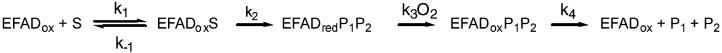Abstract
The flavoenzyme nitroalkane oxidase catalyzes the oxidation of primary and secondary nitroalkanes to the respective aldehydes or ketones, releasing nitrite. The enzyme has recently been identified as being homologous to the acyl-CoA dehydrogenase family of enzymes [Daubner, S. C., Gadda, G., Valley, M. P., and Fitzpatrick, P. F. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 2702-2707]. The glutamate which acts as an active site base in that family of enzymes aligns with Asp402 of nitroalkane oxidase. To evaluate the identification of Asp402 as an active site base, the effect of mutation of Asp402 to glutamate on the rate of cleavage of the nitroalkane C–H bond has been determined. Deuterium kinetic isotope effects on steady state kinetic parameters and direct measurement of the rate of flavin reduction establish that the mutation increases the ΔG‡ for C–H bond cleavage by 1.6–1.9 kcal/mol. There is no effect on the rate of reaction of the reduced enzyme with oxygen. These results support the assignment of Asp402 as the active site base in nitroalkane oxidase.
The flavoenzyme nitroalkane oxidase (NAO)1 catalyzes the oxidation of nitroalkanes to the respective aldehydes or ketones with consumption of oxygen and release of nitrite and hydrogen peroxide (Scheme 1) (1). While several other flavoprotein oxidases are able to oxidize nitroalkanes, they require the preformed anion as a substrate in a clearly nonphysiological reaction (2). NAO is unique in that it uses the neutral forms of nitroalkanes as substrates (3); moreover, the enzyme is induced by growth of Fusarium oxysporum on nitroethane, consistent with nitroalkane oxidation being the physiological role of the enzyme (1). NAO can oxidize a broad range of primary and secondary nitroalkanes, although primary aliphatic nitroalkanes are the best substrates (4).
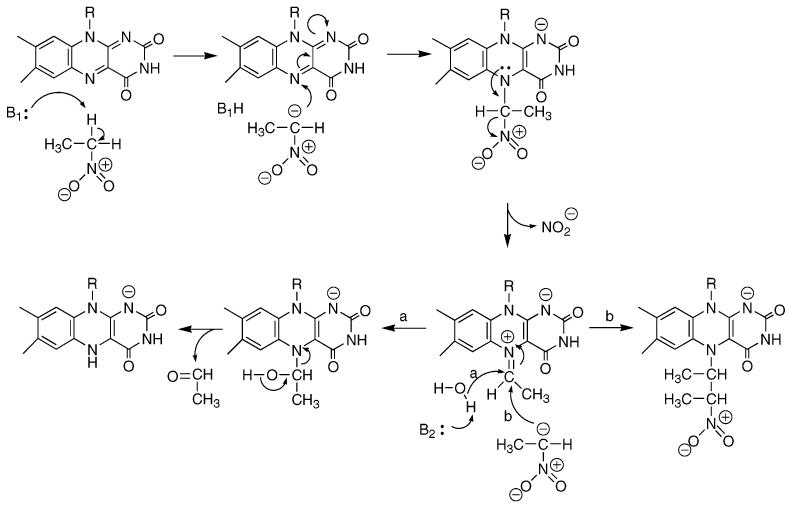
As is the case with many flavoproteins, the reaction of NAO can be divided into reductive and oxidative half-reactions (5). In the reductive half-reaction, the nitroalkane substrate reacts with the oxidized flavin to generate the oxidized product and reduced flavin. The mechanism of this half-reaction (Scheme 2) has been proposed to involve removal of the substrate α-hydrogen as a proton, followed by attack of the resulting carbanion on the flavin (6). Loss of nitrite from the resulting adduct would generate an electrophilic species to which hydroxide adds (path a); this flavin adduct then decomposes to form reduced flavin and aldehyde or ketone. Evidence for this mechanism comes from the demonstration that an inactive 5-nitrobutyl–FAD adduct can be formed when turnover is carried out in the presence of nitroethane anion, which intercepts the initial electrophilic intermediate (path b) (6). In the oxidative half-reaction, the reduced flavin reacts with molecular oxygen to regenerate the oxidized flavin, forming hydrogen peroxide.
Scheme 1.
Formation of a substrate carbanion in the active site of NAO requires a base to abstract the α-proton. The gene for nitroalkane oxidase from F. oxysporum has recently been cloned (7). The amino acid sequence of the cloned enzyme revealed that NAO is homologous to members of the acyl-CoA dehydrogenase family of flavoenzymes. Although these enzymes catalyze hydride transfer rather than formation of a carbanion as a discrete intermediate, they still require a catalytic base to remove the α-proton of the substrate as the β-hydrogen is transferred to the flavin as a hydride (8). The base in the medium and short chain acyl-CoA dehydrogenases is a conserved glutamate which aligns with Asp402 in NAO. This aspartate is in the proximity of two residues (Tyr398 and Cys397) that have been independently identified as being located near the active site of NAO by chemical modification (9, 10). In the report presented here, we describe the reductive half-reaction of NAO and evaluate the effect of replacement of Asp402 with glutamate, the residue that acts as the catalytic base in the acyl-CoA dehydrogenases.
EXPERIMENTAL PROCEDURES
Materials
All materials were purchased from Sigma-Aldrich unless otherwise specified. 1-[1,1-2H2]Nitrobutane was synthesized as previously described (11). Recombinant nitroalkane oxidase was expressed and purified as previously described (7). The D402E mutation was generated with the QuikChange site-directed mutagenesis kit (Stratagene), and the mutant enzyme was expressed and purified following the wild-type enzyme protocol. DNA sequencing was performed at the Gene Technology Laboratory of Texas A&M University. Enzyme concentrations were determined using an ε446 value of 14.2 mM−1 cm−1, determined as previously described (12).
