Abstract
Barrier-to-autointegration factor (BAF) is a highly conserved cellular protein that was identified by its activity in protecting retroviral DNA against autointegration. We show that BAF has the property of bridging double-stranded DNA in a highly ordered nucleoprotein complex. Whereas BAF protein alone is a dimer in solution, upon binding DNA, BAF forms a dodecamer with DNA bound at multiple discrete sites in the complex. The interactions between BAF and DNA are entirely nonspecific with respect to DNA sequence. The dual interaction of BAF with DNA and LAP2, a protein associated with the nuclear lamina, suggests a role for LAP2 in chromosome organization. Consistent with this idea, RNA interference experiments with Caenorhabditis elegans reveal a defect in mitosis.
Mobile genetic elements have evolved diverse mechanisms to reduce the frequency of self-destructive insertion into their own genomes. Examples include Mu B-mediated target immunity in phage Mu transposition (1) and the utilization of highly preferred target sites that are not present on the transposon itself (2). Retroviruses appear to have exploited yet another strategy to accomplish the same end. Synthesis of retroviral DNA occurs within a nucleoprotein complex derived from the core of the infecting virion (3). The DNA within preintegration complexes efficiently integrates into exogenously added DNA in vitro, but autointegration into itself is much less efficient. Barrier-to-autointegration factor (BAF) was identified as a cellular protein that blocks autointegration of murine retroviral DNA (4, 5). Stripping of BAF from preintegration complexes removed the block to autointegration, and subsequent incubation with BAF restored the barrier to autointegration (5). BAF also restores the integration activity of salt-stripped HIV preintegration complexes (6).
BAF is an 89-aa protein whose amino acid sequence exhibits no obvious similarity to other known proteins (5). It is highly conserved among species, with 86 of 89 residues identical between human and mouse and a 60% identity between the human and Caenorhabditis elegans homologues; furthermore, essentially all of the substitutions are conservative (7). BAF is a dimer in solution, and the structure of this dimer has been determined by NMR (7) and by x-ray crystallography (8). The three-dimensional structure of BAF also is dissimilar to any known protein.
We previously proposed that BAF blocks autointegration by compacting the viral DNA, thereby making it inaccessible as a target for integration (5). The ability of BAF to aggregate DNA, together with its dimeric structure in solution, suggested the possibility that each BAF monomer may bind one DNA (7). In contrast to this simple model for DNA bridging by BAF, in this paper we show that BAF forms discrete, higher-order structures with DNA bound at multiple sites in the complex. The role of BAF for the host cell remains to be elucidated. However, the ability of BAF to compact DNA suggests a possible role in chromosome organization. The finding that lamina-associated protein 2 (LAP2), a protein associated with the nuclear lamina, interacts with BAF in a two-hybrid screen (9) also supports this possibility.
Materials and Methods
Cloning, Expression, and Purification of BAF.
BAF cDNA was cloned from a human liver cDNA library by PCR by using primers designed from the murine BAF sequence (5). The BAF coding sequence was inserted into pET15B (Novagen), and the protein was expressed and purified by Ni2+-affinity chromatography under denaturing conditions as described (7). Subsequent to this step the purification procedure was modified as follows. The fractions containing BAF were pooled, and EDTA was added to 10 mM. The pooled fractions were dialyzed against 50 mM potassium acetate, pH 5.5/200 mM NaCl/10 mM EDTA/10% glycerol/5 mM 2-mercaptoethanol and then against 50 mM potassium phosphate, pH 6.5/200 mM NaCl/1 mM EDTA/10% glycerol/5 mM DTT. The histidine tag was cleaved off with thrombin, and the thrombin was removed by passage over benzamidine-Sepharose (7). Finally, BAF was applied to a Superdex 200 column (Pharmacia) equilibrated with 20 mM Tris, pH 7/150 mM NaCl/10% glycerol/5 mM DTT/0.1 mM EDTA. Fractions containing BAF dimers were pooled and stored at −80°C.
