Abstract
Tyrosine kinases of the Janus kinase family initiate cellular responses through their association with receptors for α-helical cytokines. In addition to a tyrosine kinase domain, these enzymes possess a kinase-like (KL) domain, whose function remains elusive. To investigate the role of the KL domain of Tyk2 in interferon-α/β signaling, we transfected a library of Tyk2 cDNAs containing random point mutations in KL into Tyk2-negative cells and selected for loss-of-function Tyk2 mutants. Four such mutants, V584D, G596V, H669P, and R856G, were identified through this screen. Like the wild-type Tyk2, the mutant proteins were able to sustain the level of IFNAR1 receptor protein. However, all four mutants were incapable of restoring high-affinity interferon-α binding in Tyk2-negative cells and were also catalytically impaired, even when transiently overexpressed. Interferon-α induced phosphorylation, and gene expression could be detected in V584D- or G596V-expressing cells, but not in H669P- or R856G-expressing cells. Furthermore, H669P and R856G proteins were constitutively highly phosphorylated. All together, our findings demonstrate that an intact KL domain is essential for the intrinsic catalytic activity of Tyk2 and for the establishment of a high-affinity interferon-α receptor complex.
In mammals, the four Janus kinases (JAKs) (Tyk2, JAK1, JAK2, and JAK3) have been shown to participate in the early steps of signaling cascades originating from cell surface receptors engaged with α-helix-bundled cytokine ligands (1–3). One major pathway triggered by these cytokines involves the catalytic activation of the receptor-associated JAK proteins, tyrosine phosphorylation of the cytoplasmic regions of the receptors, recruitment of SH2-containing signal transducers and activators of transcription (STAT) proteins onto the phosphorylated motifs, and phosphorylation of these transcription factors. Activated STATs then translocate into the nucleus and induce the transcription of target genes (1, 4, 5). Little is known about the molecular mechanisms regulating the JAK enzymes that play a critical role in this signaling pathway.
Sequence alignment of the JAKs reveals seven regions of homology, called JH1 to JH7. The amino-terminal region comprises JH3 through JH7 and has been implicated in receptor interaction and stability (3). JH1, located at the carboxyl terminus, is a tyrosine kinase (TK) domain that contains an activation loop, the phosphorylation of which is required for catalytic activation (6–9). The centrally located JH2 or kinase-like (KL) domain exhibits high sequence identity (up to 30%) with kinase domains but lacks intrinsic catalytic activity. Differences between the KL domain and canonical kinase domains are evident in conserved sequences defining the ATP- and substrate-binding cleft, including the glycine-rich loop; the HRDL motif, which is HGNV in KL; and the DFG motif, which is DPG (10). No tyrosine residues are present in the segment of KL corresponding to the activation loop of active kinase domains.
Functional consequences of the deletion of the KL domain in JAKs have previously been studied. Loss of function was shown for ΔKL mutants of Tyk2 and Hopscotch, the Drosophila JAK homolog (11, 12). In contrast, partial or unregulated activity of one JAK2 ΔKL mutant has been observed in a number of independent studies (13–15). The possibility of a negative regulatory role for the KL domain was raised by the study of the Hopscotch mutant, HopT42, shown to be responsible for a leukemia-like phenotype in Drosophila larvae (12). This hyperactive mutant bears a lysine substitution of a glutamate residue conserved in all KL domains. JAK2 containing the corresponding mutation, E665K, was found to be hyperactive as well. Recently, the KL domain of JAK3 was shown to inhibit the activity of the isolated TK domain when both domains were coexpressed in COS cells (16). Interestingly, an increased inhibitory effect was observed when two mutated KL domains deriving from severe combined immunodeficiency patients were assayed (17). Another role that has been proposed for the KL domain is the recruitment of substrates into the vicinity of the TK domain. A yeast two-hybrid screen using the KL domain of JAK1 as bait resulted in the identification of STAT5 as a potential interacting partner (18). This interaction was extended to the KL domains of JAK2 and JAK3.
In this paper we have investigated the role of the KL domain of Tyk2 in the context of IFNα/β signaling. The IFNα/β receptor complex is composed of two receptor subunits, IFNAR1 and IFNAR2, which are associated with Tyk2 and JAK1, respectively (19, 20). In human cells, the presence of these intracellular kinases is essential for high-affinity ligand binding (1, 21). STAT1 and STAT2 are the immediate transcription factors that are recruited onto the activated receptor complex and required for the transcriptional induction of IFNα/β-responsive genes (22). From a genetic screen of randomly point-mutated Tyk2 cDNAs, we have identified point mutations in the KL domain that are responsible for loss-of-function phenotypes. Analysis of the IFNα signaling pathway in cell lines expressing these mutants has revealed that the KL domain is crucial for both Tyk2 catalytic activity and the establishment of high-affinity IFNα binding sites.
Materials and Methods
Cell Culture and Plasmids.
11,1 cells have previously been described (23). Cells were maintained in DMEM, 10% heat-inactivated FCS, and 250 μg/ml hygromycin. Stable and transient transfections were performed with calcium phosphate (24).
