexuality if male homosexuals have low fitness.
(e) Maternal versus direct effects
If heritable maternal effects influence homosexuality, then the distinction between autosomal versus sex-linked inheritance is eliminated and the parameter space supporting polymorphism is reduced. As a consequence, the fact that empirical evidence indicates that maternal effects can influence the expression of at least male homosexuality (Blanchard & Klassen 1997; Blanchard 2004) does not appear to create a context that expands the opportunity for polymorphism of genes influencing homosexuality.
In the 1990s, there was a surge of interest in finding and mapping genes that influence homosexuality in humans, and the recent work by Mustanski et al. (2005) extends this work to more modern QTL analysis. Our theoretical work described here shows that the genetic characteristics of these genes can be informative in resolving different models of natural selection that maintain their polymorphism. Although our work does not provide a unique prediction that will differentiate between overdominance versus sexual antagonism in all cases, it does identify diagnostic patterns. Lastly, the fact that homosexuality appears to be common in many other species (e.g. Resko et al. 1999; Bagemihl 2000) should provide substantial opportunity to utilize these patterns via the comparative method. In sum, population genetic models can provide useful predictions for evaluating the selective factors maintaining polymorphism contributing to the homosexual behaviour.
Acknowledgments
Supported by National Institutes of Health grant GM56693 (S.G.) and by National Science Foundation grants DEB-0111613 (S.G.) and DEB-0128780 and DEB-0410112 (W.R.R.).
Appendix A
Here, we give the dynamic equations describing the models studied in the main body of the paper. The conditions for local stability of monomorphic equilibria in these models can be found using standard methods.
(a) Direct selection on an autosomal locus
Let female fitnesses be f1, f2 and f3 and male fitnesses be m1, m2 and m3. Let u and 1−u be the frequencies of alleles A and a in male offspring and let v and 1−v be the corresponding frequencies in female offspring. The population genetic state is conveniently defined in terms of x=u/(1−u) and y=v/(1−v). In the next generation,
(e.g. eqn (2.6) in Karlin 1972).
(b) Direct selection on an X-linked locus
Let f1, f2 and f3 be the fitnesses of females AA, Aa and aa and m1 and m2 be the fitnesses of males A and a. Using the ratios of allele frequencies in males, x, and females, y, the dynamics are described by equations
(e.g. eqn (6.2.3) in Edwards 1977).
(c) Models of maternal and mixed selection
Let u be the frequency of A in adult males and p, q and r be the frequencies of AA, Aa and aa in adult females. Let m1, m2 and m3 be the fitnesses of sons of mothers AA, Aa and aa, respectively. If the locus under consideration is X-linked, then in the next generation and up to a normalizing factor
| A1a |
If the locus under consideration is autosomal, then
| A1b |
In both cases, the normalizing factor is , i.e. the average fitness of males.
Assume that female fitness is determined maternally. Let f1, f2 and f3 be the fitnesses of daughters of mothers AA, Aa and aa. Then, in the next generation and up to a normalizing factor, the adult female frequencies are
| A2a |
| A2b |
| A2c |
Assume that female fitness is determined genetically. Let f1, f2 and f3 be the fitnesses of females AA, Aa and aa. Then, the above equations take form
| A3a |
| A3b |
| A3c |
where v=p+q/2 is the frequency of A in mature females. In both cases, the normalizing factor is , i.e. the average fitness of females.
To find conditions for the stability of monomorphic equilibria, we analysed the systems of recurrence equations for u, v and q.