DNA Substrates.
Oligonucleotides were purified by electrophoresis in a denaturing 6% polyacrylamide gel. They were 5′ end-labeled with [γ-32P]ATP by using T4 polynucleotide kinase and annealed with their complementary strand. Unincorporated label was removed by using a Micro Bio-Spin column (Bio-Rad). Except where stated otherwise, the substrate for DNA binding (substrate A) was RZ132 (5′-GTGTGGAAAATCTCTAGCAGT) annealed to RZ61 (5′-ACTGCTAGAGATTTTCCACAC). Substrates B and C in Fig. 1 were RZ159 (5′-GGAAGAGGGAGAAGAGGGAGG)/RZ160 (5′-CCTCCCTCTTCTCCCTCTTCC) and RZ161 (5′-TTAATATTTATAATATTTATT)/RZ162 (5′-AATAAATATTATAAATATTAA), respectively. The “long” DNA of Fig. 3 was RZ132 with a 20-base poly(dA) tail added at the 5′ end annealed with RZ61.
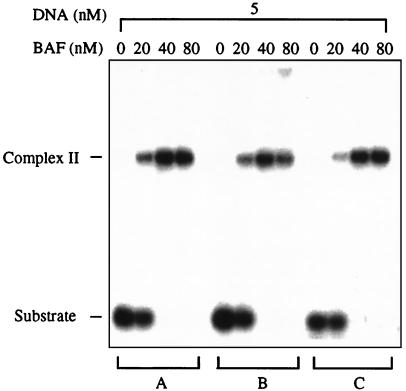
Figure 1.
BAF binds DNA nonspecifically. Gel mobility-shift assay of BAF binding to three different 21-mer duplex oligonucleotides (A, B, and C). Binding was carried out at 30°C for 30 min. Samples then were loaded onto a 4–20% polyacrylamide gel in TBE buffer, electrophoresed at 8 V/cm for 3 h on ice, and imaged by using a Molecular Dynamics PhosphorImager.
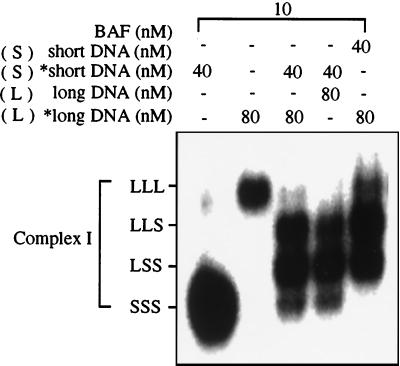
Figure 3.
Complex I contains three DNA molecules. BAF was bound to short, long, or a mixture of long and short duplex oligonucleotides as indicated. The short oligonucleotide is the substrate A shown in Fig. 1. The long oligonucleotide is the same duplex as substrate A with a 20-base, single-strand tail added to retard its electrophoretic mobility. An asterisk indicates that the DNA is labeled with 32P. The gel was electrophoresed for an extended period to resolve the multiple species of complex I with different electrophoretic mobilities. The presence of both short and long DNA results in the splitting of complex I into four bands. We interpret this to indicate the presence of three DNA molecules in complex I, with all four possible combinations of short and long DNA molecules.
DNA-Binding Assays.
Each reaction mixture (10 μl) contained 10 mM Tris⋅HCl, pH 7.0/20 mM NaCl/100 ng/ml BSA/5 mM DTT and various BAF and DNA concentrations as indicated; concentrations of BAF and DNA are expressed as molar concentrations of BAF monomer and duplex oligonucleotide, respectively. Binding was carried out at 30°C for 30 min, except where indicated otherwise. Samples were loaded onto a 4–20% polyacrylamide gel in TBE buffer (89 mM Tris/89 mM boric acid/2 mM EDTA, pH 8.3) and electrophoresed at 8 V/cm for 3 h on ice unless otherwise indicated. The signal was detected by a Molecular Dynamics PhosphorImager and analyzed by imagequant software. The experiment shown in Fig. 3 differed from the standard conditions in that binding was carried out in buffer containing 100 mM NaCl on ice for 10 min; these conditions favor formation of complex I. The gel was electrophoresed for 17 h to resolve the different species of complex I.
