Abstract
Heterogeneity in the parameters governing the spread of infectious diseases is a common feature of real-world epidemics. It has been suggested that for pathogens with basic reproductive number R0>1, increasing heterogeneity makes extinction of disease more likely during the early rounds of transmission. The basic reproductive number R0 of the introduced pathogen may, however, be less than 1 after the introduction, and evolutionary changes are then required for R0 to increase to above 1 and the pathogen to emerge. In this paper, we consider how host heterogeneity influences the emergence of both non-evolving pathogens and those that must undergo adaptive changes to spread in the host population. In contrast to previous results, we find that heterogeneity does not always make extinction more likely and that if adaptation is required for emergence, the effect of host heterogeneity is relatively small. We discuss the application of these ideas to vaccination strategies.
Keywords: emerging infectious diseases, superspreaders, mathematical model, multi-type branching process
1. Introduction
During the recent outbreak of SARS, we heard a great deal about so-called superspreaders, defined as infected individuals who passed on the infectious agent to many more people than average (Shen et al. 2004). The phenomenon of superspreading can be viewed as an extreme case of variation or heterogeneity in key epidemiological parameters. An analysis of epidemic data has indicated that the presence of such heterogeneities is a robust feature of many infectious diseases (Hethcote & Yorke 1984; May & Anderson 1987; Grenfell et al. 1995; Woolhouse 1997; Galvani & May 2005; Lloyd-Smith et al. 2005).
The emergence of a disease combines two elements: the introduction of the pathogen in a new host population and its subsequent spread and maintenance within the population. Mathematical models have been used to show that, given a successful introduction, an epidemic spreads more rapidly if there are heterogeneities in contact between individuals than in a homogeneous host population with the mean contact rate (the so-called ‘mean-field’ model; Hethcote & Van Ark 1987; Hasibeder & Dye 1988; May & Anderson 1988; Gupta et al. 1989; Diekmann et al. 1990; Marschner 1992; Dobson & Foufopoulos 2001). This phenomenon is the result of a synergy in susceptibility and infectivity in these models: by having higher susceptibilities as well as higher infectivities, some individuals contribute disproportionately to the spread of disease. The characterization of the sources of such heterogeneity is important for the design of targeted treatment strategies (Becker & Hall 1996; Murphy et al. 2003). Heterogeneities in epidemiological parameters also affect the probability that a pathogen can establish itself in a new population. Generally, for a given basic reproductive number, increasing heterogeneity leads to a decrease in the probability of emergence (Galvani & May 2005; Lloyd-Smith et al. 2005; Xiao et al. 2006). The intuitive reason for this effect is an increase in extinctions of the pathogen owing to stochastic effects in the early stages of the epidemic.
Pathogen adaptation to its new host population may also play a role in emergence. Theoretical studies of species invasion (Gomulkiewicz & Holt 1995) and disease emergence (Antia et al. 2003) have shown that adaptation, if sufficiently fast, can rescue the pathogen population from extinction. Thus, adaptability may facilitate emergence, which is indirectly supported by findings that protozoa and viruses emerge more frequently than helminths (Taylor et al. 2001).
In this paper, we systematically investigate the impact of heterogeneity on the emergence of a pathogen. The mathematical model we develop enables us to consider heterogeneity in susceptibility, infectivity and mixing patterns. Unlike earlier studies on this topic, we also include pathogens that cannot transmit efficiently and need to adapt to their new hosts to establish themselves, i.e. pathogens that have basic reproductive number R0<1 when they are introduced into the new host population. Finally, we will investigate the effect of different vaccination strategies on the probability of disease emergence in heterogeneous host populations.
2. Material and methods
(a) Disease spread as a branching process
We model disease spread in a heterogeneous host population as a multi-type branching process in discrete time (Harris 1963; Metz 1978; Ball 1983). In our framework, the population is subdivided into homogeneous subgroups with different epidemiological characteristics (figure 1). In this section, we describe how we represent transmission of a pathogen in a heterogeneous population as a branching process, and how this framework can then be used to determine the probability that an epidemic occurs given the introduction of a single infected individual, i.e. the probability of emergence.
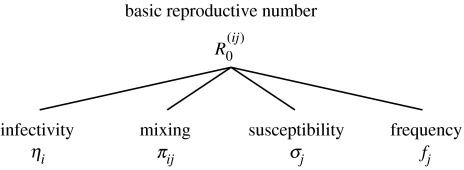
Figure 1.
Implementation of heterogeneity in our model. We partition the host population into subsets with uniform infectivities and susceptibilities. The number of secondary cases, , an infected individual in subpopulation i causes in an entirely susceptible subpopulation j is the product of the infectivity of an individual of type i, ηi, the mixing matrix of the two subsets, πij, and the susceptibility, σj, and frequency, fj, of the individuals of type j (see equation (2.2)). The dominant eigenvalue of is the basic reproductive number of the pathogen, R0.