Plasmid pIRES-Tyk2: A 3.6-kb ClaI–NotI fragment containing the Tyk2 cDNA from pBS-Tyk2 was subcloned into pIRES-neo (CLONTECH).
pBS-Δ21Tyk2B: pBS-Δ21Tyk2 (25) was digested with SphI and partially digested with AflIII to remove a 327-bp fragment, which was replaced with the corresponding fragment containing two mutations, introducing a BsiWI site and a M564V substitution.
pIRES-Tyk2B: A 2.9-kb BspEI–BstEII fragment from pIRES-Tyk2 was replaced with the corresponding fragment from pBS-Δ21Tyk2B containing the BsiWI site. The M564V mutation did not alter IFNα-induced gene expression or Tyk2 in vitro kinase activity when compared with wild-type (WT) Tyk2 (data not shown).
pIRES-V584D, -G596V, -H669P, -R856G: Each point mutation was introduced into a 1-kb BsiWI–Eco47III Tyk2 fragment by PCR, using mutagenic primers (sequences available on request) and the overlap extension method. These fragments were subcloned into pIRES-Tyk2B.
pIRES-R856G/Y1054F/Y1055F and pIRES-R856G/K930R: A 660-bp Eco47III–BstEII fragment from pIRES-R856G was replaced with the corresponding fragment from pRc-Y1054F/Y1055F or pRc-K930R (6). All fragments originally derived from PCR were fully sequenced.
Generation and Screening of the Tyk2 cDNA Library.
PCRs were carried out in 7 mM MgCl2/0.5 mM MnCl2/0.3 μM primers/200 μM dATP/200 μM dGTP/1 mM dCTP/1 mM dTTP, containing 5 units of Amplitaq DNA polymerase (Perkin-Elmer), using a previously described protocol (26). Given the expected average mutation rate per cycle of 4.4 × 10−4 and a fragment size of 1.1 kb, 4, 7, and 14 cycles of amplification were tested. As a template, 500 ng, 100 ng, and 1 ng of a pIRES/Tyk2B fragment containing the KL domain were used. Amplified products were subcloned. Eight clones from each condition were sequenced with an ABI 373A DNA Sequencer (Perkin-Elmer). From this analysis, we determined that 4, 7, and 14 cycles of amplification resulted in up to two, six, and eight point mutations, respectively. To generate the randomly mutated Tyk2 cDNA library, amplified products from a four-cycle amplification were digested with BsiWI and Eco47III and subcloned into pIRES-Tyk2B. The library was used to transform competent XL1–10 cells (Stratagene). Screening of the Tyk2 library was performed by transfecting 11,1 cells with plasmid DNA and selecting colonies in 430 μg/ml neomycin. After 10 days, the medium was supplemented with 6-thioguanine (6TG) at 30 μg/ml and 500 international units (IU)/ml human recombinant IFNα2b (kindly provided by D. Gewert, Wellcome). Surviving colonies were ring-cloned 3–5 days later. Genomic DNA was isolated and used to amplify the 1.1-kb KL coding region by high-fidelity PCR (Vent; New England Biolabs).
IFNα Binding and Sensitivity Assays.
Binding experiments with 125I-labeled human recombinant IFNα2c (kindly provided by G. Adolf, Ernst Boehringer Institute) were performed as previously described (24). IFN sensitivity was determined by seeding 6 × 104 cells into six-well plates in the presence of 30 μg/ml 6TG and various doses of IFNα2b or IFNβ (provided by Biogen). After 3 days, cells were fixed and stained.
Immunoblot Analysis and in Vitro Kinase Assay.
Tyk2 was immunoprecipitated with polyclonal R5–7 antibodies (27) and detected with monoclonal T10–2 (6) or C-20 antibodies (Santa Cruz Biotechnology). The anti-phosphotyrosine 4G10 monoclonal antibody was from Upstate Biotechnology (Lake Placid, NY). The JAK1 M7 antiserum was from A. Ziemiecki (University of Bern, Switzerland); the anti-IFNAR1 64G12 monoclonal antibody was from P. Eid (Institut de Recherches sur le Cancer, Villejuif, France); and the STAT1 antiserum was from C. Schindler (Columbia University, New York). Cells were routinely treated with 103 IU/ml IFNα2b for 15 min. Immunoprecipitates from 1–1.5 mg of cell lysates were analyzed by Western blotting (6). Kinase assays were performed as previously described (6), using 1 μg of glutathione S-transferase-IFNAR2 as the exogenous substrate and a reaction time of 5 min at 30°C.
Structural Localization of KL Residues.
The atomic coordinates of the inactive insulin receptor kinase (IRK) (28) were taken from the Protein Data Bank (entry code 1IRK) and used to locate the IRK residues that correspond, by sequence alignment, to the mutated residues in the KL domain. From this analysis, Tyk2 KL residues V584, G596, E611, S613, E657, H669, Y689, E726, and R856 correspond, respectively, to IRK residues V991, G1003, I1018, K1020, N1046, H1058, L1078, N1124, and R1253.
Results
Isolation of Loss-of-Function Tyk2 Mutants and Identification of KL Point Mutations.