Sedimentation Equilibrium Analysis.
Fresh samples of BAF were purified by gel filtration and dialyzed exhaustively against 0.2 M NaCl/0.1 M sodium phosphate buffer, pH 6.5/5 mM DTT at room temperature. Dialysis was carried out under an inert atmosphere with degassed buffer solutions. Samples of BAF were loaded into the ultracentrifuge cells under an inert nitrogen atmosphere and studied at nominal loading concentrations of 0.50 A280. Experiments also were carried out by diluting a concentrated stock solution of BAF in 20 mM NaCl/10 mM Tris⋅HCl, pH 7/5 mM 2-mercaptoethanol (buffer A). Loading BAF concentrations correspond to an A280 of 0.50. Double-stranded 21-mer DNA (RZ132/RZ61) was dissolved in buffer A to a final A260 of 0.65. Mixtures of DNA and BAF were prepared by mixing aliquots of the corresponding stock solutions in buffer A. These corresponded to 1:1 (2.70 μM) and 1:2 (1.35 and 2.70 μM) DNA/BAF ratios at the DNA concentrations indicated. Before analysis by analytical ultracentrifugation, the mixtures were incubated at 30°C for 30 min.
Sedimentation equilibrium experiments were conducted at 20°C in a Beckman Optima XL-A analytical ultracentrifuge. Individual DNA and BAF samples were studied at various rotor speeds, 14,000, 18,000, 20,000, and 22,000 rpm. Data were acquired as an average of 25 absorbance measurements at a nominal wavelength of 260 nm (DNA) and 280 nm (BAF) and a radial spacing of 0.001 cm. Equilibrium was achieved within 26 h. Data were analyzed in terms of a single ideal solute to obtain the buoyant molecular mass, M1(1 − v1ρ), using the Optima XL-A data analysis software (Beckman) running under microcal origin 3.78, by fitting data from each scan to
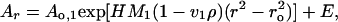 |
where Ao,1 is the absorbance at a reference point ro, Ar is the absorbance at a given radial position r, H represents ω2/2RT, ω is the angular speed in rad⋅s−1, R is the gas constant, T is the absolute temperature, and E is a small baseline correction determined experimentally by collecting data at 42,000 rpm. Residuals were calculated. A random distribution of the residuals around zero (±0.02) was obtained as a function of the radius. Values of M were obtained from the buoyant molecular mass, M1(1 − v1ρ), and calculated by using densities, ρ, at 20.0°C obtained from standard tables. A value of v = 0.7355 ml⋅g−1 was calculated for BAF based on the amino acid composition by using the consensus data for the partial specific molar volumes of amino acids (10).
Sedimentation equilibrium experiments on 1:1 and 1:2 DNA/BAF mixtures were carried out at 20.0°C and rotor speeds of 6,000, 8,000, and 10,000 rpm. Data were acquired at 260 and 285 nm, and equilibrium was reached within 48 h. Data were analyzed in terms of two ideal solutes to obtain the buoyant molecular mass, M2(1 − v2ρ), by fitting data from each scan to
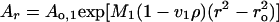 |
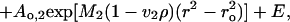 |
where Ao,2 is the absorbance at a reference point ro of species 2. The value of M1(1 − v1ρ) was set to the buoyant molecular mass of the DNA. Residuals were calculated. A random distribution of the residuals around zero (±0.02) was obtained as a function of the radius.
BAF RNA Interference (RNAi).