We consider heterogeneity in the following host properties:
susceptibility to infection,
infectiousness or infectivity, and
the mixing preferences of the different host types for each other.
We assume these properties to be independent, but correlations can be imposed if appropriate for any particular disease. For instance, infectiousness and susceptibility may be related through factors specific to the individual (e.g. someone with an STD may be both more likely to contract HIV per encounter and subsequently more likely infect later partners). In other situations, apparent correlations in infectivity and susceptibility may be explained by mixing patterns (e.g. differences in the incidence of disease among children attending day care and those who do not), rather than any host-specific factors. The following framework allows us to disentangle the contributions of these processes to disease spread.
Infection events take place between a donor and recipient, and in our model, the probability of an infection event occurring in a given encounter is proportional to the infectivity of the donor and the susceptibility of the recipient. This assumption is known as proportionate mixing (Hethcote & Van Ark 1987). Host types may also mix with different preferences for one another, and so the number of potentially infective encounters between two types per round of transmission is determined by the frequency of each in the population and the mixing preference of the donor for the recipient (figure 1).
(b) Defining the basic reproductive number for heterogeneous populations
These host properties determine the transmission dynamics of a disease. Consider a population with n host types. Let f={f1, f2, …, fn} be the set of frequencies of each host type in the population , be the mean number of secondary cases caused by one infected type i host and be the mean number of transmission events to recipient type j caused by one infected donor host of type i, such that
| 2.1 |
We assume that is proportional to the product of the following factors:
A normalization factor N proportional to the basic reproductive number R0 of the population as a whole. We describe how this is calculated below.
The relative infectivity ηi of type i hosts. The quantities ηi could be interpreted as the relative efficacies of infected hosts of each type in generating secondary cases, with all other factors held constant. For example, in a pathogen spread by coughing, η for a particular host type might be a measure of the virion load in each cough or the frequency of coughing. The sum of the ηi does not have to be one, as it cancels out in the calculation of the normalization factor N.
The contact matrix πij describing the relative preferences that type i hosts have for contact with type j hosts. More precisely, we define the quantity πijfj to be proportional to the number of contacts that may lead to transmission events to a host of type j during the infectious period of a single host of type i. The overall normalization of π is arbitrary and is absorbed into the constant N, and π need not be symmetric.
The relative susceptibility σj of type j hosts. The quantities σj are proportional to the probabilities of each susceptible host type being infected in an encounter with a given infected host. This could reflect genetic predisposition, physiological factors (e.g. age or degree of immunosuppression) or behavioural traits. The sum of σj is again arbitrary.
The population frequency fj of type j hosts.
In summary,
| 2.2 |
where is the analogue of the contact matrix in deterministic epidemiological models. We note some degeneracy in this parameterization (see electronic supplementary material), but it allows us to discuss how the conceptually distinct factors interact to determine the probability of disease emergence. As described earlier, we do not assume any correlation between susceptibility, infectivity and mixing, although this can be imposed, if appropriate.1 The matrix completely defines the branching process and thus the dynamics of disease spread after the introduction event.
The basic reproductive number, R0, is defined as the mean number of secondary infections resulting from a ‘typical’ infected individual. Epidemiological theory tells us that this tends to the largest eigenvalue of the matrix (see, for example, Diekmann et al. (1990), Becker & Hall (1996) and Hyman & Li (2000)). Here we briefly describe why this is so. In a heterogeneous population, the notion of a typical infection must involve the frequency of each infected host type in the population during the outbreak, rather than the frequencies f of each susceptible host type. Let gi be the proportion of the infected individuals that are of type i (i=1 … n). Since the susceptible hosts are unlimited in our model, the expected values of the proportions gi will be asymptotically invariant (i.e. they will approach constant values after transient behaviour depending on the proportions of each spreader type that are initially infected). Then, we define R0 of the whole population at any time as
| 2.3 |
To calculate gi, we note that the branching process takes place in discrete time and consider one round of infection. The invariance of the proportions of infected host types gi implies that asymptotically, the mean number of new cases in type j hosts resulting from a single randomly chosen infected host will be
| 2.4 |
or
| 2.5 |
Thus, formally R0 is the largest eigenvalue of the matrix and has eigenvector g. This is the rate of growth or decay that the epidemic approaches after an initial transient.
This defines the normalization factor N in equation (2.2). We set the overall R0 for the population and choose the factor N to scale the matrix R0 so that equation (2.5) is satisfied. This definition of R0 ensures that emergence is not possible for a pathogen in any population with R0<1.
