Abstract
The thyroid hormone 3,3′,5-triiodo-l-thyronine (T3) is essential for growth, differentiation, and development. Its biological activities are mediated by T3 nuclear receptors (TRs). At present, how T3 regulates TR proteins and the resulting functional consequences are still unknown. Immunofluorescence analyses of endogenous TR in the growth hormone-producing GC cells showed that the T3-induced rapid degradation of TR was specifically blocked by lactacystin, a selective inhibitor of the ubiquitin–proteasome degradation pathway. Immunoblots demonstrated that the transfected TRβ1 was ubiquitinated and that the ubiquitination was T3 independent. Studies with a series of truncated TRβ1 showed that the hormone-binding domain was sufficient for the T3-induced rapid degradation of TRβ1 by the proteasome degradation pathway. T3 also induced rapid degradation of TRβ2 and TRα1. In contrast, the stability of the non-T3-binding TRα2 and naturally occurring TRβ1 mutants that do not bind T3 was not affected by T3 treatment, indicating that hormone binding to receptor was essential for the degradation of the wild-type receptors. In the presence of proteasome protease inhibitors, the levels of both total and ubiquitinated TRβ1 protein increased, yet T3-dependent transcriptional activation and the expression of the growth hormone gene were diminished, suggesting that proteasome-mediated degradation played a novel role in modulating transcriptional activation by TR. The present study reveals a role of T3 in modulating the functions of TR by regulating its receptor level via the ubiquitin–proteasome degradation pathway.
The thyroid hormone 3,3′,5-triiodo-l-thyronine (T3) is essential in metabolic-energetic homeostasis, development, and differentiation. Its actions are mediated by thyroid hormone nuclear receptors (TRs), which regulate the expression of T3-targeted genes. TRs belong to a superfamily of hormone nuclear receptors functioning as ligand-activated transcription factors, which include receptors for steroid hormones, vitamin D3, and the retinoids (1). TR consists of domains including the N-terminal A/B domain, the DNA-binding domain C, and the hormone-binding domain (domains D and E). Recent studies indicate that the transcriptional activity of TR depends not only on the type of the thyroid hormone response elements located on the promoter regions of T3 target genes but also on a host of corepressors and coactivators (2). In the absence of T3, TR binds to corepressors, such as N-CoR. Binding of T3 leads to the release of N-CoR from TR and recruitment of coactivators leading to gene activation (2). In this model, how TR proteins are regulated and the role of T3 in this process are unknown.
Using biochemical methods, Samuels and Casanova have previously reported that T3 down-regulates its endogenous TR in growth hormone (GH)-producing GC cells (3). However, the underlying molecular mechanisms by which T3 down-regulates the TR have not been elucidated. In this report, we demonstrated that the T3-induced degradation of TR was via the ubiquitin–proteasome pathway. This degradation pathway has already been shown to be responsible for the regulation of many short-lived regulatory proteins, including transcription regulators (e.g., NK-κB, IκB, YY1), cell-cycle proteins (e.g., cyclins and cyclin-dependent kinases), oncogenes, and tumor suppressors (e.g., c-Jun, c-Fos, E2A proteins, and p53) and enzymes (e.g., DNA topoisomerase, receptor-associated protein kinases) (4, 5). In this pathway, proteins are targeted for degradation by covalent ligation to ubiquitin, a highly conserved 76-aa peptide, via the lysine residues. The ubiquitinated proteins are recognized specifically by a huge protease complex, known as 26S proteasome, and are degraded (6, 7). In the present study, we showed that both liganded and unliganded TR were ubiquitinated and degraded by the proteasome pathway. However, binding of T3 triggered a rapid degradation of TR with a reduction of t1/2 for the liganded TR. We further showed that the extent of T3-dependent transcriptional activation and the expression of the GH gene in GC cells were inversely correlated with the level of TRβ1 protein, suggesting that proteasome-mediated degradation could modulate transcriptional activation by TR. We also found that the stability of naturally occurring TRβ1 mutants from the patients with thyroid hormone resistance syndrome (RTH) was not affected by T3. This stability of mutant TRβ1 in the presence of high-thyroid hormones may play a critical role in mediating the dominant–negative action of the mutant TRβ in RTH.
Materials and Methods
Plasmids.