The cDNA clone yk333d11 was kindly provided by Y. Kohara and colleagues (National Institute of Genetics, Mishima, Japan) and used to make double-stranded RNA after linearization as described previously (11). Double-stranded RNA was resuspended at approximately 0.5 mg/ml and injected into the gonads of adult hermaphrodite C. elegans. The animals were allowed to recover overnight and then transferred to new plates daily. Progeny embryos arising on the second day after injection were arrested with near-100% penetrance at stages less than 100 cells. Wild-type and arrested embryos were collected, fixed in methanol and acetone by standard methods (12), and stained with 4′,6-diamidino-2-phenylindole (DAPI) and a mouse monoclonal anti-α-tubulin antibody (Sigma) followed by rhodamine-conjugated goat-anti-mouse IgG secondary antibody (Jackson ImmunoResearch). All images are at ×400 and were captured with a Photometrics SenSys charge-coupled device camera (Roper Scientific) by using ip labs software (Scanalytics, Billerica, MA) and compiled with Adobe photoshop.
Results
BAF Binds DNA Nonspecifically.
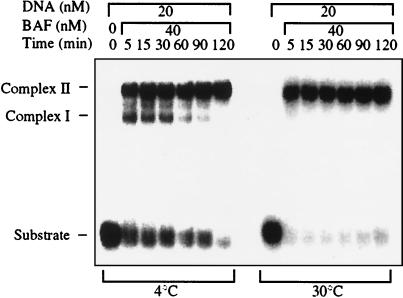
We assayed the DNA-binding activity of BAF by using a gel-shift assay. Double-stranded 21-mer oligonucleotides formed discrete complexes with slower electrophoretic mobility upon incubation with BAF. All double-stranded sequences tested bound BAF with indistinguishable affinities (Fig. 1). No detectable binding to single-strand DNA or RNA was observed (data not shown). DNA substrates longer than 25–30 bp formed large aggregates that did not enter the gel, and the affinity for BAF decreased markedly as the DNA length was reduced to less than about 18 bp (data not shown). The kinetics of binding at 4°C and 30°C are shown in Fig. 2. At 4°C two distinct complexes were present at early time points, the faster-migrating complex I and the more slowly migrating complex II. After 2 h all of the DNA substrate was converted into complex II. The kinetics were much faster at 30°C, and all of the substrate was converted to complex II within 5 min. These kinetics suggest that complex I may be an intermediate in the pathway to formation of complex II. However, at a higher stoichiometry of DNA/BAF, some complex I remains even after extended incubation (data not shown).
Figure 2.
BAF forms two types of complexes with DNA. Time course of binding of BAF to oligonucleotide A (Fig. 1) at 4°C and at 30°C was assayed by gel mobility shift. At the lower temperature, complexes with two different mobilities (complex I and complex II) are observed at early time points.
BAF Bundles Multiple DNAs in a Discrete Nucleoprotein Structure.
Because BAF is a dimer in solution we initially speculated that complex I contains two DNA molecules, one bound to each monomer within the dimer, and that complex II may be a multimer of complex I. To test this hypothesis, complexes were formed with a mixture of two DNAs that differ in their electrophoretic mobility. Binding was carried out with a stoichiometry of BAF/DNA that results in the formation of both complex I and complex II. The “short” DNA was the same as that shown in Fig. 2, and a single-strand tail was added to this DNA to make the “long” DNA substrate; the single-strand tail retards the electrophoretic mobility but does not allow multiple units of BAF to aggregate the DNA as would occur if the duplex were extended. We predicted that complex I would split into three bands with this mixed DNA substrate, corresponding two long, one long plus one short, and two short DNAs. Contrary to our expectation, complex I split into four bands with the mixed DNA substrate (Fig. 3). We interpret this result to indicate that complex I contains three DNA molecules; one intermediate band contains two long DNAs plus one short DNA, and the other intermediate band contains two short DNAs plus one long DNA. The dependence on whether the long, the short, or both DNAs were labeled, and the relative abundance of the products, is also fully consistent with this interpretation. We also attempted to use the same strategy to determine the number of DNA molecules in complex II. Although complex II clearly splits into more than four bands, the resolution was insufficient to unambiguously count the number (data not shown); however, repeated experiments indicated the presence of between five and seven bands, indicating the presence of four to six DNA molecules.