(c) Calculating the probability of emergence
We model the spread of a pathogen in a population as a multi-type branching process in discrete time. We assume a constant infectious period for each case, and during this period, the number of secondary cases in type j hosts originating from one infected type i host is a Poisson-distributed random variable with mean . This can be expressed with the probability-generating function (PGF) Fi(s), which specifies the distribution of secondary cases generated by a case of type i in each of the n host types, and its argument s={s1, s2, … sn} is an n-dimensional vector of dummy variables. Specifically, Fi(0) is the extinction probability after one generation, given a starting condition of one infected host of type i. Using standard multi-type branching process theory (see, for example, Harris (1963)), from the assumption of Poisson-distributed secondary cases between hosts of each type, it follows that
| 2.6 |
The extinction probability after m generations starting with one infected host of type i is , where the superscript denotes the mth iterate of the PGF. The ultimate extinction probability, Pi, the limit as m→∞, is the solution to
| 2.7 |
as F is a strictly increasing function of its arguments. This can be seen graphically for a simple model with one host type (n=1), and it is extended easily to arbitrary n. We then calculate the probability of emergence in the population as
| 2.8 |
where we assume that the probability of the first infection event (i.e. the introduction of the disease into the host population) occurring in the host type i is proportional to both that type's susceptibility and its frequency in the population.
In this paper, we compare the probability of emergence in a heterogeneous population with the reference case of the probability of emergence in a homogeneous population with the same R0. In our framework, disease spread in a homogeneous population can be described with a simple single-component probability-generating function , and the probability of emergence is then2
| 2.9 |
where s* is the solution to .
(d) Incorporating pathogen evolution
This framework is easily extended to allow us to consider multiple pathogen strains and pathogen evolution. Assume that the pathogen can mutate sequentially and irreversibly through strains 1 … ω, each with distinct population average R0. In each transmission event, there is a probability μ of an adaptation occurring and the newly infected host acquiring this new strain. The PGFs are now indexed by both the host type and the pathogen strain:
| 2.10 |
where Fiα is the PGF for the spreading of type i hosts infected with strain type α(=1 … ω−1) and is the mean number of secondary cases caused in hosts of type j by a host of type i infected with strain α. Secondary cases are still Poisson distributed within each host type, but now there is a probability μ per transmission event of an adaptation into the strain α+1. As earlier, we can use these PGFs to calculate the probability of disease emergence, given a single introduction of a host of a given type infected with a given strain.
3. Results
First, we deal with the spread of a single non-evolving pathogen in a heterogeneous host population. Later, we introduce the possibility of pathogen adaptation.
We emphasize that the results in this section relate to calculating how, for a given value of R0, the probability of emergence is influenced by the different forms of host heterogeneity. In other words, we are comparing the risk of disease emergence in populations with different levels of heterogeneity, but the same average R0.
(a) Emergence of non-evolving pathogens
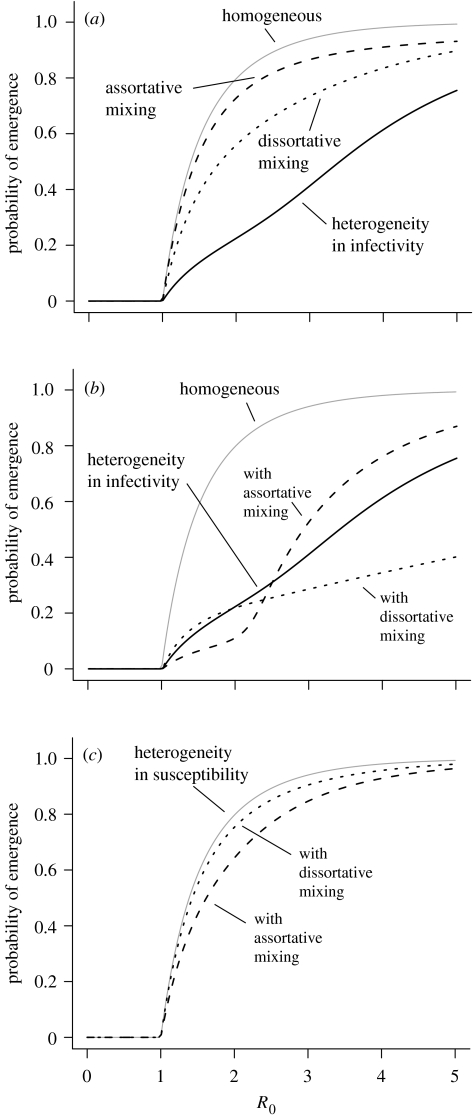
In figure 2, we illustrate how the three forms of heterogeneity (susceptibility, infectivity and mixing) affect the probability of emergence of a single pathogen strain. In each panel, the grey line shows the reference case of the probability of emergence of disease in a homogeneous population, as calculated in equation (2.9).
Figure 2.
The influence of host heterogeneity on the probability of emergence of a non-evolving pathogen. Grey line, the homogeneous case (equation (2.9)). (a) Each form of heterogeneity individually. Solid black line, a population comprising 10% superspreaders with 20-fold higher infectivity than normal; dotted line, dissortative mixing (q=0.05; see equation (3.1)) in a population split of 10–90%; and the dashed line, assortative mixing (q=0.95). Multiple subpopulations with different susceptibilities give rise to the same probability of emergence as the homogeneous case with the same R0 (grey line). (b) Combining heterogeneity in infectivity (solid black line) with heterogeneous mixing patterns. (c) Combining heterogeneity in susceptibility (grey line, which coincides with the homogeneous case) with heterogeneous mixing patterns.