The mammalian expression plasmid (pCLC51) and the T7 expression plasmid (pCJ3) for TRβ1 were obtained as described (8, 9). The mammalian expression vector for the truncated form of TRβ1ΔA/B [pYZ(CMV)JL08] was obtained as described (10). pCMV-PV and pSV2-S are the expression vectors for the naturally occurring mutants, PV and S, respectively, which are gifts from C. A. Meier (University Hospital Geneva, Geneva). PTK28 m-CAT with two palindromic repeats in tandem of the consensus TRE was a gift from G. Brent (University of California Los Angeles West Los Angeles Veterans Affairs Medical Center, Los Angeles). The same Pal-TRE fused to the luciferase gene reporter (pdouble-Pal-TK-Luc) is a generous gift from J. L. Jameson (Northwestern University School of Medicine, Chicago). Plasmids containing Lys-TRE and DR4-TRE fused to Luc reporter are gifts from P. Yen (National Institutes of Health, Bethesda, MD). The expression vector ΔA/B/C (pCMV06) was prepared by PCR by using the following 5′ primer, 5′-AGACCCAAGCTTAAAATGGCAACAGATTTGGTGCTG-3′ and the 3′ primer, 5′-GGGGCATTCCACCTTCAT-3′ by using pCLC51 as a template (8). The resulting cDNA was restricted with HindIII and BglII and cloned into the pCLC51. The sequence of ΔA/B/C (pCMV06) was confirmed by DNA sequencing and restriction analysis. pTRE-TRα1, pTRE-TRβ1, and pTRE-TRβ2 are expression plasmids used for the selection of stable cell lines. They were prepared by ligation of cDNA of hTRα1 (from pCLC61), hTRβ1 (from pCLC51), and hTRβ2 [from pKCR2; a gift from F. Wondisford, (Beth Israel–Deaconess Medical Center, Boston, MA)], respectively, to pTRE plasmid (CLONTECH).
Confocal Fluorescence Microscopy.
Confocal microscopic images of GC cells were taken under a confocal laser scanning fluorescence microscope (Zeiss, LSM-410). The detection path of the confocal microscope had a 35-μm pinhole aperture, and we used a glycerine-immersed 1.4-numerical aperture ×63 Zeiss Planapo objective (11). For the confocal measurements, GC cells were harvested from culture dishes and transferred to an observation chamber from Elekon (Chiba, Japan). Twenty-four hours later, T3 (100 nM) was added to the cells in the presence or absence of lactacystin (Calbiochem). After that, the cells were processed for observation as described (12). The primary antibody was the monoclonal anti-TRβ1 antibody (mAb C4; 10 μg/ml; ref. 13).
Transient Transfection Assay.
GC cells were trypsinized and placed in an electroporation cuvette. Cell suspension (5 × 106 cells/cuvette) in DMEM was mixed with 20 μg of pdouble-Pal-TK-Luc and incubated on ice for 10 min. Cells were electroporated at 450 V and 950 μF by using the Gene Pulser apparatus (Bio-Rad). After electroporation, cells were incubated on ice for 10 min and seeded in 35-mm dishes. Cells were cultured in DMEM containing 10% thyroid hormone-deficient calf serum (Td medium). Twenty-four hours later, 100 nM of T3 with or without lactacystin (25 μM) was added. After incubation for an additional 12 h, cells were harvested. Cells were lysed, and luciferase activity was determined according to the manufacturer's protocols.
Transient transfection experiments were also carried out by using CV1 cells as described similarly by Lin et al. (8). Briefly, cells were transfected with plasmids containing the Pal-TRE-TK-CAT (0.4 μg) and the expression vector for TRβ1 (pCLC51; 0.2 μg). Five hours after transfection, cells were incubated in Td medium. Twenty-four hours after transfection, T3 (100 nM) were added in the presence or absence of lactacystin, MG132 (Calbiochem) or leupeptin (Calbiochem) with the indicated concentrations for the time marked. Cells were lysed, and CAT (and/or Luc) assays were carried out by using 50 μg of cellular lysates. The values were normalized against the protein concentrations, which were determined by the Bradford method.
Western Blot Analysis.
CV1 cells were transfected as described above. Forty-eight hours after transfection, cells were lysed, and the blots were prepared as described by Bhat et al. (14). The primary antibody for the detection of TRβ1 was either mAb C4, C3 (15), J51, or J52 (2 μg/ml; ref. 16), and the signal was developed by using the enhanced chemiluminescence detection kit.
Preparation of Stable Cell Lines Expressing TR Isoforms.
HeLa Tet-on cells (2 × 106 cells/10-cm dish) were cotransfected with pTK-Hyg (CLONTECH) and pTRE-TRβ1, β2, or α1 expression plasmids. The clones stably expressing TRβ1, β2, or α1 were selected in the presence of 100 μg/ml of G418 and hygromycin (200 μg/ml). Drug-resistant clones were obtained after 10–14 days. Clones stably expressing TR isoforms were identified by Western blot analysis as described above.
Immunocytochemical localization of TR isoforms in stable clones was carried out similarly as described for the localization of TRs in GC cells by using mAb C4 (10 μg/ml; ref. 13). Quantitative analysis of immunofluorescence intensity was performed on images obtained from a Dage intensified charge-coupled device at a constant gain value. Both data acquisition and average pixel intensity of the resulting micrographs were calculated by using the National Institutes of Health image analysis program.
Determination of the GH mRNA in GC Cells by Northern Blot Analysis.
GC cells (2 × 106 cells/10-cm dish) were plated in DMEM containing 10% calf serum. After 24 h, cells were cultured in medium containing 10% Td calf serum. Two days later, cells were cultured with or without T3 in the presence of increasing concentrations of lactacystin for 16 h. Total RNA was prepared by using TRIzol reagent (GIBCO/BRL). Ten micrograms of RNA was analyzed by gel, transferred onto the blots, and probed with 32P-labeled GH cDNA, as described (14).