References
- Arnqvist G, Rowe L. Princeton University Press; Princeton, NJ: 2005. Sexual conflict. [Google Scholar]
- Bagemihl B. Stonewall Inn Editions; New York, NY: 2000. Biological exuberance: animal homosexuality and natural diversity. [Google Scholar]
- Bailey J.M, Pillard R.C, Dawood K, Miller M.B, Farrer L.A, Trivedi S, Murphy R.L. A family history study of male sexual orientation using three independent samples. Behav. Genet. 1999;29:79–86. doi: 10.1023/a:1021652204405. doi:10.1023/A:1021652204405 [DOI] [PubMed] [Google Scholar]
- Bearman P.S. Opposite-sex twins and adolescent same-sex attraction. Am. J. Sociol. 2005;107:1179–1205. doi:10.1086/341906 [Google Scholar]
- Bell A, Weinberg M, Hammersmith S. Indiana University Press; Bloomington, IL: 1981. Sexual preference: its development in men and women. [Google Scholar]
- Blanchard R. Quantitative and theoretical analyses of the relation between older brothers and homosexuality in men. J. Theor. Biol. 2004;230:173–187. doi: 10.1016/j.jtbi.2004.04.021. doi:10.1016/j.jtbi.2004.04.021 [DOI] [PubMed] [Google Scholar]
- Blanchard R, Bogaert A.F. Homosexuality in men and the number of older brothers. Am. J. Psychiat. 1996;153:27–31. doi: 10.1176/ajp.153.1.27. [DOI] [PubMed] [Google Scholar]
- Blanchard R, Klassen P. H–Y antigen and homosexuality in men. J. Theor. Biol. 1997;185:373–378. doi: 10.1006/jtbi.1996.0315. doi:10.1006/jtbi.1996.0315 [DOI] [PubMed] [Google Scholar]
- Camperio-Ciani A, Corna F, Capiluppi C. Evidence for maternally inherited factors favouring male homosexuality and promoting female fecundity. Proc. R. Soc. B. 2004;271:2217–2221. doi: 10.1098/rspb.2004.2872. doi:10.1098/rspb.2004.2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawood K, Pillard R.C, Horvath C, Revelle W, Bailey J.M. Familial aspects of male homosexuality. Arch. Sex. Behav. 2000;29:155–163. doi: 10.1023/a:1001955721992. doi:10.1023/A:1001955721992 [DOI] [PubMed] [Google Scholar]
- Devlin R.H, Nagahama Y. Sex determination and sex differentiation in fish: an overview of genetic, physiological, and environmental influences. Aquaculture. 2002;208:191–364. doi:10.1016/S0044-8486(02)00057-1 [Google Scholar]
- Diamond M. Homosexuality and bisexuality in different populations. Arch. Sex. Behav. 1993;32:291–310. doi: 10.1007/BF01542119. doi:10.1007/BF01542119 [DOI] [PubMed] [Google Scholar]
- Edwards A.W.F. Cambridge University Press; Cambridge, UK: 1977. Foundations of mathematical genetics. [Google Scholar]
- Gavrilets S. One-locus two-allele models with maternal (parental) selection. Genetics. 1998;149:1147–1152. doi: 10.1093/genetics/149.2.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhard P. Incidence of overt homosexuality in the United States and Western Europe. In: Livingood J, editor. National institute of mental health task force on homosexuality: final report and background papers. US Department of Health, Education, and Welfare: DHEW Publication (HMS) 72-9116; Washington, DC: 1972. pp. 22–29. [Google Scholar]
- Getz W.M. Invasion and maintenance of alleles that influence mating and parental success. J. Theor. Biol. 1993;162:515–537. doi: 10.1006/jtbi.1993.1102. doi:10.1006/jtbi.1993.1102 [DOI] [PubMed] [Google Scholar]
- Hamer D.H, Hu S, Magnuson V.L, Hu N, Pattatucci A.M.L. A linkage between DNA markers on the X-chromosome and male sexual orientation. Science. 1993;261:321–327. doi: 10.1126/science.8332896. [DOI] [PubMed] [Google Scholar]
- Hammer D, Copeland P. Simon & Schuster; New York, NY: 1994. The science of desire: the search for the gay gene and biology of behavior. [Google Scholar]
- Hu S, Pattatucci A.M.L, Patterson C, Li L, Fulker D.W, Cherny S.S, Kruglyak L, Hamer D.H. Linkage between sexual orientation and chromosome Xq28 in males but not in females. Nat. Genet. 1995;11:248–256. doi: 10.1038/ng1195-248. doi:10.1038/ng1195-248 [DOI] [PubMed] [Google Scholar]
- Hutchinson G.E. A speculative consideration of certain forms of sexual selection in man. Am. Nat. 1959;93:81–91. doi:10.1086/282059 [Google Scholar]
- Karlin S. Some mathematical models of population genetics. Am. Math. Mon. 1972;79:699–739. doi:10.2307/2316262 [Google Scholar]