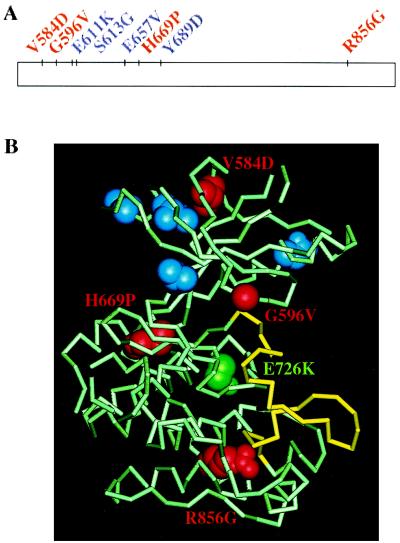
To study the role of the KL domain of Tyk2, we took a genetic approach in mammalian cells, where we screened for KL point mutations that inactivate the function of the Tyk2 protein. Human fibrosarcoma 11,1 cells, which lack endogenous Tyk2 and are unresponsive to IFNα, were transfected with a human Tyk2 cDNA library containing randomly generated point mutations in the KL domain. The library was estimated to contain around 3,400 independent clones, each with 0–3 point mutations in the KL coding region. Because 11,1 cells contain an integrated IFNα/β-responsive promoter driving the bacterial guanine phosphoribosyltransferase gene (gpt), neomycin-resistant colonies were assayed for IFNα/β-induced gpt expression in 6TG or hypoxanthine/aminopterin/thymidine-containing medium as a measurement of their transcriptional response to IFNα/β (6, 11). A total of 128 IFNα-unresponsive colonies were selected on the basis of their ability to survive in 6TG and IFNα (500 IU/ml). Of the 26 colonies that were found to express full-length Tyk2 protein, 7 were chosen for further analysis. To retrieve the mutated KL sequences, genomic DNA was isolated and used to amplify the 1.1-kb KL coding region, which was then directly sequenced. One point mutation from each of the 7 KL sequences was chosen for further study. For sequences carrying more than one point mutation, we chose the mutation that was most likely to be responsible for the loss-of-function phenotype, based on the nature of the substitution and the conservation of the mutated residue within the JAK family. From this analysis, we identified the following point mutations: V584D, G596V, S613G, E657V, H669P, Y689D, and R856G (Fig. 1). Four of the seven residues, G596, E657, H669, and R856, are found in all KL domains of vertebrate JAKs. R856 is also conserved in Drosophila Hopscotch.
Figure 1.
(A) Point mutations in the KL domain of Tyk2 analyzed in this study. The KL domain is represented by the rectangle. Residues are color-coded as described below. (B) Predicted locations of Tyk2 KL residues on the structure of the IRK. IRK residues corresponding to the Tyk2 KL residues analyzed here are highlighted on the crystal structure of the inactive form of IRK (PDB code 1IRK), the activation loop of which is shown in yellow. Residues whose substitutions in Tyk2 had no functional consequences (E611K, S613G, E657V, Y689D) are in blue. Residues whose substitutions in Tyk2 led to loss of function are in red and are labeled. The residue corresponding to the HopT42 gain-of-function mutation (E726K) is in green.
These point mutations were recreated individually into the pIRES-Tyk2 vector with a PCR-based method. During this procedure, a second point mutation, E611K (Fig. 1), was inadvertently introduced into the pIRES-S613G plasmid. The constructs were transfected into 11,1 cells. The IFN response of each pool of neomycin-resistant colonies was tested by assaying their phenotype in 6TG and IFNα. Cells transfected with pIRES-Tyk2 did not survive, nor did cells transfected with pIRES-E611K/S613G, -E657V, or -Y689D. Based on this criterion, these four point mutations did not affect Tyk2 function. In contrast, cells transfected with pIRES-V584D, -G596V, -H669P, or -R856G survived in the selective medium, demonstrating that these four point mutations impaired Tyk2 function.
Point Mutations in KL Lead to Loss of High-Affinity IFNα Binding.
Individual clones expressing comparable levels of the WT, V584D, G596V, H669P, or R856G proteins were generated. Two clones were routinely used for further characterization. Sensitivity to IFNα was assayed by monitoring cell survival in 6TG and increasing doses of IFNα (Table 1). V584D-expressing and G596V-expressing cells were at least 100-fold less sensitive than WT-expressing cells, whereas R856G-expressing and H669P-expressing cells behaved like 11,1 cells. The response to IFNβ was similarly affected (Table 1), suggesting that the KL mutants exert comparable effects on signaling events induced by either IFNα or IFNβ.
Table 1.
Sensitivity to IFN of 11,1 cells stably reconstituted with Tyk2 point mutants
| Cell line | IFN conc. for 50% cell death, IU/ml
|
|
|---|---|---|
| IFNα2 | IFNβ | |
| 11,1 | >10,000 | 400 |
| WT | <10 | <40 |
| V584D | 1,000 | 40 |
| G596V | 1,000 | 40 |
| H669P | >10,000 | 400 |
| R856G | >10,000 | 400 |
Transcriptional response to IFNs was measured by using the 6TG cytotoxicity assay.