Scheme 2.
Methods
Enzyme activity was typically measured in air-saturated 50 mM Hepes and 0.1 mM FAD at pH 8.0 and 30 °C by monitoring oxygen consumption with a computer-interfaced Hansatech Clark oxygen electrode (Hansatech Instruments, Pentney King's Lynn, U.K.). The steady state kinetic parameters were determined at ambient oxygen concentrations, since the Km value for oxygen ranges between 20 and 42 μM with various substrates (3). To prevent the formation of the anionic form of the substrate, stock solutions of nitroalkanes were prepared in DMSO and assays were initiated by the addition of substrate. Oxidation of acyl-CoA substrates was assayed by either oxygen consumption or reduction of ferrocenium hexafluorophosphate, as described previously (7). Rapid reaction kinetic measurements were performed with an Applied Photophysics SX-18MV stopped-flow spectrophotometer. For anaerobic experiments, the instrument was incubated with 20 mM dithionite and flushed with anaerobic buffer immediately prior to use. Solutions were initially made anaerobic by placing them under vacuum and then flushing them with oxygen-scrubbed argon; this was repeated twice. Afterward, glucose and glucose oxidase were added at final concentrations of 5 mM and 36 nM, respectively, to maintain anaerobic conditions. To minimize formation of the nitroalkane anion, small volumes (e.g., 0.1 mL) of concentrated substrate were added to solutions (e.g., 3 mL) that had already been made anaerobic, and the final substrate solution was used within 15 min.
Data Analysis
Data were fit using the programs Kaleidagraph (Adelbeck Software, Reading, PA) and Igor Pro (WaveMetrics, Inc., Lake Oswego, OR). Steady state kinetic parameters were determined by fitting the data to the Michaelis–Menten equation or eq 1, which includes the effect of substrate inhibition.
| (1) |
Here v is the initial velocity, Vmax is the maximal velocity, Km is the Michaelis constant, S is the substrate concentration, and Kai is the substrate inhibition constant. Transient changes in absorbance were fit to eq 2
| (2) |
where A is the measured absorbance, A∞ is the final absorbance, t is time, and An and λn are the amplitude and rate, respectively, of the nth exponential phase. The concentration dependence of the rate of reduction by nitroethane was fit to eq 3
| (3) |
where kobs is the observed rate, k2 is the limiting rate of reduction, and KD is the dissociation constant of the enzyme–substrate complex. Steady state kinetic isotope effects were calculated from eq 4
| (4) |
where Fi is the fraction of deuterium in the substrate and EV and EVK are the isotope effects minus one for Vmax and V/K, respectively. The same equation, without the inhibition term, was used to calculate the transient kinetic isotope effects.
RESULTS
Steady State Kinetic Measurements
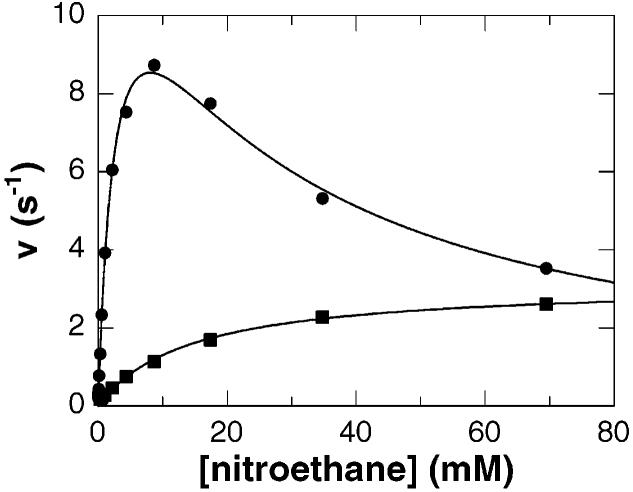
The steady state kinetic parameters for NAO with a number of substrates have been reported previously for the enzyme purified from F. oxysporum (4). The flavin cofactor of the enzyme isolated from its native source is in the form of an inactive nitrobutyl adduct and consequently requires reactivation, which may be deleterious to the enzyme. The recent availability of the recombinant enzyme provides a source of NAO that is expressed and purified in the absence of nitroalkanes, generating an enzyme which does not require activation. In addition, nitroalkanes are not soluble at high concentrations in aqueous solution. This has necessitated the use of an organic solvent for preparation of stock solutions of the nitroalkane substrates. Ethanol was previously used as the organic solvent for substrate manipulation, but we have recently observed that ethanol inhibits NAO. A survey of water miscible solvents showed that 3% ethanol causes a 45% decrease in activity, whereas 3% DMSO causes only a 6% decrease in activity (results not shown). Consequently, DMSO was chosen as the cosolvent for stock solutions of substrate, and the concentration of DMSO in assays was maintained at ≤1%. The steady state kinetic parameters for the recombinant enzyme with both nitroethane and nitrobutane as substrates under these conditions are given in Tables 1 and 2, and representative data with nitroethane are shown in Figure 1. They agree qualitatively with the results with the nonrecombinant enzyme, but the Vmax and V/K values for the recombinant enzyme average 50% more (11), consistent with the greater activity of this enzyme due to either the source or the changes in the assay.
Table 1.
Steady State Kinetic Parameters for Nitroalkane Oxidasea
| wild type | D402E | |
|---|---|---|
| nitroethane | ||
| Vmax (s−1) | 15 ± 1 | 3.07 ± 0.06 |
| V/K (mM−1 s−1) | 6.3 ± 0.4 | 0.21 ± 0.01 |
| Km (mM) | 2.3 ± 0.2 | 14.6 ± 0.8 |
| Kai (mM) | 25 ± 3 | —b |
| 1-nitrobutane | ||
| Vmax (s−1) | 6.1 ± 0.4 | 8.4 ± 0.2 |
| V/K (mM−1 s−1) | 220 ± 70 | 47 ± 5.0 |
| Km (mM) | 0.03 ± 0.01 | 0.18 ± 0.02 |
| Kai (mM) | 12 ± 2 | 120 ± 20 |
Initial rates of oxygen consumption were measured as described in Experimental Procedures. The conditions were 50 mM Hepes, 0.1 mM FAD, pH 8.0, and 30 °C.