Results
Specific Inhibitors of Proteasome-Mediated Proteolysis Block T3 Induced Degradation of Endogenous TR.
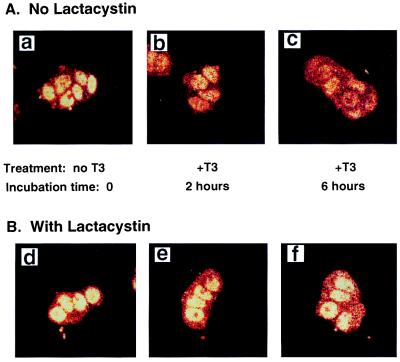
To study the regulation of TR proteins by T3, we first visualized the effect of T3 on the stability of the endogenous TR in GC cells by confocal fluorescence microscopy. We chose GC cells because they have functional endogenous TR (3). Fig. 1 shows the representative confocal fluorescence microscopic images of TR under different conditions. Fig. 1A-a shows that by using monoclonal anti-TR antibody C4 (mAb C4; ref. 13), TR was localized in the nuclei of cells cultured in the absence of T3. Fig. 1A-b and -c show that treating of cells with T3 led to a reduction of nuclear fluorescence in a time-dependent manner.
Figure 1.
Confocal fluorescence microscopic images of TR proteins. TRs were detected by using mAb C4 in GC cells before and after T3 treatment. Images of GC cells without pretreatment with lactacystin before addition of T3 to the cell culture (a) and after 2 h (b) and 6 h (c) of treatment with 100 nM T3 at 37°C. Images of GC cells pretreated with lactacystin (25 μM) for 12 h before T3 addition (d) and after 2 h (e) and 6 h (f) of T3 treatment (100 nM) at 37°C.
A different picture emerged when cells were pretreated with lactacystin, a specific protease inhibitor for the ubiquitin-mediated proteasome degradation pathway (Fig. 1B; refs. 4, 7, 17). The fluorescence intensities of TR in Fig. 1B-e and -f remained similar to that seen in Fig. 1B-d, even though the cells were treated with T3 for 2 and 6 h, respectively, indicating that lactacystin prevented the T3-induced degradation of TR. As a control, similar experiments to those of Fig. 1B were carried out by pretreating the cells with a nonspecific protease inhibitor, leupeptin. No inhibition of the T3-induced degradation was detected (data not shown). These results showed that the T3-induced rapid degradation of endogenous TR occurred via a proteasome degradation pathway.
Inhibitors of Proteasome-Mediated Proteolysis Block T3-Induced Degradation of the Transfected TRβ1.
To further characterize this pathway, we used the transfected TRβ1 in CV1 cells because the amounts of the endogenous TRs in GC cells are too low to be conveniently detected by either Western blotting or immunoprecipitation. Cells transfected with TRβ1 expression plasmid were incubated with T3 for increasing lengths of time, and the amounts of TRβ1 protein were determined by Western blotting by using mAb C4. A representative result of such experiments is shown in Fig. 2A. Lane 1 shows the molecular size of TRβ1 by using the in vitro translated receptor. Consistent with that seen for the endogenous TR in GC cells, T3 induced a time-dependent degradation of TRβ1 protein by T3 (lanes 7–10). After 12 h, only ≈10% of TRβ1 remained. The intensities of the bands were quantified and the apparent t1/2 was calculated to be 2.59 ± 0.51 h (mean ± SD; n = 3). In the absence of T3, the degradation of TRβ1 was also detected (lanes 1–5; Fig. 2A); however the degradation was much slower with an apparent t1/2 of 10.9 ± 2.9 h (mean ± SD; n = 3). Lane 11 (Fig. 2A) was the negative control from cells transfected only with empty vector. No TRβ1 was detected, indicating that the TRβ1 seen in lanes 1–10 was specific.
Figure 2.
Time-dependent degradation of transfected TRβ1 by T3. (A) Cellular lysates (50 μg) from CV1 cells transfected with TRβ1 expression plasmid were analyzed by Western blot analysis by using mAb C4. Lanes 1–6: no T3 treatment; lanes 7–11: with T3 (100 nM) for time as indicated; lane 11: control from cells transfected with empty vectors only. (B) Cellular lysates (50 μg) from CV1 cells transfected with TRβ1 expression plasmid were analyzed by Western blot analysis by using different monoclonal antibodies as indicated. MOPC, a control antibody. Lane 1 in A and B shows TRβ1 from the in vitro translation as a molecular weight marker.
The epitope for mAb C4 was mapped to the conserved C-terminal E (457)VFED (461) (13). Fig. 2A shows that no smaller degraded TRβ1 fragments were detected by using mAb C4. Attempts were made to see whether degraded fragments could be detected by using mAb J51 and 52, whose epitopes are located in the amino A/B domain (16) and mAb C3, whose epitopes are located in E (248)-V (256) (15). The results shown in Fig. 2B indicate no stable intermediate fragments were detected in the presence of T3 (lanes 2, 4, 6, and 8).