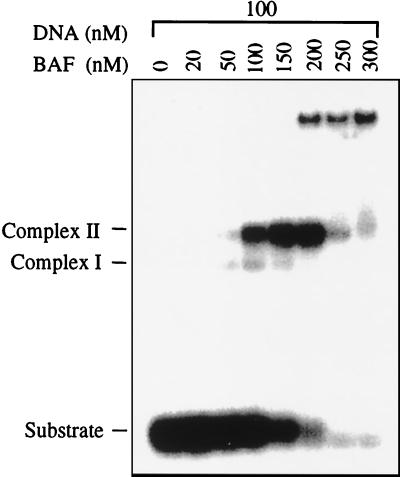
Stoichiometry of BAF/DNA in Complex II.
The stoichiometry of BAF/DNA in complex II was examined by titrating BAF at a fixed concentration of DNA (Fig. 4). At a stoichiometry of two BAF monomers per DNA molecule essentially all of the DNA formed complex II. DNA was lost at higher concentrations of BAF, presumably because of formation of aggregates that did not enter the gel. The stoichiometry of BAF/DNA in complex I could not be determined by this method because we were unable to find conditions in which all of the DNA substrate could be driven into complex I without the formation of a significant fraction of complex II.
Figure 4.
Stoichiometry of BAF/DNA in complex II. BAF was titrated with oligonucleotide A kept at a fixed concentration of 100 nM. This is well above the Kd under these binding conditions (data not shown), and, therefore, the ratio of BAF/DNA at which all of the DNA is incorporated into complex II reflects the stoichiometry of BAF/DNA in the complex. This titration suggests that the molar ratio of BAF monomer/duplex DNA in complex II is close to 2:1.
The mass of Complex II was determined by equilibrium ultracentrifugation. Because sedimentation equilibrium experiments yield the buoyant molecular mass M(1 − vρ), rather than the molecular mass itself, experiments first were performed to determine the values of M(1 − vρ) of the components. Both the BAF and DNA were found to be monodisperse with buoyant molecular masses of 5,400 ± 120 and 6,030 ± 100 g/mol, respectively (Fig. 5 and Table 1). These values showed that BAF is dimeric, having an experimentally determined molecular mass of 21,200 ± 500 g/mol, whereas the DNA is monomeric and characterized by a v of 0.557 ml/g. Sedimentation equilibrium data collected for different loading ratios and concentrations of DNA/BAF were best analyzed in terms of two single, ideal solutes corresponding to free DNA [M1(1 − v1ρ] = 6,030 g/mol and the protein–nucleic-acid complex (see Fig. 5 legend). In all cases, excellent fits were obtained and identical values of M2(1 − v2ρ) were returned at different rotor speeds. The buoyant mass of complex II was calculated to be 65,300 ± 3,900 g/mol, corresponding to a DNA/BAF stoichiometry of 5:12 or 6:12, assuming that at least one BAF dimer is required for DNA binding. No evidence for an intermediate complex was observed in equilibrium sedimentation experiments.