Substrate inhibition was not observed with the D402E enzyme.
Table 2.
Steady State Kinetic Isotope Effects for Nitroalkane Oxidasea
| wild type |
D402E |
|||
|---|---|---|---|---|
| DVmax | D(V/K) | DVmax | D(V/K) | |
| nitroethane | 1.4 ± 0.2 | 9.2 ± 1.1 | 3.6 ± 0.7 | 9.6 ± 2.3 |
| 1-nitrobutane | 1.04 ± 0.09 | 1.7 ± 0.6 | 1.17 ± 0.05 | 6.8 ± 1.0 |
Conditions as described in footnote a of Table 1.
Figure 1.
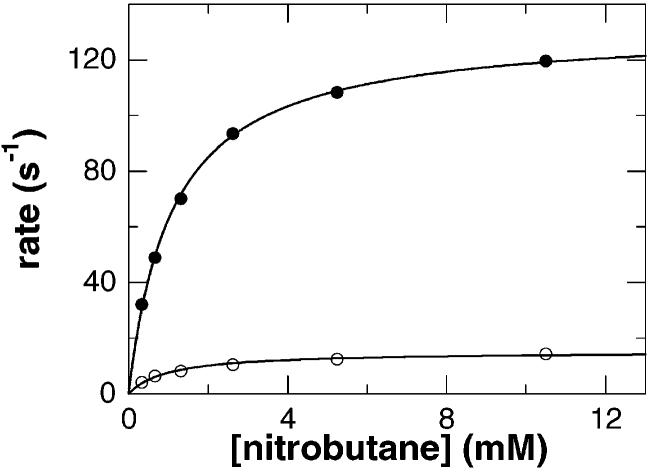
Steady state kinetics of nitroalkane oxidase with nitroethane as the substrate. The initial velocity of the wild-type enzyme (●) or D402E enzyme (■) at pH 8.0 and 30 °C is plotted vs nitroethane concentration. The lines are fits of the data to either the Michaelis–Menten equation or eq 1.
Reductive Half-Reaction with Nitroethane
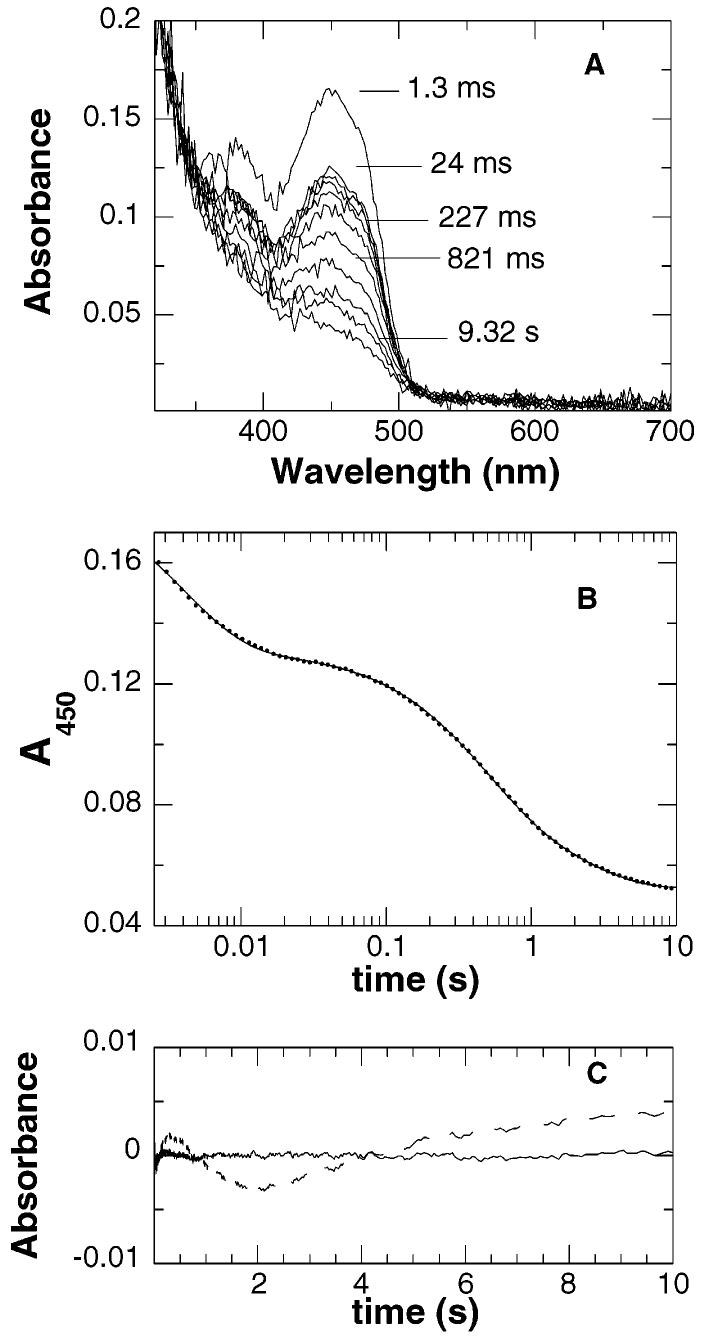
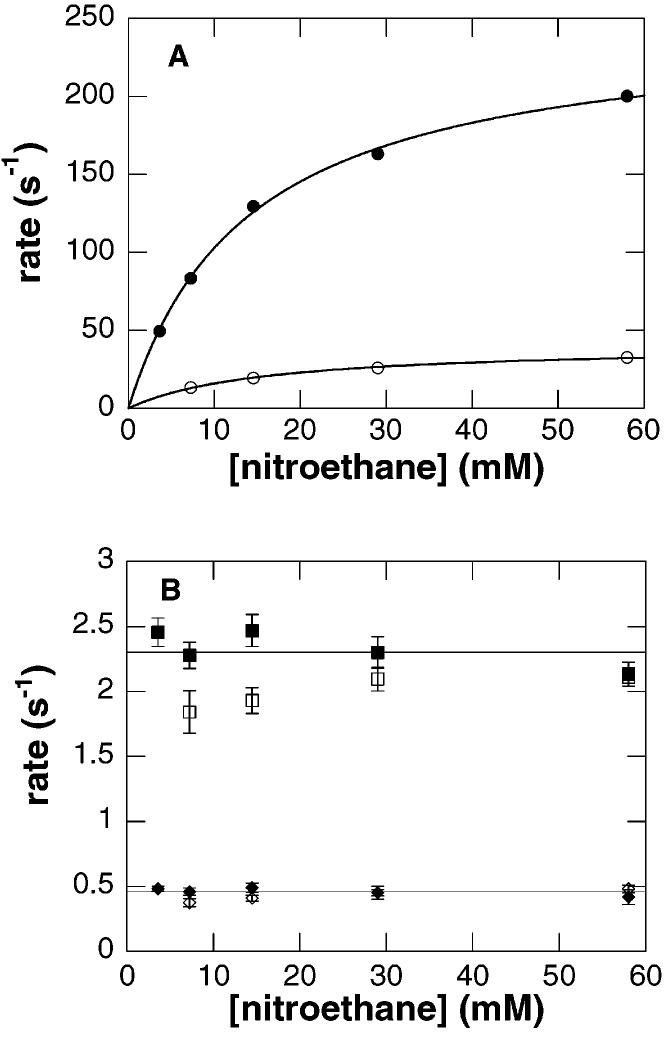
The reductive half-reaction of wild-type nitroalkane oxidase was studied by rapidly mixing the enzyme with nitroethane in the absence of oxygen in a stopped-flow spectrophotometer. The changes in the visible absorbance spectrum of the enzyme as followed by diode array detection are shown in Figure 2A. The largest change in absorbance occurs at 450 nm; consequently, more extensive analyses were carried out by monitoring this wavelength. At 450 nm, the reaction occurs in three phases, with no change in absorbance observed within the dead time of the instrument (Figure 2B,C). The effect of substrate concentration on the rates of the three phases is consistent with the kinetic mechanism of Scheme 3, where P1 and P2 are the products acetaldehyde and nitrite, respectively. The rate of the first phase exhibits a hyperbolic dependence on the concentration of nitroethane with a value for the y-intercept of zero (Figure 3A). This is consistent with substrate binding in a rapid equilibrium to the enzyme with no perturbation of the flavin chromophore, followed by irreversible C–H bond cleavage and reduction of the flavin. Fitting the observed rate of the