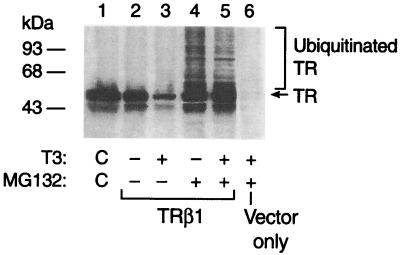
To determine whether the T3-induced degradation of the transfected TRβ1 was also mediated by the proteasome degradation pathway, we treated the cells with T3 in the presence or absence of lactacystin or MG132. The latter is also a specific inhibitor for the proteasome degradation system (4). Both inhibited the T3-induced degradation of the transfected TRβ1. Fig. 3 shows a typical result of such experiments, in which T3-induced degradation of the transfected TRβ1 (lane 3 vs. lane 2) was prevented in the presence of MG132 (lane 5 of Fig. 3), resulting in the accumulation of the characteristic “laddering” of the ubiquitinated high molecular-weight species (lane 5). In the absence of T3, the degradation of the unliganded TRβ1 was also prevented by MG132 (compare lanes 2 and 4). Furthermore, the characteristic “laddering” of the ubiquitinated high molecular-weight species was also detected in the absence of T3 (lane 4), indicating that the degradation of the unliganded TRβ1 was also via the ubiquitin-mediated pathway. Lane 6 shows the control in which only the empty vectors were transfected. No characteristic high molecular-weight species were detected, indicating the specific bands in lanes 4 and 5 were the ubiquitinated high molecular-weight species of TRβ1. Taken together, these results indicate that both the liganded and unliganded TRβ1 were ubiquitinated, and binding of T3 triggered the rapid degradation of TRβ1.
Figure 3.
Proteasome inhibitor MG132 blocks the T3-induced degradation of TRβ1. Cellular lysates (75 μg) from CV1 cells transfected with TRβ1 expression plasmid that were treated with (lanes 4 and 5) or without (lanes 2 and 3) MG132 (50 μM for 12 h) were analyzed by Western blotting similarly as described for Fig. 2A. For control, lane 1 is the marker TRβ1 from the in vitro translation.
The Hormone-Binding Domain Is Essential for the Degradation of TRβ1.
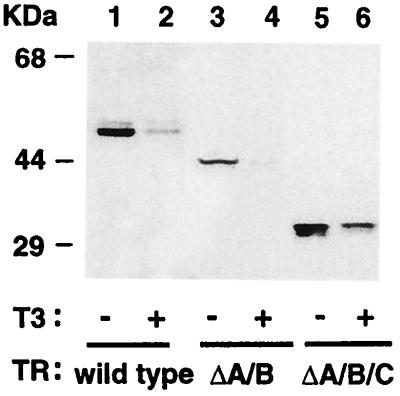
TRβ1 consists of functional domains including the N-terminal A/B domain, the DNA-binding domain C, and the hormone-binding domain (domains D and E). To locate the domain that was essential for mediating its degradation, we transfected truncation mutants of TRβ1 into cells and evaluated the amounts of these truncated TRβ1 by Western blotting (Fig. 4). Lanes 1 and 2 were positive controls to show that T3 induced the degradation of the intact TRβ1. Lanes 3/4 and 5/6 show that the lack of domain A/B (ΔA/B) or A/B/C (ΔA/B/C), respectively, had no effect on the T3-induced degradation of the remainder molecule of TRβ1. These results indicate that the hormone-binding domain was sufficient for the T3-induced degradation of TR.
Figure 4.
The hormone-binding domain is sufficient for T3-induced degradation. The expression plasmids for the wild-type TRβ1 (lanes 1–2), truncation mutant ΔA/B (lanes 3–4), or ΔA/B/C (lanes 5–6) were transfected into CV1 cells. Cells were treated with (lanes 2, 4, and 6) or without (lanes 1, 3, and 5) 100 nM T3. Fifty micrograms of total cell lysates was analyzed by Western blotting similarly as described for Fig. 2A.
T3 Induces Degradation of TRβ2, TRα1, but Not TRα2.
The above results predicted that other T3-binding TR isoforms should undergo T3-induced degradation. Similar to that seen for TRβ1, lanes 2–5 of Fig. 5A show that in the absence of T3, the transfected TRα1 was slowly degraded but underwent T3-induced rapid degradation with a t1/2 of 2.3 ± 0.4 h (mean ± SD, n = 4). Similar results were found for TRβ2 (Fig. 5B). The three bands detected by mAb C4 in lane 1 of Fig. 5B represented the initiation of translation of TRβ2 at different ATG sites. The intact and two smaller truncated TRβ2 were induced to be degraded by T3 with similar rates (lanes 6–9; Fig. 5B). Fig. 5C shows that the non-T3-binding TRα2 was stable and no T3-induced degradation was detected, supporting further the essential role of T3 binding in the rapid degradation of TRs.
Figure 5.