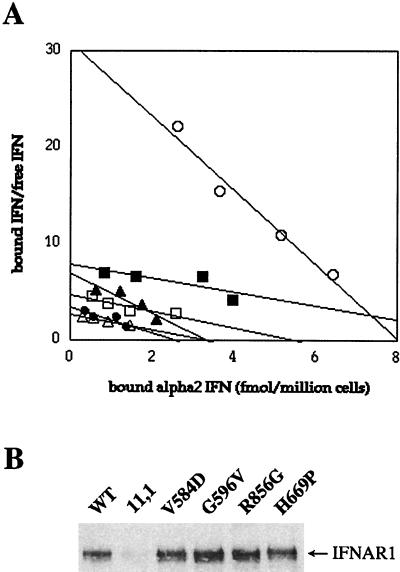
To understand how the point mutations in the KL domain might alter the function of Tyk2, we looked at major events along the IFN signaling pathway in cells expressing the mutants. Because, in human cells, the binding of IFNα is severely impaired in the absence of Tyk2 (23, 29), a comparative binding analysis of iodinated IFNα2 was performed. Fig. 2A shows that cells expressing the mutant Tyk2 proteins behaved like 11,1 cells, whereas only WT cells exhibited a binding curve characteristic of high-affinity IFNα binding. We have previously demonstrated that Tyk2 sustains the level of the IFNAR1 receptor subunit (24, 25). Therefore, we analyzed the steady-state level of IFNAR1 in the various clones to determine if the lack of high-affinity IFNα binding sites might be due to reduced IFNAR1 protein. All four Tyk2 mutants were as capable as WT Tyk2 at restoring IFNAR1 levels (Fig. 2B). Thus the mutations in the KL domain did not impinge on the ability of the protein to associate with and stabilize IFNAR1, although the reconstitution of high-affinity ligand binding was impaired.
Figure 2.
(A) Scatchard plot analysis of the IFNα binding activity of cells expressing the KL mutants. The binding of 125I-labeled IFNα2c on 11,1 cells (●) and 11,1 cells expressing WT Tyk2 (○) is compared with the binding on 11,1 cells expressing the V584D mutant (▴), G596V (■), R856G (▵), and H669P (□). (B) Steady-state level of IFNAR1. IFNAR1 was immunoprecipitated from cells, as indicated, and immunoblotted with an anti-IFNAR1 antibody.
Point Mutations in KL Impair Tyk2 Kinase Activity.
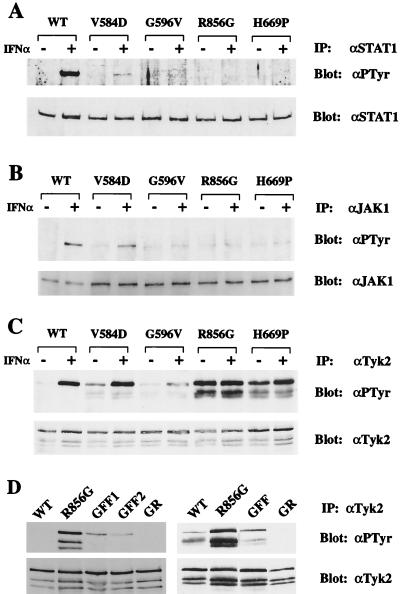
IFNα-induced gene expression, which is the basis of the 6TG cytotoxicity assay, is dependent on the phosphorylation and activation of STAT1 and STAT2. We therefore measured the phosphorylation state of these transcription factors. In WT cells, IFNα induced STAT1 phosphorylation (Fig. 3A). In IFN-treated V584D-expressing cells, weak STAT1 phosphorylation could be seen. In G596V-, H669P-, or R856G-expressing cells, no STAT1 phosphorylation was detected. Similar results were obtained for STAT2 (data not shown). Because STAT1 and STAT2 are targets of JAK1 and Tyk2, we next looked at the phosphorylation state of these kinases. In WT cells, IFNα induced the phosphorylation of JAK1 (Fig. 3B). In V584D-expressing cells, JAK1 phosphorylation was detectable but reduced. In contrast, no detectable JAK1 phosphorylation was observed in G596V-, H669P-, or R856G-expressing cells.
Figure 3.
In vivo phosphorylation levels of STAT1, JAK1, and Tyk2. (A) Cell lines expressing WT or mutant Tyk2 were treated with 103 IU/ml IFNα2 for 15 min. STAT1 was immunoprecipitated (IP) with an anti-STAT1 antibody (αSTAT1) and immunoblotted with an anti-phosphotyrosine antibody (αPTyr). The membrane was stripped and reprobed with an anti-STAT1 antibody to determine the level of immunoprecipitated STAT1. (B) JAK1 immunoprecipitates were immunoblotted with an anti-phosphotyrosine antibody. The membrane was stripped and reprobed with an anti-JAK1 antibody. In this experiment, more lysate was used from mutant-expressing cells than from WT cells to optimize the detection of phosphorylated JAK1. (C) Tyk2 immunoprecipitates were immunoblotted with an anti-phosphotyrosine antibody. The membrane was stripped and reprobed with an anti-Tyk2 antibody. The two lower bands, which have consistently been observed, correspond to Tyk2 degradation products. (D) Phosphorylation levels of the R856G/Y1054F/Y1055F and R856G/K930R mutants. (Left) Tyk2 was immunoprecipitated from stable clones expressing WT Tyk2, R856G, R856G/Y1054F/Y1055F (GFF, clones 1 and 2), or R856G/K930R (GR) proteins. Phosphorylation levels were detected by immunoblotting with an anti-phosphotyrosine antibody. The membrane was stripped and reblotted with an anti-Tyk2 antibody to determine protein levels. (Right) Immunoprecipitates of WT or mutant Tyk2 from transiently transfected 11,1 cells were analyzed as above.