first phase to eq 3 yields k2, the limiting rate constant for reduction of the flavin, and KD, the dissociation constant for nitroethane (Table 3). With [1,1-2H2]nitroethane as the reducing substrate, the k2 and k2/KD values exhibit comparable large isotope effects (Table 4). This identifies the first phase in the reductive half-reaction as the step in which the substrate C–H bond is cleaved.
Figure 2.
Reduction of wild-type nitroalkane oxidase by nitroethane. NAO (final concentration of 14 μM) was rapidly mixed with nitroethane (final concentration of 58 mM) under anaerobic conditions. The reaction was monitored first at multiple wavelengths (A) and then specifically at 450 nm (B). The line in panel B is a fit of the data to eq 2, with every 10th data point shown. Panel C shows the residuals from a fit of the data to eq 2 using three (—) or two (– – –) phases. Conditions were 100 mM Hepes, pH 8.0, and 30 °C.
Figure 3.
Concentration dependence of reduction of wild-type nitroalkane oxidase by nitroethane. Conditions as described in the legend of Figure 2. The rates of the fast phase (A) and the two slower phases (B) are plotted vs the concentration of nitroethane (filled symbols) or [1,1-2H2]nitroethane (empty symbols). The lines in panel A are individual fits of the data for each substrate to eq 3; the error in each data point in panel A is smaller than the symbols.
Table 3.
Rapid Reaction Kinetic Parameters for Nitroalkane Oxidasea
| wild type | D402E | |
|---|---|---|
| nitroethaneb | ||
| k2 (s−1) | 247 ± 5 (66 ± 3)c | 16.3 ± 0.2 |
| KD (mM) | 14.0 ± 0.7 | 22.8 ± 0.7 |
| k2/KD (mM−1 s−1) | 17.6 ± 0.8 | 0.72 ± 0.02 |
| 1-nitrobutanec | ||
| k2 (s−1) | >4000 | 132 ± 1 |
| KD (mM) | ndd | 1.10 ± 0.03 |
| k2/KD (mM−1 s−1) | ndd | 120 ± 3 |
Determined from the rate of change in the flavin absorbance at 450 nm as described in Experimental Procedures. Conditions were 50 mM Hepes and pH 8.0.
At 30 °C.
At 5 °C.
Not determined.
Table 4.
Transient Kinetic Isotope Effects for Nitroalkane Oxidasea
Scheme 3.