T3 induces rapid degradation of TRα1 and TRβ2, but not TRα2. CV1 cells transfected with the expression plasmids of TR isoforms were treated with or without T3 for the duration, as indicated. Cellular lysates (50 μg) were analyzed by Western blotting similarly as described for Fig. 2A. The primary antibody for TRα1 (A) and TRβ2 (B) was mAb C4 but for TRα2 (C) was mAb C3. “M” in lane 1 (A-C) represents the in vitro-translated TRβ1 as a molecular weight marker.
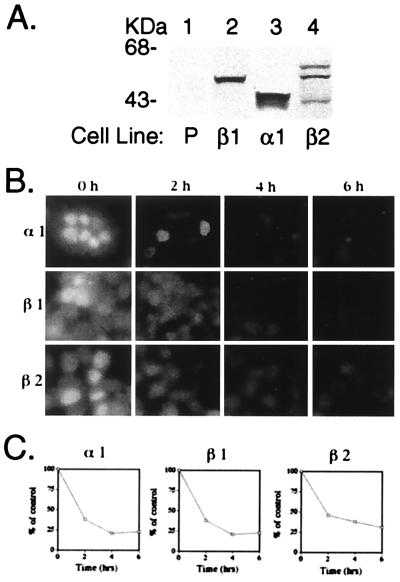
To visualize theT3-induced rapid degradation of TR isoforms, we prepared cell lines stably expressing TRα1, TRβ1, or TRβ2 (Fig. 6A). Fig. 6B Upper shows that on treatment of cells with T3, TRα1 underwent T3-induced degradation in a time-dependent manner. Similar time-dependent T3-induced degradation of TRβ2 was also seen (Fig. 6B Lower). The time-dependent T3-induced degradation of TRβ1 was also included for comparison (Fig. 6B Middle). In the absence of T3, very few changes in the intensities of the nuclear fluorescence for each of the three TR isoforms were seen during the 6-h time span (data not shown). Quantitative image analysis of the intensities of nuclear fluorescence indicates that TR isoforms degraded with a similar rate in the presence of T3 (Fig. 6C), which are consistent with that seen by Western blot analysis for the transfected TRs (see above).
Figure 6.
Immunofluorescence detection of TR in cell lines stably expressing each of the TR isoforms after treatment with T3 for the indicated times. (A) Detection of TR isoform in cell lines stably expressing TR isoform. P, parental cell line. (B) Immunofluorescence was performed as described in Materials and Methods. In C, fluorescence intensity of each of the isoforms after T3 treatment was monitored by measuring the mean pixel intensity of four micrographs each to obtain the average values given. Data are expressed as a percentage of the total fluorescence in the absence of T3.
Inhibition of Proteasomal Degradation Causes Reduced T3 Responsiveness.
The accumulation of TRβ1 in the presence of proteasome inhibitor provided an opportunity to address the role of the proteasome-mediated proteolysis in the T3-dependent TR-mediated transcriptional activity. We assessed the transcriptional activity of TRβ1 in the presence of increasing amounts of lactacystin, which causes accumulation of the total including the ubiquitinated forms of TR. Fig. 7A shows the results of experiments in which increasing concentrations of lactacystin were used to correlate the transcriptional activity with the protein levels of TRβ1. At each concentration of lactacystin (5, 10, and 25 μM), both the CAT activities and the receptor protein concentrations were determined. Fig. 7A clearly shows that increasing concentrations of lactacystin led to increases in TRβ1 proteins. However, the T3-dependent transcriptional activity demonstrated by CAT activity was concomitantly decreased. As a control, we have also carried out similar experiments by using leupeptin. No effect was detected on the transcriptional activity of TRβ1 by leupeptin (data not shown). In the presence of lactacystin, similar reduction in the transcriptional activity of TRβ1 mediated by other TREs (Lys-TRE and DR4-TRE; data not shown) was observed. These results indicate that normal function of the proteasome pathway is essential for optimal T3-dependent transcriptional activity, suggesting that proteasomal function may remove proteins that inhibit activation by TR, or more intriguingly, that proteasome-mediated degradation of TR could be one of the critical steps leading to transcriptional activation. The reverse correlation could be TR specific, because no effect of lactacystin on the retinoic acid-dependent transcriptional activation mediated by the retinoic acid receptor was detected (data not shown).
Figure 7.
Transcriptional activity of TRβ1 is inversely correlated with TRβ1 protein level. (A) Correlation of the transcriptional activity with the TRβ1 protein level. CV1 cells were transfected with pTK28mCAT plasmid and TRβ1-expressing plasmid as described in Materials and Methods. The amounts of TRβ1 protein were determined by Western blot analysis [50 μg of cellular lysates by using mAb C4 (2 μg/ml)]. CAT assays were performed as described in Materials and Methods. The data were from four different experiments, each with duplicates. Data are expressed as mean ± SD (n = 4). (B) Twenty-four hours after GC cells were electroporated with pdouble-pal-TKLuc, cells were incubated with 100 nM T3 in the presence or absence of lactacystin (25 μM) for 12 h. At the end of incubation, cells were lysed, and luciferase assays were performed as described in Materials and Methods. (C) GC cells were treated with or without T3 and lactacystin, as indicated. Northern blot analysis was carried out as described in Materials and Methods. Data are expressed as mean ± SD (n = 3).