We illustrate the influence of host heterogeneity with simple examples. We divide the population into two host types with frequencies of 10 and 90%. For the case of heterogeneity in infectivity, members of the smaller population are 20 times more infectious than the majority (superspreading). For heterogeneous susceptibility, the smaller population is 100 times more susceptible to contracting the disease per contact with an infected individual. Heterogeneity in contact is represented by the following: assortative mixing, where each type has a preference for contacts with hosts of the same type; and dissortative mixing, in which hosts prefer contact with hosts of the other type. These contact patterns can be represented with a simple one-parameter mixing matrix,
| 3.1 |
so that q=0.5 corresponds to equal mixing of the two types, q>0.5 assortative and q<0.5 dissortative mixing.
Figure 2a shows how the probability of emergence is influenced by each of these forms of heterogeneity alone, as a function of R0 of the whole population. We note that a homogeneous population and a population with heterogeneous susceptibility but the same R0 have identical probabilities of emergence. This follows from equations (2.6) and (2.7), since Fi(s) does not depend on i in the case of heterogeneous susceptibility alone. We also note that variation in the contact preferences of otherwise identical hosts influences disease emergence.
In figure 2b,c, we illustrate how these forms of heterogeneity can interact. We find that the addition of multiple forms of heterogeneity results in complex outcomes. Our results confirm that a non-evolving pathogen with R0<1 is guaranteed to go extinct and suggest that:
Host heterogeneity does not increase the probability of disease emergence, and in many, but not all, cases it decreases it. Our results are consistent with those of Lloyd-Smith et al. (2005), who considered the case of heterogeneity in infectivity alone.
The influence of heterogeneity depends on the nature of the host variation. For example, we show that variation in either susceptibility or infectivity alone gives very different outcomes. The former does not affect emergence, and the latter reduces its risk. We find that the addition of multiple forms of heterogeneity generates complex results—for example, while variation in susceptibility alone gives the same effect as a homogeneous population with the same R0, when combined with heterogeneity in mixing, it reduces the risk of emergence compared to the homogeneous case.
(b) Emergence of evolving pathogens
We now turn to the case where the emergence of a pathogen requires adaptation to its new host. Here, the introduced pathogen has R0<1 and it will go extinct unless it evolves during the chains of transmission to have R0>1. Previous studies have shown how, in the absence of heterogeneity in epidemiological parameters, the probability of emergence depends on: (i) the rate of adaptation, (ii) the number of adaptation events needed to generate a strain with R0>1 and (iii) the fitness (as measured by R0) of the intermediate strains (Antia et al. 2003). Since heterogeneity in infectivity, susceptibility and contact will influence the distribution of lengths of the chains of transmission, they are likely to impact the probability of emergence. In this section, we explore how host heterogeneity and pathogen evolution interact.
As mentioned in §1, the emergence of the evolving pathogen can be considered as a two-step process. The first step is evolution, i.e. the generation of the adapted pathogen (which is defined as the one having an R0>1). The second step, i.e. the emergence of this adapted pathogen, has been considered in §2. Consequently, we focus on how heterogeneity affects the probability of evolution by setting R0 of the evolved pathogen to much greater than 1 (we use a value of 10 000 in the simulations we present here).
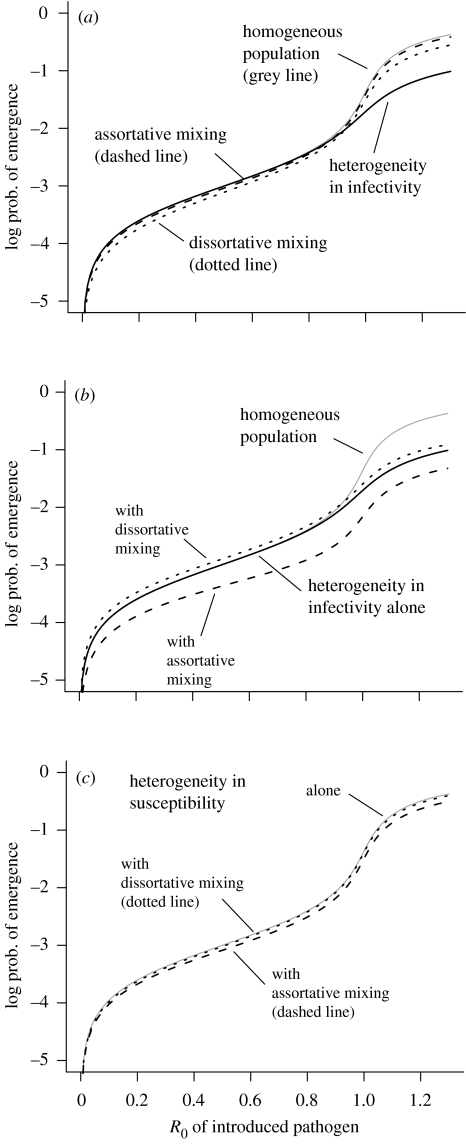
In figure 3 we examine the case where a single adaptation event is sufficient to bring R0 from below to well above 1. Figure 3a illustrates that as for non-evolving pathogens, heterogeneity in either infectivity or contact decreases the probability of emergence with respect to a homogeneous population with the same R0, and heterogeneity in susceptibility alone has no effect. Note that the effect of heterogeneity declines at low values of R0 of the introduced pathogen. We see that even relatively large degrees of host heterogeneity have only a modest influence on emergence, when evolution is necessary for emergence (i.e. when R0<1).