The rates of the two subsequent phases (k13 and k14) are independent of both nitroethane concentration and isotopic substitution, with average values of 2.3 ± 0.2 and 0.46 ± 0.09 s−1, respectively (Figure 3B). Since both of these rates are slower than the Vmax value for nitroethane (15 s−1), neither is likely to be relevant to catalysis. A reasonable explanation for these absorbance changes is that they reflect the stepwise release of the products acetaldehyde and nitrite from the reduced enzyme. In such a case, the apparent rates are the sums of the forward and reverse rate constants (k13 + k−13 for P1 and k14 + k−14 for P2) for each product release step. In the absence of product, the observed rate of each phase is simply equal to the forward rate constant. However, in the presence of a specific product, the apparent rate of the phase associated with release of that product should increase. To test this experimentally, the reduction of wild-type NAO by nitroethane was repeated in the presence of nitrite. Qualitatively, the spectral changes at 450 nm observed in the presence of nitrite were similar to those observed in the absence of nitrite (data not shown). As the concentration of nitrite increased, there was no effect on the rate of the second phase, but the rate of the third phase increased. The magnitude of the absorbance change at 450 nm also decreased with increasing concentrations of nitrite until this phase was no longer seen at high concentrations (results not shown). This supports the assignment of the last phase in the reductive half-reaction to the release of nitrite. The wild-type enzyme was also reduced by nitroethane in the presence of either acetaldehyde or butyraldehyde, but no effect on the rates or magnitudes of either of the two slow phases was observed. Consequently, the second phase can only be tentatively assigned as aldehyde release.2 Since both k13 and k14 are less than Vmax, nitrite and aldehyde do not appear to be released from the active site until after the flavin is reoxidized. Retention of products formed during the reductive half-reaction until after flavin reoxidation is a phenomenon that has been described previously with a number of flavoprotein oxidases (13-15).
Reductive Half-Reaction with 1-Nitrobutane
When wild-type NAO is anaerobically mixed with 1-nitrobutane at 30 °C, the spectral changes are qualitatively similar to those seen with nitroethane. However, the observed spectral changes occur in only two phases (Figure 4), whose rates are both independent of 1-nitrobutane concentration and isotopic substitution. The values of these rates (2.1 ± 0.32 and 0.42 ± 0.032 s−1 at 30 °C) are comparable to those of the two slower phases observed upon reduction by nitroethane and are thus likely to reflect product release from the reduced enzyme. In contrast to the reaction with nitroethane, there is a large change in absorbance within the dead time of the stopped-flow instrument in the reaction with 1-nitrobutane. The magnitude of this absorbance change is approximately one-third of the total absorbance change, roughly equivalent to the fractional change in absorbance of the first phase with nitroethane. In an attempt to measure the rate of the initial phase with nitrobutane, this experiment was repeated at 5 °C with 1-[1,1-2H2]nitrobutane. After mixing had been carried out, a small decrease in absorbance was observed in the first 5 ms. Thus, the reaction was still too rapid for accurate measurement even under these conditions. However, if one assumes that the total absorbance change occurs within five half-lives (∼97% complete) and account for the 2 ms dead time and 6-fold decrease in rate with the deuterated substrate, it is possible to estimate a lower limit of 4000 s−1 for the value of k2 with 1-nitrobutane at 5 °C. The magnitude of this rate constant is consistent with the V/K value for 1-nitrobutane being significantly larger than that for nitroethane (Table 1).
Figure 4.
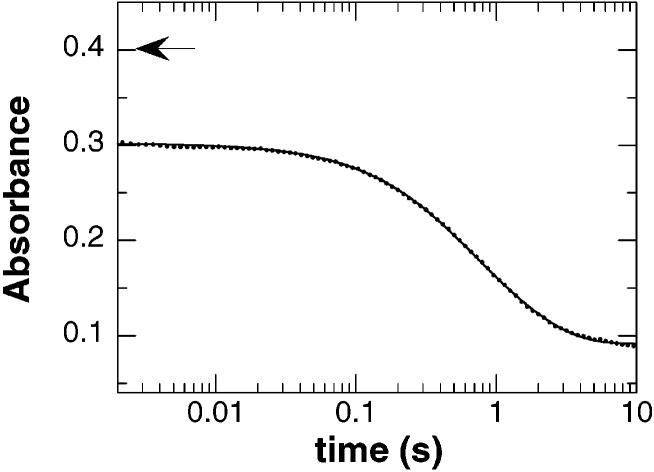
Reduction of wild-type nitroalkane oxidase by 1-nitrobutane. NAO (final concentration of 30 μM) was rapidly mixed with 1-nitrobutane (final concentration of 10 mM) under anaerobic conditions, and the reaction was monitored at 450 nm. The line is a fit of the data to eq 2 containing only two phases, with every 10th data point shown. The initial absorbance of the oxidized enzyme is indicated by the arrow. Conditions as described in the legend of Figure 2.
Steady State Kinetics of the D402E Enzyme
To probe the role of Asp402 in catalysis, this residue was replaced with glutamate, the residue found in the acyl-CoA dehydrogenases, and the steady state parameters of the mutant enzyme were determined (Table 1). With nitroethane as the substrate, both the Vmax and V/K values are significantly lower. With 1-nitrobutane as the substrate, the Vmax value is roughly the same for the mutant enzyme, but the V/K value is decreased severalfold. The level of substrate inhibition seen with the wild-type enzyme is significantly diminished in the D402E enzyme (Table 1). With 1-nitrobutane, the Kai value increases to ∼3 times the wild-type value, while substrate inhibition is no longer observed with nitroethane (Figure 1). Since the mutant enzyme has the same catalytic residue as the acyl-CoA dehydrogenases, it was assayed to determine if it could oxidize acyl-CoA substrates. At concentrations as high as 532 μM butyryl-CoA or 295 μM octanoyl-CoA, no activity was detected with either oxygen or ferrocenium hexafluorophosphate as the electron acceptor. Thus, the lack of acyl-CoA dehydrogenase activity in NAO is not simply due to a change in the active site base.