The phosphorylation state of the Tyk2 mutants was then analyzed (Fig. 3C). Mutant V584D was as inducibly phosphorylated as WT Tyk2, whereas mutant G596V was weakly phosphorylated in response to IFNα. What was especially remarkable was the elevated constitutive phosphorylation of the H669P and R856G proteins, which approached that of the IFNα-induced WT Tyk2. We therefore investigated whether this hyperphosphorylation was occurring on Y1054 and/or Y1055, residues that were previously shown to be major sites of induced phosphorylation (6). These two tyrosines are located in the activation loop of the TK domain and are required for the catalytic activation of Tyk2 by IFNα. We generated two stable clones expressing the triple mutant R856G/Y1054F/Y1055F. As shown in Fig. 3D Left, the phosphorylation content of the R856G/Y1054F/Y1055F protein was significantly reduced compared with the R856G protein. To confirm this finding, WT Tyk2, R856G, and R856G/Y1054F/Y1055F proteins were overexpressed by transient transfection in 11,1 cells. As with the stable clones, the R856G mutant was more phosphorylated than WT Tyk2, and the R856G/Y1054F/Y1055F triple mutant was considerably less phosphorylated than the R856G mutant (Fig. 3D Right). These results demonstrate that Y1054 and/or Y1055 are required for the hyperphosphorylation of the R856G protein and suggest that these residues themselves may be constitutively phosphorylated.
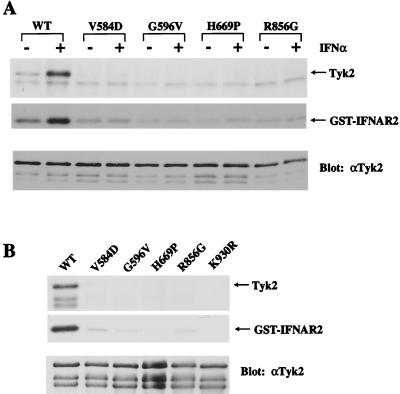
To determine whether this hyperphosphorylation depended on the intrinsic catalytic activity of the R856G mutant, we introduced the kinase-dead K930R mutation in the TK domain, which prevents ATP binding (6). The in vivo phosphorylation of this double mutant was abolished, both when expressed transiently and in stable clones (Fig. 3D), suggesting that intrinsic kinase activity was required for the phosphorylation. Next we measured the ability of the mutants to autophosphorylate and to phosphorylate an exogenous substrate in an in vitro kinase assay. Tyk2 immunoprecipitates from untreated or IFNα-treated cells were incubated with [γ-32P]ATP and a glutathione S-transferase fusion protein containing the cytoplasmic region of IFNAR2. In contrast to WT Tyk2, no autophosphorylation and only minimal substrate phosphorylation were seen in all of the mutants, regardless of IFNα treatment (Fig. 4A). Furthermore, in the absence of IFNα, the level of substrate phosphorylation was consistently lower for the KL mutants than for WT Tyk2. This was further demonstrated by measuring the kinase activity of overexpressed Tyk2 proteins transiently expressed in 11,1 cells (Fig. 4B). All four KL mutants were catalytically impaired and, in fact, behaved like the kinase-dead K930R mutant (6).
Figure 4.
In vitro kinase activity of Tyk2 KL mutants. (A) Stable cell lines were untreated or treated with 103 IU/ml IFNα2 for 15 min. Tyk2 immunoprecipitates were then assayed for autophosphorylation (Tyk2) and substrate [glutathione S-transferase (GST)-IFNAR2] phosphorylation activities. (Bottom) The membrane was immunoblotted with an anti-Tyk2 antibody to determine protein levels. (B) Tyk2 proteins were transiently overexpressed in 11,1 cells and assayed for kinase activity, as above. The faint GST-IFNAR2 background band can also be detected in 11,1 immunoprecipitates.
Discussion
From screening a library of Tyk2 cDNAs containing randomly mutated KL sequences, we identified eight KL point mutations, four of which (E611K, S613G, E657V, and Y689D) did not affect the ability of the protein to rescue IFNα signaling in Tyk2-deficient cells. The other four mutations (V584D, G596V, H669P, and R856G) gave rise to catalytically compromised proteins that could not rescue high-affinity IFNα binding. This latter defect cannot be directly attributed to the catalytic impairment of the KL mutants because we previously showed that the catalytically inactive mutant, K930R, is able to reconstitute high-affinity ligand binding, although with altered down-phase dynamics (6, 11). Like the K930R mutant, all four KL mutants were capable of stabilizing IFNAR1. Because expression of the amino-terminal region alone is sufficient for restoration of IFNAR1 at the cell surface (ref. 24 and unpublished observation), it is more likely that the KL domain is involved, instead, in maintaining the configuration of the high-affinity receptor-kinase complex through its interaction with receptor components (Fig. 5). Catalytic impairment resulting from point mutations in the KL domain was also shown recently for the JAK3 C759R mutant as well as for two JAK mutants containing the corresponding HopT42 mutation (Fig. 1B), JAK3 E639K and Tyk2 E726K (ref. 16 and unpublished observation).