Figure 3.
The probability of one-step pathogen evolution and emergence occurring in the presence of host heterogeneity. The adaptation probability μ per transmission is 0.01. (a) The effect of the different forms of heterogeneity alone. Note that heterogeneity in susceptibility alone gives identical probability of emergence to the homogeneous case (grey line). Heterogeneous infectivity was modelled with 10% of the population as superspreaders with 20-fold higher infectivity than normal; susceptibility with 10% of the population 20 times more likely to contract the infection per contact; and assortative and dissortative mixing modelled with the parameter q=0.95 and 0.05, respectively. (b) Combining heterogeneity in infectivity and in mixing. (c) Combining heterogeneity in susceptibility and mixing.
Figure 3b,c illustrates the effect of combining different forms of heterogeneity. We obtain the surprising result that a host population with a small proportion of superspreaders with high infectivity but who mix dissortatively (i.e. superspreaders have a preference for mixing with ‘normal’ spreaders, and vice versa), has a higher probability of emergence than in the homogeneous case. While the magnitude of this effect is small, it is surprising in the light of the observation of Lloyd-Smith et al. (2005) and the results in §2, which showed that pathogen emergence is always less likely in heterogeneous host populations.
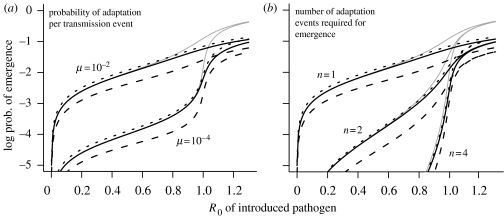
For completeness, we also consider the effects of changing the adaptation rate or the number of adaptation events needed for emergence in the presence of superspreaders with different mixing patterns (figure 4). Here, we see that the probability of emergence is much more sensitive to changes in R0 or the properties of the pathogen (such as changes in the rate of adaptation or the number of adaptation events required) than to heterogeneity in the host population.
Figure 4.
Emergence of pathogens with different evolutionary characteristics in heterogeneous host populations. (a) One-step evolution (n=1) for different values of μ, the probability of adaptation per transmission. (b) Varying the number of adaptation events, n, required to reach the adapted strain with R0≫1. Intermediate strains have the same R0 as the original (introduced) strain. The probability of adaptation per transmission is fixed at μ=0.01. Grey line, the reference case of a homogeneous population. Solid black line, heterogeneity in infectivity (10% superspreaders with 20-fold higher infectivity than normal). Dashed line, heterogeneity in infectivity with assortative mixing (q=0.95) of the two spreader types. Dotted line, heterogeneity in infectivity with dissortative mixing (q=0.05).
If the evolved pathogen strain has R0 only slightly greater than 1, the probability of emergence is approximately equal to the ‘probability of evolution’ calculated earlier multiplied by the probability of emergence of the evolved strain, which we calculated in §2.
In summary, we see two regimes; evolution is considerably more important than host heterogeneity for determining the risk of emergence at low R0. However, heterogeneity becomes an important influence on emergence, when R0>1.
4. Consequences of targeted vaccination
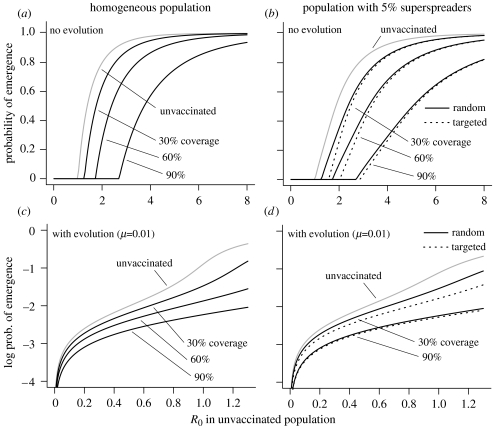
Is it always beneficial to target superspreaders for vaccination? The framework we use in this paper can be used to predict the levels of protection that different vaccine targeting strategies provide against disease emergence. Here, we compare two classes of treatment—a ‘leaky’ vaccine that reduces susceptibility by x% in all recipients and an ‘all-or-none’ vaccine that provides 100% protection to x% of recipients and no protection to the remainder. Our analysis shows that these two vaccines give an identical reduction in the probability of emergence in a homogeneous population (figure 5a).