- King M, Green J, Osborn D.P.J, Arkell J, Hetherton J, Pereira E. Family size in white gay and heterosexual men. Arch. Sex. Behav. 2005;34:117–122. doi: 10.1007/s10508-005-1006-8. doi:10.1007/s10508-005-1006-8 [DOI] [PubMed] [Google Scholar]
- Kinsey A.C, Pomeroy W.B, Martin C.E. W. B. Saunders; Philadelphia, PA: 1948. Sexual behavior in the human male. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsey A.C, Pomeroy W.B, Martin C.E, Gebhard P.H. W. B. Saunders; Philadelphia, PA: 1953. Sexual behavior in the human female. [Google Scholar]
- Kirsch J.A.W, Weinrich J.D. Homosexuality, nature, and biology: is homosexuality natural? Does it matter? In: Gonsiorek J, Weinrich J, editors. Homosexuality: research implications for public policy. Sage Publications; Newbury Park, CA: 1991. pp. 13–31. [Google Scholar]
- MacIntyre F, Estep K.W. Sperm competition and the persistence of genes for male homosexuality. Biosystems. 1993;31:223–233. doi: 10.1016/0303-2647(93)90051-d. doi:10.1016/0303-2647(93)90051-D [DOI] [PubMed] [Google Scholar]
- McKnight J. Routledge; London, UK: 1997. Straight science? Homosexuality, evolution and adaptation. [Google Scholar]
- Miller E.M. Homosexuality, birth order, and evolution: Toward an equilibrium reproductive economics of homosexuality. Arch. Sex. Behav. 2000;29:1–34. doi: 10.1023/a:1001836320541. doi:10.1023/A:1001836320541 [DOI] [PubMed] [Google Scholar]
- Miller P.M, Gavrilets S, Rice W.R. Sexual conflict via maternal-effect genes in ZW species. Science. 2006;312:73. doi: 10.1126/science.1123727. doi:10.1126/science.1123727 [DOI] [PubMed] [Google Scholar]
- Mustanski B.S, DuPree M.G, Nievergelt C.M, Bocklandt S, Schork N.J, Hamer D.H. A genomewide scan of male sexual orientation. Hum. Genet. 2005;116:272–278. doi: 10.1007/s00439-004-1241-4. doi:10.1007/s00439-004-1241-4 [DOI] [PubMed] [Google Scholar]
- Pattatucci A.M.L. Molecular investigations into complex behavior: lessons from sexual orientation studies. Hum. Biol. 1998;70:367–386. [PubMed] [Google Scholar]
- Pillard R.C, Bailey J.M. Human sexual orientation has a heritable component. Hum. Biol. 1998;70:347–365. [PubMed] [Google Scholar]
- Pillard R.C, Poumadere J, Carretta R.A. Is homosexuality familial? A review, some data, and a suggestion. Arch. Sex. Behav. 1981;10:465–475. doi: 10.1007/BF01541437. doi:10.1007/BF01541437 [DOI] [PubMed] [Google Scholar]
- Pillard R.C, Poumadere J, Carretta R.A. A family study of sexual orientation. Arch. Sex. Behav. 1982;11:511–520. doi: 10.1007/BF01542476. doi:10.1007/BF01542476 [DOI] [PubMed] [Google Scholar]
- Resko J.A, Perkins A, Roselli C.E, Stellflug J.N, Stormshak F.K. Sexual behavior in rams: male orientation and its endocrine correlates. J. Reprod. Fertil. Suppl. 1999;54:259–269. [PubMed] [Google Scholar]
- Rice W.R. Sex-chromosomes and the evolution of sexual dimorphism. Evolution. 1984;38:735–742. doi: 10.1111/j.1558-5646.1984.tb00346.x. doi:10.2307/2408385 [DOI] [PubMed] [Google Scholar]
- Rice W.R, Holland B. The enemies within: intergenomic conflict, interlocus contest evolution (ICE) and the intraspecific Red Queen. Behav. Ecol. Sociobiol. 1997;41:1–10. doi:10.1007/s002650050357 [Google Scholar]
- Rice G, Anderson C, Risch N, Ebers G. Male homosexuality: absence of linkage to microsatellite markers at Xq28. Science. 1999;284:665–667. doi: 10.1126/science.284.5414.665. doi:10.1126/science.284.5414.665 [DOI] [PubMed] [Google Scholar]
- Sell R.L, Wells J.A, Wypij D. The prevalence of homosexual behavior and attraction in the United States, the United Kingdom, and France: results of national population-based samples. Arch. Sex. Behav. 1995;24:235–248. doi: 10.1007/BF01541598. doi:10.1007/BF01541598 [DOI] [PubMed] [Google Scholar]
- Spencer H.G. Further properties of Gavrilets' one-locus two-allele model of maternal selection. Genetics. 2003;164:1689–1692. doi: 10.1093/genetics/164.4.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivers R.L. Parent–offspring conflict. Am. Zoo. 1974;14:249–264. [Google Scholar]
- Weinrich J.D. Scribner's; New York, NY: 1987. Sexual landscapes. [Google Scholar]
- Wilson E.O. Harvard University Press; Cambridge, MA: 1975. Sociobiology: the new synthesis. [Google Scholar]