We also evaluated the transcriptional activity of endogenous TR as a result of blocking of the T3-induced degradation by lactacystin in GC cells. Bar 2 of Fig. 7B shows that T3-dependent activation mediated by the endogenous TR was ≈10-fold. When GC cells were pretreated with lactacystin (25 μM), the T3-dependent transcriptional activity of endogenous TR was decreased ≈50%. We could not accurately determine the level of the endogenous TR by Western blotting because of low levels of the endogenous TR proteins. However, the results of fluorescence experiments shown in Fig. 1 indicate that the level of TR was high because of prevention of degradation by lactacystin (see Fig. 1B-e and -f). Therefore, the inverse correlation between the transcriptional activity and TR protein level was also observed for the endogenous TR.
This reverse correlation was further supported by the expression of the endogenous GH gene (GH) in GC cells. Fig. 7C demonstrates that the T3-induced expression of GH mRNA was repressed by lactacystin in a concentration-dependent manner. At 25 μM of lactacystin, ≈50% T3-induced expression of GH mRNA was repressed, which was consistent with that detected by using the reporter system (see Fig. 7B).
The Important Role of T3-Induced Degradation in the Dominant–Negative Effect of TRβ1 Mutants Derived from Patients with Thyroid Hormone Resistance Syndrome (RTH).
RTH is a genetic disease caused by mutations in the TRβ gene located on chromosome 3 (18). Patients have normal or elevated level of thyroid-stimulating hormone, in spite of the elevated levels of circulating thyroid hormones. TRβ mutants derived from RTH patients have reduced T3-binding affinities and transcriptional capacities and act in a dominant–negative fashion to cause the clinical phenotype. At present, the molecular basis of the dominant–negative action of TRβ mutants is unknown. One of the unresolved key issues relates to the relative level of mutant and the wild-type TR to produce a clinical phenotype. We therefore ascertained the effect of T3 on the stability of TRβ mutants.
We used two TRβ1 mutants, S (Δ337T) or PV (frame-shift mutation in the last C-terminal 16 amino acids; refs. 18, 19), which were isolated from RTH patients. Both mutants have completely lost T3-binding activity and have potent dominant–negative action (20). When we transfected mutants S and PV into CV1 cells and determined the amounts of mutants S and PV in the presence or absence of T3 by Western blotting, little degradation was observed (data not shown). Under these conditions, high molecular-weight ubiquitinated “laddering” mutant S and PV were detected when cells were treated with MG132, whether T3 was present or not (data not shown). We also transfected TRβ1 into SK-Hep-1-PV, in which mutant PV is stably expressed (14), and determined the ratios in the expression of TRβ1 with PV in the same cells after treating the cells with T3. The coexpression of both the wild-type TRβ1 and mutant PV proteins in the same cells would mimic RTH. The ratios of PV/TRβ1 proteins were ≈0.3 and ≈1.2 in the absence or presence of T3, respectively, indicating a higher PV/TRβ1 protein ratio in the presence of T3 (data not shown).
Discussion
In the present study, we demonstrated that T3 induced rapid degradation of TR via the ubiquitin-mediated proteasome-degradation pathway. Thus, TR becomes a new member of transcription regulators in which the biological functions are regulated by the proteasome-degradation pathway. Moreover, the signaling mechanism by TR leading to their ultimate degradation by the proteasome has added a new dimension to the paradigm. Similar to other regulatory proteins degraded by this pathway, TR was ubiquitinated. However, ubiquitination was not sufficient for rapid degradation of TR by this pathway. Binding of T3 triggered the proteolysis of TR by the proteasome. That binding of T3 was obligatory for the degradation of TR is analogous to that observed for G1 cyclins in that in addition to ubiquitination, degradation is coupled to the phosphorylation of G1 cyclins (5). Furthermore, the important role of ligand in the signaling of degradation has also been observed for Met tyrosine kinase receptor (21). However, the role of T3 in the signaling of TR for degradation differs from that of Met tyrosine kinase receptor in that its ligand, hepatocyte growth factor/scatter factor, enhances the ubiquitination of Met tyrosine kinase receptor. For TR, T3 did not enhance the ubiquitination, but rather triggered the degradation of ubiquitinated TR. At present, it is not known what led to the ubiquitination of TR. It was previously observed that many rapidly degraded regulatory proteins contain PEST elements, regions enriched in proline, glutamic acid, serine, and threonine residues (5). We found an imperfect element 323PESe (326) located in the hormone-binding domain. However, at present, its possible role in the ubiquitination of TR is uncertain. It is also known that phosphorylation of proteins may serve as a signal for ubiquitination and subsequent proteasomal degradation. Indeed, regulatory proteins such as yeast and mammalian G1 cyclins, Cut2, p27, Sic1, Rum1, I-κBα, STAT1, IRAK, and β-catenin, are known to be phosphorylated proteins before degradation (5, 22). TRβ1 was found to be a phosphoprotein (23). Even though the precise locations of phosphorylation sites have not been identified, sequence analysis revealed that TRβ1 contains two S/TP sequences. One is located in domain A/B (55SP56), and the other is located in domain E (277TP278). This S/TP motif was found to be the minimum consensus phosphorylation site required for the ubiquitination and degradation of the yeast G1 cyclins Cln3, Cln2, as well as the Gcn4 transcriptional activator (5). The verification of the involvement of these sites in the ubiquitination of TR requires future studies.