Figure 5.
Vaccination strategies in (a, c) homogeneous and (b, d) heterogeneous populations, without (a, b) and with (c, d) pathogen adaptation. Grey lines, the probability of disease emergence as a function of R0 in the unvaccinated population. Solid black lines, the effect of random vaccination programmes with different levels of coverage. Dotted black lines in (b, d), the effect of targeted vaccination with the same overall coverage, i.e. vaccinating all the superspreaders and applying the remaining vaccine resources randomly to the rest of the population. The heterogeneous population in (b, d) comprises 5% superspreaders with 10-fold higher infectivity than the remaining 95%. In (c, d) the pathogen requires a single adaptation step to gain a high value of R0 (here 10 000). In this case, the probability of emergence becomes the probability of evolution (see main text). In (a-d) we simulated both a ‘leaky’ vaccine, which reduces susceptibility in all recipients by 70%, and an ‘all-or-none’ vaccine that confers complete immunity on 70% of recipients and none on the remainder. These vaccines give identical results.
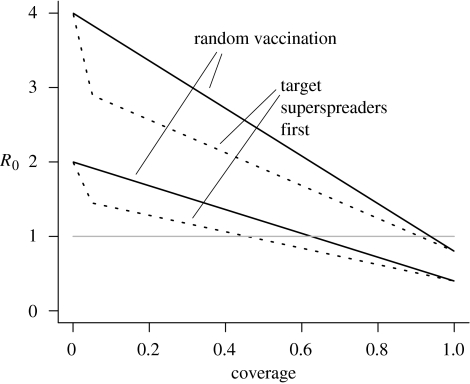
Given the constraint of an available level of coverage (the proportion of the population that can be vaccinated), we compare two strategies in a population with heterogeneity in infectiousness: random vaccination and targeted vaccination of previously identified or anticipated ‘superspreaders’, with the random assignment of vaccine resources to the remaining population (figure 5b,d). Figure 5 illustrates the effect of vaccination strategies on the probability of emergence, and the effect on R0 itself for different levels of coverage is shown in figure 6.
Figure 6.
Comparing random and targeted vaccination strategies. We take a population comprising 5% superspreaders (10-fold more infectious than normal). For two values of R0 of the untreated population (2 and 4), we calculate the resulting values of R0 after vaccination with different levels of coverage (horizontal axis). Solid black line, the results of vaccinating randomly chosen individuals; dotted line, the effect of targeted vaccination, in which superspreaders are identified and vaccinated before normal spreaders. Grey horizontal line, the threshold, R0=1, below which emergence is not possible without pathogen evolution.
Three simple observations can be made here. First, irrespective of heterogeneity, vaccine coverage or the targeting strategy, leaky and all-or-none vaccines provide identical levels of protection. Second, perhaps more obvious, targeted vaccination provides a bigger reduction in the probability of emergence than random treatment. Third, however, targeting is only of substantially increased benefit if vaccine resources are limited, and otherwise the effort of identifying and vaccinating potential superspreaders may be unnecessary.
5. Discussion
In this paper, we propose a simple framework for characterizing variation in host epidemiological parameters and describing disease transmission among these hosts with stochastic (multi-type branching process) models. We show that with some exceptions, variation in infectiousness, susceptibility to infection and contact patterns makes the extinction of chains of disease transmission more likely, i.e. epidemics are less likely to occur in epidemiologically diverse populations than in homogeneous ones (with the same basic reproductive number R0).
We also find that the degree of reduction in risk is sensitive to the nature of the host variability. If adaptation of the pathogen is required, then under some circumstances, the risk of emergence may even be increased by heterogeneity. However, this is a small effect. Our main result is that the rate of adaptation or evolution is considerably more important for determining the risk of emergence at low R0, and heterogeneity becomes a more important influence on emergence, if R0>1.
Branching processes are appropriate for describing the early stages of disease outbreak, when susceptible ones are not limited and stochastic effects are most important (Metz 1978; Ball 1983). This is precisely the time during which extinctions may occur. Discrete time branching process models have been used to estimate R0 from epidemic data (Becker 1977; Farrington et al. 2003). Multi-type branching processes have also been used to predict the outcome of vaccination strategies in populations with different levels of mixing (Ball & Becker 2006). Lloyd-Smith et al. (2005) used single-type branching processes to study the influence of heterogeneity on the emergence of non-evolving pathogens. They take a phenomenological approach, using inference with branching processes to assess the degree of heterogeneity in real epidemics. They assume that secondary cases from each infected individual are Poisson distributed, but with a mean drawn from a distribution (itself with mean R0) that represents host variation. Superspreading individuals are then defined statistically and belong to the tail of this distribution. We take a complementary approach, explicitly describing the heterogeneity on three levels (susceptibility, infectivity and mixing) and modelling transmission between homogeneous subpopulations as independent Poisson processes. In our framework, continuous variation of epidemiological parameters in a population can be modelled with arbitrarily large numbers of compartments. Thus, our approach is less empirical and also not suited to inference using epidemiological datasets; however, it is more systematic, in that it allows us to describe the contributions of different forms of heterogeneity to disease emergence, with and without pathogen evolution.