The steady state deuterium kinetic isotope effects for the D402E enzyme are given in Table 2. With nitroethane as the substrate, the D(V/K) value is identical to that of the wild-type enzyme but the DVmax value increases ∼3-fold. With 1-nitrobutane as the substrate, the DVmax value is still low, but the D(V/K) value is considerably larger than the wild-type value. The kinetic isotope effects establish that C–H bond cleavage is more rate-limiting in the mutant enzyme, consistent with mutagenesis of an active site base.
Reductive Half-Reaction of the D402E Enzyme
Rapid mixing of the D402E enzyme with nitroethane in the absence of oxygen produced spectral changes similar to those observed with the wild-type enzyme, although the absorbance change occurred in only two phases. At 450 nm, the rate of the first phase was dependent on the concentration of nitroethane, yielding the kinetic parameters listed in Table 3. The rate of this phase decreased with [1,1-2H2]nitroethane as the substrate, yielding the Dk2 and D(k2/KD) values listed in Table 4. As with the wild-type enzyme, these values confirm that the first phase is due to C–H bond cleavage. The rate of the second phase (0.13 ± 0.05 s−1) is independent of the concentration of nitroethane, much slower than turnover, and again likely to be due to product release from the reduced enzyme.
The D402E enzyme was also rapidly mixed at 5 °C with 1-nitrobutane in the absence of oxygen. In this case, more than 90% of the absorbance change occurred in a single phase. The change in absorbance was best fit as two phases, but the rate of the second phase was very slow and could not be accurately determined because of its small magnitude. Reduction of the D402E enzyme was not complete within the dead time of the instrument, in contrast to the reaction of the wild-type enzyme. Instead, the rate of the fast phase was clearly dependent on the concentration and isotopic substitution of the substrate (Figure 5 and Table 3). As with nitroethane, the isotope effects on the parameters with 1-nitrobutane are large with the mutant enzyme (Table 4), indicating that this phase is indeed due to C–H bond cleavage
Figure 5.
Concentration dependence of reduction of D402E nitroalkane oxidase by 1-nitrobutane. The mutant enzyme (final concentration of 18 μM) was rapidly mixed with the indicated concentrations of 1-nitrobutane (●) or 1-[1,1-2H2]nitrobutane (○) under anaerobic conditions, and the reaction was monitored at 450 nm. Conditions were 100 mM Hepes, pH 8.0, and 5 °C. The error in each data point is smaller than the symbol.
Oxidative Half-Reaction
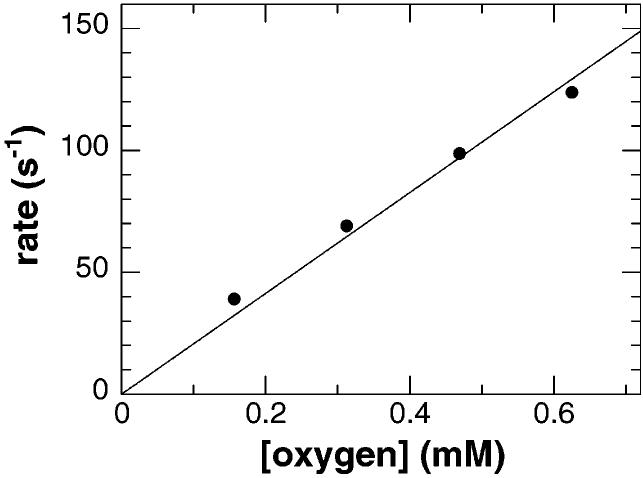
To study the reaction of reduced NAO with oxygen, the wild-type enzyme was reduced anaerobically with a slight excess of nitroethane. The reduced enzyme was then mixed with different concentrations of oxygen. The appearance of the spectrum of the oxidized enzyme occurred very rapidly and in a single phase. The rate of this phase was linearly dependent on the concentration of oxygen with a second-order rate constant of 207 ± 7 mM−1 s−1 (Figure 6). This value is comparable to the rate of oxidation observed for other flavoenzyme oxidases (16, 17). When this experiment was repeated with the D402E enzyme, the results were similar, with a second-order rate constant of 158 ± 7 mM−1 s−1. Thus, the mutation does not alter significantly the rate of reaction of the reduced enzyme with oxygen.
Figure 6.
Reaction of reduced wild-type nitroalkane oxidase with oxygen. The reduced wild-type enzyme (at a final concentration of 13 μM) was rapidly mixed with varying concentrations of oxygen, and the reaction was monitored at 450 nm. The rate of the absorbance change at each oxygen concentration was obtained by fitting the data to eq 2 with only one phase. The solid line is a simple linear fit of the data. The error in each data point is smaller than the symbol. Conditions as described in the legend of Figure 2.
Scheme 4.
DISCUSSION
The initial chemical step in the reductive half-reaction of NAO is formation of the substrate carbanion by abstraction of the nitroalkane α-hydrogen as a proton (Scheme 2). Although no three-dimensional structure of NAO is yet available, the recent identification of the enzyme as being homologous to the acyl-CoA dehydrogenases has allowed identification of Asp402 as the probable active site base in NAO (7). The studies described here of the effects of mutating Asp402 to glutamate, the residue found in the acyl-CoA dehydrogenases, support the identification of this residue as the active site base and strengthen the case for homology between NAO and the acyl-CoA dehydrogenases.
Nitroethane is the most extensively characterized substrate for NAO (5, 18), while nitrobutane is the best substrate described to date based on relative V/K values (4). The kinetic parameters of these two substrates with both the wild-type and D402E enzyme can be rationalized by reference to the minimal kinetic mechanism of Scheme 4. Here, the step with the rate constant k2 includes the chemical events in the oxidation of enzyme-bound nitroalkane to enzyme-bound aldehyde and nitrite, and the step with rate constant k4 includes the product release steps and any conformational changes required to convert the oxidized enzyme product complex to the active free oxidized enzyme.