We have shown that the binding of T3 triggered the degradation of the ubiquitinated TR. However, the molecular basis underlying this triggering event is not clear. It has been suggested earlier that the time course of the change in receptor half-life is consistent with induction by hormone of a protease for receptor (24). This event could be mediated by the T3-induced conformational changes of TR. The ubiquitinated proteins are recognized by a large 26S proteolytic complex consisting of 19 S and 20 S complexes (17). Even though the molecular details in proteasome-mediated proteolysis have not been completely elucidated, it is generally believed that the ubiquitinated substrates are unfolded and translocated through the 19S complex to be degraded by the 20 S proteasome (25). Binding of T3 is known to induce dramatic structural reorganization of TRα1 and TRβ1 (26). The structural reorganization could facilitate the docking of the ubiquitinated TR onto the proteasome and facilitate the translocation, unfolding, and/or substrate/proteasome interaction. The precise molecular steps in the triggering of degradation of TR by T3 will await future studies.
The present study shows that the transcriptional activity of TRβ1 was inversely correlated with the receptor protein level. This observation raises an interesting possibility in that proteasome-mediated degradation plays a critical role in the transcriptional activation of TR. The current model suggests that T3-bound TR is in a conformation that can recruit an array of coactivators for transcriptional activation (2). However, the nature of the transcriptionally active TR is unclear. The present results raise the possibility that the transcriptionally active TR is a truncated form resulting from specific proteolysis by proteasome triggered by T3 binding. The generation of functionally active moiety by specific proteolysis is not without precedent. It has been shown that the ubiquitin–proteasome pathway is required for processing the NF-κB1 precursor protein and for the activation of NF-κB (27). However, at present, we cannot rule out the possibility that binding of T3 signifies the rapid degradation of TR for receptor turnover. It is unclear how these two T3-induced paradoxical events, if coexisting, are kinetically coordinated to fulfill the functional role of TR. However, one could imagine that rapid degradation of transcriptionally active TR is required, or the consequence of perpetual activation of genes by TRs could lead to deleterious effects on cells.
An important finding from the present study was the observation that mutants PV and S were as stable as the wild-type TR in the presence of T3, but unlike the wild-type receptor, these mutants are not degraded in the presence of T3. This observation has important clinical implications. RTH is a syndrome characterized by refractoriness of the pituitary and/or peripheral tissues to the action of thyroid hormones. One of the unresolved issues in understanding the molecular basis of RTH has related to the relative levels of mutant to wild-type TRβ1 protein to produce a clinical phenotype in different organs. The only two studies that address this issue reported conflicting results (28, 29). In the first study, the severity of growth delay in a kindred with RTH correlated with the increased ratio of mutant to wild-type TRβ1 mRNA in fibroblasts (28). However, in a second study, no correlation was found between the relative ratios of mutant and normal TRβ1 mRNA in fibroblasts and the clinical phenotype of RTH patients (29). Neither of these studies had addressed the expression and stability of the wild-type and mutant TRβ1 at the protein level. The present study clearly shows that T3 induced the degradation of the wild-type TRβ1 but not the mutant receptors. By transfecting TRβ1 into SK-Hep-1-PV cells, we demonstrated that the ratio of PV/TRβ1 was higher in the presence than in the absence of T3. Because RTH patients have high thyroid hormones, these results suggest the higher PV/TRβ1 ratios exist in the heterozygous RTH patients, thereby favoring the dominant–negative activity of mutant TRβ1. Furthermore, it is entirely possible that the factors involved in the multistep proteasome degradation pathway are expressed in a tissue-dependent manner. This could lead to different efficiency in the T3-induced degradation of the wild-type TR in a tissue-dependent manner, resulting in the varied ratios of mutant to the wild-type TR among tissues. This could lead to heterogeneous phenotypic features in patients. Therefore, the proteasome-mediated degradation pathway plays a very important role in functions of TR in the normal and pathological state.