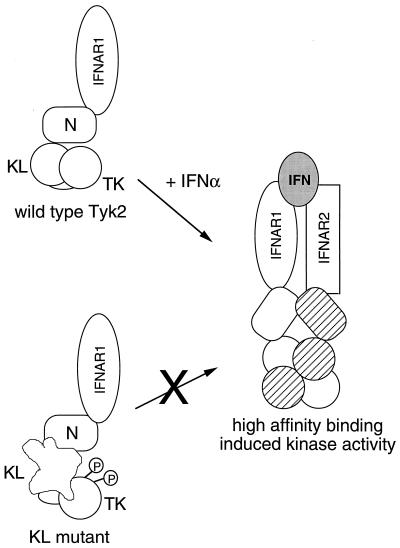
Figure 5.
A model of Tyk2 regulation in the IFNα receptor-kinase complex. Tyk2 is associated with IFNAR1 through its N region. The KL domain maintains the TK domain in an unphosphorylated and resting conformation. Severe disruption of KL leads to hyperphosphorylation of the protein. Upon IFNα binding, KL-dependent conformational changes allow transition of the receptor-kinase complex to a high-affinity binding state where Tyk2 and JAK1 are stabilized in their activated conformations.
Although the crystal structure of the KL domain has yet to be solved, its high degree of sequence identity with tyrosine kinase domains allowed us to localize the KL residues of interest, using the structure of the inactive IRK domain as a framework (28) (Fig. 1B). V584 is an internal residue, the substitution of which may affect the local structure within the small lobe. The fully exposed G596 residue corresponds to the first invariant glycine of the ATP-binding glycine-rich loop present in protein kinases (30, 31). Whether this divergent region in the KL domain binds nucleotides remains to be established. H669 is an internal residue located at the interface of the two lobes. R856 corresponds to an invariant arginine in the protein kinase superfamily that stabilizes the large lobe by forming an internal salt bridge with the glutamate of the highly conserved APE motif, which is also conserved in KL domains (31).
The four KL mutants can be separated into two classes based on the predicted structural locations of their mutated residues, their ability to weakly restore IFNα responsiveness, and their phosphorylation states. The V584D and G596V mutants, which bear mutations unlikely to perturb the large lobe of the KL domain, are able to rescue sensitivity to high doses of IFNα and are not hyperphosphorylated. G596V-expressing cells behave like V584D-expressing cells in the 6TG cytotoxicity assay despite undetectable JAK1 and STAT1/2 phosphorylation, suggesting that induction of the 6–16 promoter does not require high amounts of phosphorylated JAK1 and STAT1/2. In contrast, the H669P and R856G mutants, which carry mutations likely to cause major structural alterations of the KL large lobe, cannot rescue IFNα sensitivity and are hyperphosphorylated. Thus the KL domain serves to keep the protein in a low-phosphorylated conformation (Fig. 5). In the V584D and G596V mutants, this function of KL is preserved, whereas in severely disrupted mutants, such as R856G and H669P, this control is relieved and the protein becomes prone to nonactivating hyperphosphorylation. That severe disruption of KL leads to an increase in the phosphorylation of the protein is supported by other studies. Tyk2 and JAK2 ΔKL mutants were reported to be constitutively phosphorylated (11, 15), as were two JAK3 KL mutants from severe combined immunodeficiency patients, one bearing a small deletion and the other, the C759R substitution (16, 17). Like R856 in Tyk2, C759 in JAK3 corresponds to a highly conserved internal residue in the large lobe of the kinase domain.
These concordant findings raise the questions of what mechanism and what kinase are responsible for this hyperphosphorylation. KL may provide an interaction surface for a tyrosine phosphatase that maintains the protein in a low phosphorylated state (32, 33). Alternatively, the structural alteration of KL may have repercussions on the conformation of other regions of the protein, leading to the increased accessibility of target tyrosines. Our finding that hyperphosphorylation of the R856G protein was significantly reduced upon phenylalanine substitution of the activation loop tyrosines suggests that these tyrosines are principal targets of hyperphosphorylation or that they are at least essential for phosphorylation at other sites. The inability to detect in vitro autophosphorylation of the mutants suggests the involvement of another kinase. Surprisingly, however, introducing the K930R mutation of the ATP-binding site into the R856G protein abolished its in vivo phosphorylation. The same result was reported for the JAK3 C759R mutant containing the corresponding K855A mutation (16). Thus intrinsic catalytic activity that is undetectable in vitro or the presence of this lysine residue is required for the hyperphosphorylation. Autophosphorylation on an inhibitory tyrosine is a possibility, although this would not account for the impairment of the V584D and G596V mutants, which themselves are not hyperphosphorylated. Another possibility is that weak residual kinase activity leads to in vivo accumulation of phosphorylation, especially if it is not counterbalanced by a KL-associated phosphatase.