With nitroethane as the substrate, the values for the isotope effects on V/K and k2 for both the wild-type and mutant enzymes are not significantly different, with values of 7–9. This agreement and the magnitude of the isotope effects suggest that proton abstraction is fully rate-limiting for reduction and effectively irreversible. The irreversibility of the chemistry is confirmed by the zero intercept in Figures 3A and 5 (19). The subsequent steps in the reductive half-reaction which lead to the reduced enzyme and products (Scheme 2) must therefore be much more rapid than C–H bond cleavage. The small DVmax value for the wild-type enzyme with nitroethane as the substrate is consistent with k4 being much smaller than k2 in the mechanism of Scheme 4.3 If 9 is taken to be the intrinsic isotope effect on k2, k2/k4 must be approximately 19 to suppress the DVmax value to the observed value. The rapid reaction data for the reductive half-reaction with this substrate provide a value for k2 directly of 247 s−1. A value of 17 s−1 for k4 will yield a Vmax value of 16 s−1 when combined with this k2 value. The resulting value for k2/k4 of 15 agrees well with the value predicted from the isotope effect on Vmax, providing support for the kinetic mechanism of Scheme 4. The data for the mutant enzyme can also be accommodated by the same kinetic mechanism. There is a decrease in the Vmax value and a corresponding increase in the DVmax value upon mutation of Asp402 to glutamate. C–H bond cleavage is still fully rate-limiting in the reductive half-reaction, given the isotope effects on the V/Knitroethane and k2 values. If 9 is again taken to be the value of the intrinsic isotope effect on k2, the increase in the DVmax value to 3.6 yields a value for k2/k4 of 1.5 ± 0.4. Simply combining the measured Vmax and k2 values yields a calculated value for k4 of 3.9 ± 0.1 s−1, for a k2/k4 of 4.2 ± 0.2, in reasonable agreement.
The primary effect of the D402E mutation is thus on the value of k2 with nitroethane as the substrate. Direct measurement of the limiting rate of reduction shows that the mutation decreases k2 by a factor of 15. This is equivalent to an increase in the ΔG‡ for C–H bond cleavage of 1.6 kcal/mol in the mutant enzyme. Comparison of the V/Knitroethane values for the wild-type and mutant enzyme yields a decrease for the mutant of 30-fold. Since the V/Knitroethane value is equal to k1k2/k−1 for this substrate, the somewhat larger effect on the V/K value is consistent with an increase in the KD value for nitroethane of slightly less than 2, the amount obtained by comparison of the KD values of the wild-type and mutant enzymes in Table 3. There is also a decrease in the value of k4 of ∼4. This is not very different from the change in the KD value, suggesting that the mutation slightly restricts the entry of the substrate and exit of products from the active site.
The V/K value with nitrobutane is much larger than that for nitroethane, while the Vmax value and the kinetic isotope effects are smaller. This is consistent with the chemical step (k2) being much faster with this substrate, but substrate and product dissociation rate constants (k−1 and k4) being comparable or slower. The intrinsic isotope effect on k2 with nitrobutane is probably close to that for nitroethane, based on the isotope effect seen in the reductive half-reaction of the D402E enzyme. This allows an estimation of the value of the forward commitment (k2/k−1) with this substrate of ∼9. The very small DVmax value with this substrate establishes that the rate constant for C–H bond cleavage (k2) is much greater than for the subsequent nonchemical steps (k4). Direct comparison of the lower limit for k2 with this substrate with the Vmax value confirms that k2 is in fact at least 200 times greater than k4 with nitrobutane. The rapid reaction data establish that the D402E mutation decreases the value of k2 with nitrobutane by ≥30-fold. This corresponds to an increase in the ΔG‡ for C–H bond cleavage of at least 1.9 kcal/mol in the mutant enzyme. The lack of a significant change in the Vmax value for the mutant enzyme is consistent with k4 being unaffected by the mutation. The intrinsic isotope effect of 9 is almost fully expressed in the DV/Knitrobutane value for the mutant enzyme. A decrease in the value of k2 of 30 would decrease k2/k−1 from ∼9 to ∼0.3, resulting in the observed increase in the DV/Knitrobutane value. Thus, the data with nitrobutane as the substrate are consistent with an effect on k2 alone in the mutant enzyme.
The increase in the V/K value for a range of primary nitroalkanes as substrates for NAO was previously used to show that each additional methylene contributes 2.6 kcal/mol (4). pH and kinetic isotope effects were used to isolate the relative contributions to binding and catalysis of this additional energy (11). This analysis yielded a value of 1.7 kcal/mol for the contribution to binding, leaving 0.9 kcal/mol available to enhance the rate of catalysis. The change per methylene group in the free energy of the transition state for proton abstraction (ΔΔG‡) can also be calculated from the rates of reduction by 1-nitrobutane and nitroethane. At 5 °C, the ΔΔG‡ value is 2.3 kcal/mol, or 1.1 kcal/mol per methylene group, which agrees well with the previously calculated value.