Our results have relevance to the ongoing concerns regarding emerging infectious diseases. First, they emphasize that the risk of emergence is strongly influenced by the adaptability of pathogens. This can be assessed quantitatively by serial passage of the pathogen in the new host (for example, avian flu in pigs). Second, for the design of vaccination or treatment strategies, our results highlight the importance of having good estimates of R0 and of heterogeneity through identification of the key spreader types. If one uses incomplete data to estimate R0, e.g. by failing to identify a class of superspreading individuals, R0 may be significantly underestimated. If R0 is known and superspreading groups identified, the most efficient use of limited treatment resources can be made.
A priori identification and location of risk groups may not always be feasible, of course. In the context of STDs, methods exist for identifying potential superspreaders by their virtue of being ‘connected’ to the contact network at all (Cohen et al. 2003), and of course for other diseases highly infectious or susceptible subpopulations might be predicted in advance (e.g. children and the elderly). In any event, the results here reinforce the notion that the primary purpose of intervention must be to decrease R0. The ‘homogenization’ that arises from vaccination or removing superspreaders, which may have been thought to increase the risk of disease emergence, is always outweighed by the associated reduction in R0.
Acknowledgments
All numerical computations were performed with R (R Development Core Team 2006). This work was supported by an NIH grant GM-U01GM70749 to Rustom Antia.
Endnotes
Typically this correlation is implicit in deterministic models in which the rate of new infections is represented a term of the form βIS, where β is a single parameter expressing a combination of transmissibility and susceptibility and I, S are the numbers of infectives and susceptibles, respectively.
In models in which individual infectious periods are exponentially distributed random variables and infectious contacts are made at the points of a Poisson process, then if R0>1 the probability of emergence increases with R0 as 1−1/R0 (May et al. 2001). In contrast, the model we describe here assumes non-random infectious periods, and no closed-form expression exists for the probability of emergence.
Supplementary Material
The degeneracies in their parametrisation of host heterogeneity
References
- Antia R, Regoes R.R, Koella J.C, Bergstrom C.T. The role of evolution in the emergence of infectious diseases. Nature. 2003;426:658–661. doi: 10.1038/nature02104. doi:10.1038/nature02104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball F. The threshold behaviour of epidemic models. J. Appl. Probab. 1983;20:227–241. doi:10.2307/3213797 [Google Scholar]
- Ball F, Becker N.G. Control of transmission with two types of infection. Math. Biosci. 2006;200:170–187. doi: 10.1016/j.mbs.2005.12.024. doi:10.1016/j.mbs.2005.12.024 [DOI] [PubMed] [Google Scholar]
- Becker N. Estimation for discrete time branching processes with application to epidemics. Biometrics. 1977;3:515–522. doi:10.2307/2529366 [PubMed] [Google Scholar]
- Becker N.G, Hall R. Immunization levels for preventing epidemics in a community of households made up of individuals of various types. Math. Biosci. 1996;132:205–216. doi: 10.1016/0025-5564(95)00080-1. doi:10.1016/0025-5564(95)00080-1 [DOI] [PubMed] [Google Scholar]
- Cohen R, Havlin S, Ben-Avraham D. Efficient immunization strategies for computer networks and populations. Phys. Rev. Lett. 2003;91:2 479 011–2 479 014. doi: 10.1103/PhysRevLett.91.247901. [DOI] [PubMed] [Google Scholar]
- Diekmann O, Heesterbeek J.A, Metz J.A. On the definition and the computation of the basic reproduction ratio R0 in models for infectious diseases in heterogeneous populations. J. Math. Biol. 1990;28:365–382. doi: 10.1007/BF00178324. doi:10.1007/BF00178324 [DOI] [PubMed] [Google Scholar]
- Dobson A, Foufopoulos J. Emerging infectious pathogens of wildlife. Phil. Trans. R. Soc. B. 2001;356:1001–1112. doi: 10.1098/rstb.2001.0900. doi:10.1098/rstb.2000.0758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrington C.P, Kanaan M.N, Gay N.J. Branching process models for surveillance of infectious diseases controlled by mass vaccination. Biostatistics. 2003;4:279–295. doi: 10.1093/biostatistics/4.2.279. doi:10.1093/biostatistics/4.2.279 [DOI] [PubMed] [Google Scholar]