We have never obtained a mutant form of Tyk2 that rescues high-affinity IFNα binding without also rescuing signaling in 11,1 cells, suggesting a tight interdependence between intracellular and extracellular events. The binding of IFNα to the receptor-kinase complex brings about the juxtaposition of Tyk2 and JAK1 and their transphosphorylation, which was previously shown to occur in a defined temporal order, in which active JAK1 is an essential prerequisite for Tyk2 activation (6, 34). Interestingly, the V584D mutant in this study was as inducibly phosphorylated as WT Tyk2, despite the absence of high-affinity ligand binding, suggesting that Tyk2 transphosphorylation by JAK1 might even precede and be required for the establishment of high-affinity binding sites. We propose that the KL domain plays a role in the assembly of a receptor-kinase complex endowed with both high-affinity IFNα binding and induced catalytic activity through its interactions with other receptor components (Fig. 5). KL may provide a dimerization surface for juxtaposed KL and/or TK domains. To this end, interaction of the two kinase domains was demonstrated for JAK1 KL and TK domains expressed in insect cells (35) and, more recently, for JAK3 KL and TK domains overexpressed in COS cells (16). Our finding that Tyk2 KL mutants are catalytically impaired is consistent with the existence of a critical interaction between the two domains.
Whether the KL domain of JAK1 would function similarly has to be addressed. IFNα/β-unresponsive U4 mutant cells, which lack endogenous JAK1, are similar to 11,1 cells in that they also lack high-affinity IFNα binding sites (ref. 1 and unpublished observation), suggesting parallel functions of Tyk2 and JAK1, and of their KL domains, in this receptor-kinase complex.
Acknowledgments
We thank P. Alzari for his help with Fig. 1B, V. Barreto for construction of pRc/E726K, H. K. Lorenzo for his help with sequence alignments, and J. Ragimbeau for technical help. We thank P. Eid, C. Schindler, and A. Ziemiecki for reagents; O. Acuto, P. Alzari, V. Di Bartolo, and M. C. Gauzzi for insightful discussions and critical reading of the manuscript; and M. Fellous and M. Tosi for use of automatic sequencing facilities. This work, initiated in the Institut National de la Santé et de la Recherche Médicale (INSERM) Unit 276, was supported by INSERM, the Centre National de la Recherche Scientifique, and the Association pour la Recherche sur le Cancer. Work at the Institut de Génétique Moléculaire, Montpellier, was supported in part by a grant from the Association pour la Recherche sur le Cancer. T.C.Y. is supported by a postdoctoral fellowship from the National Arthritis Foundation. E.D. is supported by a postdoctoral fellowship from the European Community.
Abbreviations
- JAK
Janus kinase
- STAT
signal transducers and activators of transcription
- KL
kinase-like
- TK
tyrosine kinase
- 6TG
6-thioguanine
- WT
wild type
- IU
international units
- IRK
insulin receptor kinase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.160130297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.160130297
References
- 1.Darnell J E, Jr, Kerr I M, Stark G R. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 2.Ihle J N. Nature (London) 1995;377:591–594. doi: 10.1038/377591a0. [DOI] [PubMed] [Google Scholar]
- 3.Yeh T C, Pellegrini S. Cell Mol Life Sci. 1999;55:1523–1534. doi: 10.1007/s000180050392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Darnell J E., Jr Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 5.Decker T, Kovarik P. Cell Mol Life Sci. 1999;55:1535–1546. doi: 10.1007/s000180050393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gauzzi M C, Velazquez L, McKendry R, Mogensen K E, Fellous M, Pellegrini S. J Biol Chem. 1996;271:20494–20500. doi: 10.1074/jbc.271.34.20494. [DOI] [PubMed] [Google Scholar]
- 7.Feng J, Witthuhn B A, Matsuda T, Kohlhuber F, Kerr I M, Ihle J N. Mol Cell Biol. 1997;17:2497–2501. doi: 10.1128/mcb.17.5.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu K D, Gaffen S L, Goldsmith M A, Greene W C. Curr Biol. 1997;7:817–826. doi: 10.1016/s0960-9822(06)00369-1. [DOI] [PubMed] [Google Scholar]