The effects of mutating Asp402 in NAO to glutamate described here are fully consistent with Asp402 being the catalytic base in NAO. This result provides further support for the assignment of NAO as a member of the acyl-CoA dehydrogenase superfamily and for the alignment of Asp402 with the glutamate which acts as the catalytic base in those enzymes. The average effect of the D402E mutation on the rate of proton abstraction of ∼22-fold or 1.8 kcal/mol is somewhat less than has been seen when the reverse mutation is carried out in other enzymes. In the classic case of triosephosphate isomerase, substitution of Glu165 for Asp caused a 1000-fold decrease in specific activity, equivalent to the expected result if the base is removed completely (20). In the acyl-CoA dehydrogenases, the effect of the reverse mutation varies among the different enzymes. Replacing the glutamate with an aspartate in the rat short chain acyl-CoA dehydrogenase reduces the kcat/Km values for various substrates less than 3-fold, yet the same mutation in human isovaleryl-CoA dehydrogenase renders its activity toward its native substrate barely detectable (21). In the case of human liver medium chain acyl-CoA dehydrogenase, the E376D mutation decreases the rate of flavin reduction by 800-fold (22). The greater effects of glutamate to aspartate mutations suggest that it is easier for an enzyme to accommodate an extra methylene group, possibly due to the increased flexibility of the amino acid side chain, than the loss of the methylene and the subsequent movement of the base 1.4 Å farther from the proton.
Footnotes
This research was supported in part by NIH Grant GM58698.
This paper is dedicated to the memory of Dr. Vincent Massey.
Abbreviations: NAO, nitroalkane oxidase; Hepes, N-(2-hydroxyethyl)piperazine-N′-2-ethanesulfonic acid; DMSO, dimethyl sulfoxide.
It was not possible to generate a stable ternary complex between the reduced enzyme and both the aldehyde and nitrite products. Addition of a high concentration of nitrite to the reduced enzyme slowly forms a novel nitrite complex regardless of the presence or absence of aldehyde. The rate at which this species forms is affected by the aldehyde concentration, but the same final absorbance spectrum of the nitrite complex is observed.
REFERENCES
- 1.Kido T, Hashizume K, Soda K. J. Bacteriol. 1978;133:53–58. doi: 10.1128/jb.133.1.53-58.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Porter DJT, Voet JG, Bright HJ. Z. Naturforsch. 1972;27b:1052–1053. doi: 10.1515/znb-1972-0914. [DOI] [PubMed] [Google Scholar]
- 3.Heasley CJ, Fitzpatrick PF. Biochem. Biophys. Res. Commun. 1996;225:6–10. doi: 10.1006/bbrc.1996.1122. [DOI] [PubMed] [Google Scholar]
- 4.Gadda G, Fitzpatrick PF. Arch. Biochem. Biophys. 1999;363:309–313. doi: 10.1006/abbi.1998.1081. [DOI] [PubMed] [Google Scholar]
- 5.Gadda G, Fitzpatrick PF. Biochemistry. 2000;39:1400–1405. doi: 10.1021/bi9922547. [DOI] [PubMed] [Google Scholar]
- 6.Gadda G, Edmondson RD, Russel DH, Fitzpatrick PF. J. Biol. Chem. 1997;272:5563–5570. doi: 10.1074/jbc.272.9.5563. [DOI] [PubMed] [Google Scholar]
- 7.Daubner SC, Gadda G, Valley MP, Fitzpatrick PF. Proc. Natl. Acad. Sci. U.S.A. 2002;99:2702–2707. doi: 10.1073/pnas.052527799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thorpe C, Kim JP. FASEB J. 1995;9:718–725. doi: 10.1096/fasebj.9.9.7601336. [DOI] [PubMed] [Google Scholar]
- 9.Gadda G, Banerjee A, Fitzpatrick PF. Biochemistry. 2000;39:1162–1168. doi: 10.1021/bi9921743. [DOI] [PubMed] [Google Scholar]
- 10.Gadda G, Banerjee A, Dangott LJ, Fitzpatrick PF. J. Biol. Chem. 2000;275:31891–31895. doi: 10.1074/jbc.M003679200. [DOI] [PubMed] [Google Scholar]
- 11.Gadda G, Choe DY, Fitzpatrick PF. Arch. Biochem. Biophys. 2000;382:138–144. doi: 10.1006/abbi.2000.2009. [DOI] [PubMed] [Google Scholar]
- 12.Gadda G, Fitzpatrick PF. Biochemistry. 1998;37:6154–6164. doi: 10.1021/bi973085y. [DOI] [PubMed] [Google Scholar]
- 13.Palmer G, Massey V. In: Biological Oxidation. Singer TP, editor. John Wiley and Sons; New York: 1968. pp. 263–300. [Google Scholar]
- 14.Bright HJ, Porter DJT. In: The Enzymes. 3rd ed. Boyer P, editor. XII. Academic Press; New York: 1975. pp. 421–505. [Google Scholar]
- 15.Emanuele JJ, Jr., Fitzpatrick PF. Biochemistry. 1995;34:3710–3715. doi: 10.1021/bi00011a028. [DOI] [PubMed] [Google Scholar]
- 16.Porter DJT, Voet JG, Bright HJ. J. Biol. Chem. 1977;252:4464–4473. [PubMed] [Google Scholar]
- 17.Weibel MK, Bright HJ. J. Biol. Chem. 1971;246:2734–2744. [PubMed] [Google Scholar]
- 18.Gadda G, Fitzpatrick PF. Biochemistry. 2000;39:1406–1410. doi: 10.1021/bi992255z. [DOI] [PubMed] [Google Scholar]
- 19.Strickland S, Palmer G, Massey V. J. Biol. Chem. 1975;250:4048–4052. [PubMed] [Google Scholar]
- 20.Raines RT, Sutton EL, Straus DR, Gilbert W, Knowles JR. Biochemistry. 1986;25:7142–7154. doi: 10.1021/bi00370a057. [DOI] [PubMed] [Google Scholar]
- 21.Battaile KP, Mohsen AA, Vockley J. Biochemistry. 1996;35:15356–15363. doi: 10.1021/bi961113r. [DOI] [PubMed] [Google Scholar]
- 22.Peterson KL, Galitz DS, Srivastava DK. Biochemistry. 1998;37:1697–1705. doi: 10.1021/bi972590s. [DOI] [PubMed] [Google Scholar]