- Galvani A.P, May R.M. Epidemiology: dimensions of superspreading. Nature. 2005;438:293–305. doi: 10.1038/438293a. doi:10.1038/438293a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomulkiewicz R, Holt R.D. When does evolution by natural selection revent extinction? Evolution. 1995;49:201–207. doi: 10.1111/j.1558-5646.1995.tb05971.x. doi:10.2307/2410305 [DOI] [PubMed] [Google Scholar]
- Grenfell B.T, Wilson K, Isham V.S, Boyd H.E, Dietz K. Modelling patterns of parasite aggregation in natural populations: trichostrongylid nematode-ruminant interactions as a case study. Parasitology. 1995;111(Suppl.):S135–S151. doi: 10.1017/s0031182000075867. [DOI] [PubMed] [Google Scholar]
- Gupta S, Anderson R.M, May R.M. Networks of sexual contacts: implications for the pattern of spread of HIV. AIDS. 1989;3:807–817. [PubMed] [Google Scholar]
- Harris T.E. Springer; Berlin, Germany: 1963. The theory of branching processes. [Google Scholar]
- Hasibeder G, Dye C. Population dynamics of mosquito-borne disease: persistence in a completely heterogeneous environment. Theor. Popul. Biol. 1988;33:31–53. doi: 10.1016/0040-5809(88)90003-2. doi:10.1016/0040-5809(88)90003-2 [DOI] [PubMed] [Google Scholar]
- Hethcote H.W, Yorke J.A. Lecture notes in biomathematics. Vol. 56. Springer Verlag; Berlin, Germany: 1984. Gonorrhea transmission dynamics and control. [Google Scholar]
- Hethcote H.W, Van Ark W. Epidemiological models for heterogeneous populations: proportionate mixing, parameter estimation, and immunization programs. Math. Biosci. 1987;84:85–118. doi:10.1016/0025-5564(87)90044-7 [Google Scholar]
- Hyman J.M, Li J. An intuitive formulation for the reproductive number for the spread of diseases in heterogeneous populations. Math. Biosci. 2000;167:65–86. doi: 10.1016/s0025-5564(00)00025-0. doi:10.1016/S0025-5564(00)00025-0 [DOI] [PubMed] [Google Scholar]
- Lloyd-Smith J.O, Schreiber S.J, Kopp P.E, Getz W.M. Superspreading and the effect of individual variation on disease emergence. Nature. 2005;438:355–359. doi: 10.1038/nature04153. doi:10.1038/nature04153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner I.C. The effect of preferential mixing on the growth of an epidemic. Math. Biosci. 1992;109:39–67. doi: 10.1016/0025-5564(92)90051-w. doi:10.1016/0025-5564(92)90051-W [DOI] [PubMed] [Google Scholar]
- May R.M, Anderson R.M. Transmission dynamics of HIV infection. Nature. 1987;326:137–142. doi: 10.1038/326137a0. doi:10.1038/326137a0 [DOI] [PubMed] [Google Scholar]
- May R.M, Anderson R.M. The transmission dynamics of human immunodeficiency virus (HIV) Phil. Trans. R. Soc. B. 1988;321:565–607. doi: 10.1098/rstb.1988.0108. [DOI] [PubMed] [Google Scholar]
- May R.M, Gupta S, McLean A.R. Infectious disease dynamics: what characterizes a successful invader? Phil. Trans. R. Soc. B. 2001;356:901–910. doi: 10.1098/rstb.2001.0866. doi:10.1098/rstb.2001.0866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz J.A. The epidemic in a closed population with all susceptibles equally vulnerable; some results for large susceptible populations and small initial infections. Acta Biotheor. 1978;27:75–123. doi: 10.1007/BF00048405. doi:10.1007/BF00048405 [DOI] [PubMed] [Google Scholar]
- Murphy B.M, Singer B.H, Kirschner D. On treatment of tuberculosis in heterogeneous populations. J. Theor. Biol. 2003;223:391–404. doi: 10.1016/s0022-5193(03)00038-9. doi:10.1016/S0022-5193(03)00038-9 [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: a language and environment for statistical computing R Foundation for Statistical Computing, Vienna, Austria, 2006. URL http://www.R-project.org ISBN 3-900051-07-0.
- Shen Z, Ning F, Zhou W, He X, Lin C, Chin D.P, Zhu Z, Schuchat A. Superspreading SARS events, Beijing, 2003. Emerg. Infect. Dis. 2004;10:256–260. doi: 10.3201/eid1002.030732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor L.H, Latham S.M, Woolhouse M.E. Risk factors for human disease emergence. Phil. Trans. R. Soc. B. 2001;356:983–999. doi: 10.1098/rstb.2001.0888. doi:10.1098/rstb.2001.0975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhouse M.E, et al. Heterogeneities in the transmission of infectious agents: implications for the design of control programs. Proc. Natl Acad. Sci. USA. 1997;94:338–342. doi: 10.1073/pnas.94.1.338. doi:10.1073/pnas.94.1.338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Clancy D, French N.P, Bowers R.G. A semi-stochastic model for Salmonella infection in a multi-group herd. Math. Biosci. 2006;200:214–233. doi: 10.1016/j.mbs.2006.01.006. doi:10.1016/j.mbs.2006.01.006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The degeneracies in their parametrisation of host heterogeneity