- 9.Zhou Y-J, Hanson E P, Chen Y-Q, Agnuson K, Chen M, Swann P G, Wange R L, Changelian P S, O'Shea J J. Proc Natl Acad Sci USA. 1997;94:13850–13855. doi: 10.1073/pnas.94.25.13850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernards A. Oncogene. 1991;6:1185–1187. [PubMed] [Google Scholar]
- 11.Velazquez L, Mogensen K E, Barbieri G, Fellous M, Uzé G, Pellegrini S. J Biol Chem. 1995;270:3327–3334. doi: 10.1074/jbc.270.7.3327. [DOI] [PubMed] [Google Scholar]
- 12.Luo H, Rose P, Barber D, Hanratty W P, Lee S, Roberts T M, D'Andrea A D, Dearolf C R. Mol Cell Biol. 1997;17:1562–1571. doi: 10.1128/mcb.17.3.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frank S J, Yi W, Zhao Y, Goldsmith J F, Gilliland G, Jiang J, Sakai I, Kraft A. J Biol Chem. 1995;270:14776–14785. doi: 10.1074/jbc.270.24.14776. [DOI] [PubMed] [Google Scholar]
- 14.Zhao Y, Wagner F, Frank S J, Kraft A S. J Biol Chem. 1995;270:13814–13818. doi: 10.1074/jbc.270.23.13814. [DOI] [PubMed] [Google Scholar]
- 15.Kohlhuber F, Rogers N C, Watling D, Feng J, Guschin D, Briscoe J, Witthuhn B A, Kotenko S V, Pestka S, Stark G R, et al. Mol Cell Biol. 1997;17:695–706. doi: 10.1128/mcb.17.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen M, Cheng A, Candotti F, Zhou Y, Hymel A, Fasth A, Notarangelo L, O'Shea J. Mol Cell Biol. 2000;20:947–956. doi: 10.1128/mcb.20.3.947-956.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Candotti F, Oakes S A, Johnston J A, Giliani S, Schumacher R F, Mella P, Fiorini M, Ugazio A G, Badolato R, Notarangelo L D, et al. Blood. 1997;90:3996–4003. [PubMed] [Google Scholar]
- 18.Fujitani Y, Hibi M, Fukuda T, Takahashi-Tezuka M, Yoshida H, Yamaguchi T, Sugiyama K, Yamanaka Y, Nakajima K, Hirano T. Oncogene. 1997;14:751–761. doi: 10.1038/sj.onc.1200907. [DOI] [PubMed] [Google Scholar]
- 19.Domanski P, Colamonici O R. Cytokine Growth Factor Rev. 1996;7:143–151. doi: 10.1016/1359-6101(96)00017-2. [DOI] [PubMed] [Google Scholar]
- 20.Mogensen K E, Lewerenz M, Reboul J, Lutfalla G, Uzé G. J Interferon Cytokine Res. 1999;19:1069–1098. doi: 10.1089/107999099313019. [DOI] [PubMed] [Google Scholar]
- 21.Velazquez L, Fellous M, Stark G R, Pellegrini S. Cell. 1992;70:313–322. [PubMed] [Google Scholar]
- 22.Stark G R, Kerr I M, Williams B R G, Silverman R H, Schreiber R D. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 23.Pellegrini S, John J, Shearer M, Kerr I M, Stark G R. Mol Cell Biol. 1989;9:4605–4612. doi: 10.1128/mcb.9.11.4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gauzzi M C, Barbieri G, Richter M F, Uzé G, Ling L, Fellous M, Pellegrini S. Proc Natl Acad Sci USA. 1997;94:11839–11844. doi: 10.1073/pnas.94.22.11839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richter M F, Duménil G, Uzé G, Fellous M, Pellegrini S. J Biol Chem. 1998;273:24723–24729. doi: 10.1074/jbc.273.38.24723. [DOI] [PubMed] [Google Scholar]
- 26.Cadwell R C, Joyce G F. PCR Methods Appl. 1992;2:28–33. doi: 10.1101/gr.2.1.28. [DOI] [PubMed] [Google Scholar]
- 27.Barbieri G, Velazquez L, Scrobogna M, Fellous M, Pellegrini S. Eur J Biochem. 1994;223:427–435. doi: 10.1111/j.1432-1033.1994.tb19010.x. [DOI] [PubMed] [Google Scholar]
- 28.Hubbard S R, Wei L, Ellis L, Hendrickson W A. Nature (London) 1994;372:746–754. doi: 10.1038/372746a0. [DOI] [PubMed] [Google Scholar]
- 29.Uzé G, Lutfalla G, Mogensen K E. J Interferon Cytokine Res. 1995;15:3–26. doi: 10.1089/jir.1995.15.3. [DOI] [PubMed] [Google Scholar]
- 30.Bossemeyer D. Trends Biochem Sci. 1994;19:201–205. doi: 10.1016/0968-0004(94)90022-1. [DOI] [PubMed] [Google Scholar]
- 31.Hanks S K, Hunter T. In: The Protein Kinase Facts Book: Protein-Tyrosine Kinases. Hardie G, Hanks S, editors. London: Academic; 1995. pp. 7–47. [Google Scholar]
- 32.David M, Chen H E, Goelz S, Larner A C, Neel B G. Mol Cell Biol. 1995;15:7050–7058. doi: 10.1128/mcb.15.12.7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yetter A, Uddin S, Krolewski J J, Jiao H, Yi T, Platanias L. J Biol Chem. 1995;270:18179–18182. doi: 10.1074/jbc.270.31.18179. [DOI] [PubMed] [Google Scholar]
- 34.Briscoe J, Rogers N C, Witthuhn B A, Watling D, Harpur A G, Wilks A F, Stark G R, Ihle J N, Kerr I M. EMBO J. 1996;15:799–809. [PMC free article] [PubMed] [Google Scholar]
- 35.Chang M-S, Chang G-D, Leu J-H, Huang F-L, Chou C-K, Huang C-J, Lo T-B. DNA Cell Biol. 1996;15:827–844. doi: 10.1089/dna.1996.15.827. [DOI] [PubMed] [Google Scholar]