Abbreviations
- T3
3,3′,5-triiodo-l-thyronine
- TRs
thyroid hormone nuclear receptors
- RTH
thyroid hormone resistance syndrome
- GH
growth hormone
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.160257997.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.160257997
References
- 1.Cheng S-y. J Biomed Sci. 1995;2:77–89. doi: 10.1007/BF02253060. [DOI] [PubMed] [Google Scholar]
- 2.McKenna N J, Lanz R B, O'Malley B W. Endocrine Rev. 1999;20:321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- 3.Samuels H H, Casanova J. Endocrinol Res. 1989;15:495–545. doi: 10.3109/07435808909036350. [DOI] [PubMed] [Google Scholar]
- 4.Lee D H, Goldberg A L. Trends Cell Biol. 1998;8:397–403. doi: 10.1016/s0962-8924(98)01346-4. [DOI] [PubMed] [Google Scholar]
- 5.Hershko A, Ciechanover A. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 6.Hochtrasser M. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 7.Fenteany G, Standaert R F, Lane W S, Shoi S, Corey E J, Schreiber S L. Science. 1995;268:726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- 8.Lin K-H, Ashizawa K, Cheng S-y. Proc Natl Acad Sci USA. 1992;89:7737–7741. doi: 10.1073/pnas.89.16.7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin K-H, Fukuda T, Cheng S-y. J Biol Chem. 1990;265:5161–5165. [PubMed] [Google Scholar]
- 10.Zhu X-G, Yu C-L, McPhie P, Wong R, Cheng S-y. Endocrinology. 1996;137:712–721. doi: 10.1210/endo.137.2.8593822. [DOI] [PubMed] [Google Scholar]
- 11.Furuno T, Teshima R, Kitani S, Sawada J, Nakanishi M. Biochem Biophys Res Commun. 1996;219:740–744. doi: 10.1006/bbrc.1996.0304. [DOI] [PubMed] [Google Scholar]
- 12.Lin K H, Willingham M C, Liang C- M, Cheng S-y. Endocrinology. 1991;128:2601–2609. doi: 10.1210/endo-128-5-2601. [DOI] [PubMed] [Google Scholar]
- 13.Bhat M K, McPhie P, Ting Y T, Zhu X- G, Cheng S-y. Biochemistry. 1995;34:10591–10599. doi: 10.1021/bi00033a034. [DOI] [PubMed] [Google Scholar]
- 14.Bhat M K, Dace A, Cheng S-y. Thyroid. 1999;9:411–418. doi: 10.1089/thy.1999.9.411. [DOI] [PubMed] [Google Scholar]
- 15.Bhat M K, McPhie P, Cheng S-y. Biochem Biophys Res Commun. 1995;210:464–471. doi: 10.1006/bbrc.1995.1683. [DOI] [PubMed] [Google Scholar]
- 16.Fukuda T, Willingham M C, Cheng S-y. Endocrinology. 1988;123:2646–2652. doi: 10.1210/endo-123-6-2646. [DOI] [PubMed] [Google Scholar]
- 17.Fenteany G, Schreiber S L. J Biol Chem. 1998;273:8545–8548. doi: 10.1074/jbc.273.15.8545. [DOI] [PubMed] [Google Scholar]
- 18.Refetoff S, Weiss R E, Usala S J. Endocrinol Rev. 1993;14:348–399. doi: 10.1210/edrv-14-3-348. [DOI] [PubMed] [Google Scholar]
- 19.Parrilla R A, Mixson A J, McPherson J A, McClaskey J H, Weintraub B D. J Clin Invest. 1991;88:2123–2130. doi: 10.1172/JCI115542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meier C A, Parkinson C, Chen A, Ashizawa K, Meier-Heusler S C, Muchmore P, Cheng S-y, Weintraub B D. J Clin Invest. 1993;92:1986–1993. doi: 10.1172/JCI116793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeffers M, Tayor G A, Weidner K M, Omura S, Vande Woude G F. Mol Cell Biol. 1997;17:799–808. doi: 10.1128/mcb.17.2.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanaka K. J Biochem. 1998;123:195–204. doi: 10.1093/oxfordjournals.jbchem.a021922. [DOI] [PubMed] [Google Scholar]
- 23.Lin K-H, Ashizawa K, Cheng S-y. Proc Natl Acad Sci USA. 1992;89:7737–7741. doi: 10.1073/pnas.89.16.7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raaka B M, Samuels H H. J Biol Chem. 1981;256:6883–6889. [PubMed] [Google Scholar]
- 25.Gerards W L H, De Jong W W, Bloelens W, Bloemendal H. Cell Mol Life Sci. 1998;54:253–262. doi: 10.1007/s000180050147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ribeiro R C, Apriletti J W, Wagner R L, Feng W, Kushner P J, Nilsson S, Scanlan T S, West B L, Fletterick R J, Baxter J D. J Steroid Biochem Mol Biol. 1998;65:133–141. doi: 10.1016/s0960-0760(98)00029-6. [DOI] [PubMed] [Google Scholar]
- 27.Palombella V J, Rando O J, Goldberg A L, Maniatis T. Cell. 1994;78:773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 28.Mixson A J, Hauser P, Tennyson G, Renault J C, Bodenne D L, Weintraub B D. J Clin Invest. 1993;91:2296–2300. doi: 10.1172/JCI116458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hayashi Y, Janssen O E, Weiss R E, Murata Y, Seo H, Refetoff S. J Clin Endocrinol Metab. 1993;76:64–69. doi: 10.1210/jcem.76.1.8421105. [DOI] [PubMed] [Google Scholar]