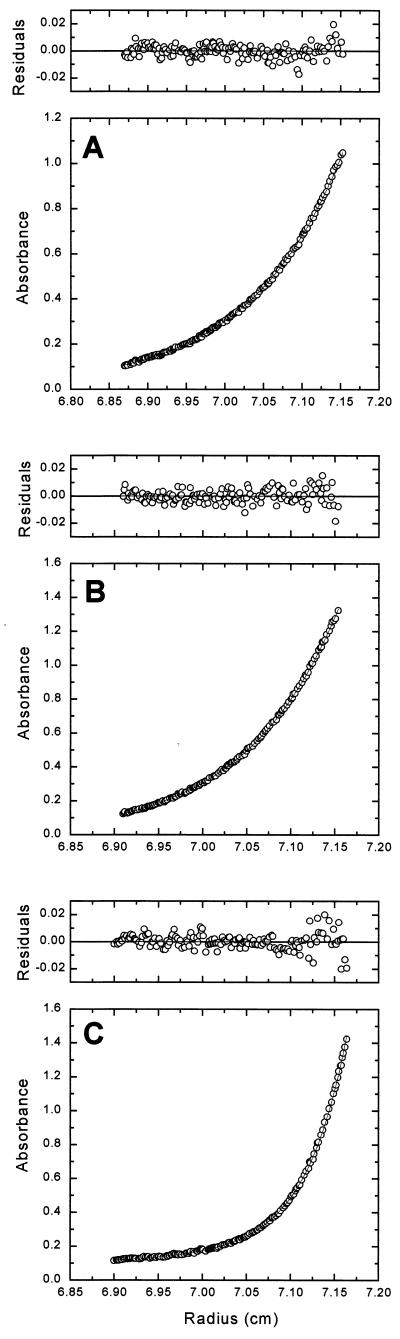
Figure 5.
(A) BAF is a single, monodisperse dimer. A sedimentation equilibrium profile at 280 nm for BAF is shown as a distribution of A280 at equilibrium. Data were collected at 20.0°C and 22,000 rpm. The results are analyzed for the best single-component M(1 − vρ) fit, shown as a line through the experimental points. The corresponding distribution of the residuals is shown above the plot. (B) The double-stranded DNA oligomer is monodisperse. A sedimentation equilibrium profile at 260 nm for 21-mer dsDNA is shown as a distribution of A260 at equilibrium. Data were collected at 20.0°C and 22,000 rpm and analyzed as described in A. (C) BAF and DNA interact to form a monodisperse and large molecular mass complex. A sedimentation equilibrium profile at 260 nm for a 1:1 (2.70 mM) DNA/BAF mixture is shown as a distribution of A260 at equilibrium. Data were collected at 20.0°C and 10,000 rpm. Data are analyzed in terms of two ideal solutes, with the value of M1(1 − v1ρ) set to 6,030 g⋅mol−1. The best component M2(1 − v2ρ) fit, shown as a line through the experimental points, corresponds to a value of 66,500 ± 400 g⋅mol−1. The corresponding distribution of the residuals is shown above the plot. We note that, because the extinction coefficient ratio of DNA/(BAF)2 is approximately 20 at 260 nm, data collected at this wavelength reflect the state of the DNA. It is noted further that the buoyant molecular masses of the DNA and (BAF)2 are similar enough that they cannot be distinguished at the low rotor speeds used to characterize the large protein—DNA complex.
Table 1.
Summary of data on composition of complexes I and II
| Species | No. of DNAs | Buoyant mass, g/mol | BAF/DNA |
|---|---|---|---|
| DNA alone | 1 | 6,030 ± 100 | NA |
| BAF dimer | NA | 5,400 ± 100 | NA |
| Complex I | 3 | ND | ND |
| Complex II | 5 ± 1 | 65,300 ± 3,900 | ≈2:1 |
The predicted buoyant mass of a complex of six DNA molecules and six BAF dimers is 68,580 g/mol. NA, not applicable; ND, not determined.
The DNA-binding and equilibrium centrifugation data are summarized in Table 1. Based on these results, we propose that complex I is a hexamer of BAF bound to three DNA molecules and complex II is a dodecamer of BAF bound to five or six DNA molecules.
BAF Is Essential in C. elegans, and Its Deficiency Leads to a Block in Mitosis.
The intriguing DNA-binding properties of BAF led us to a preliminary investigation of its cellular function. Because BAF is highly conserved between humans and C. elegans, we chose to investigate its cellular role with this system by exploiting the technique of RNAi, whereby injection of double-stranded RNA of the gene sequence of interest can result in a knockout phenotype (11). Although this phenomenon is not well understood, it has been proposed that the natural function of RNAi may be involved in transposon silencing (13, 14). One possible pathway is that RNAi induces enzymatic degradation of the corresponding mRNA and, therefore, results in depletion of the targeted protein (13).
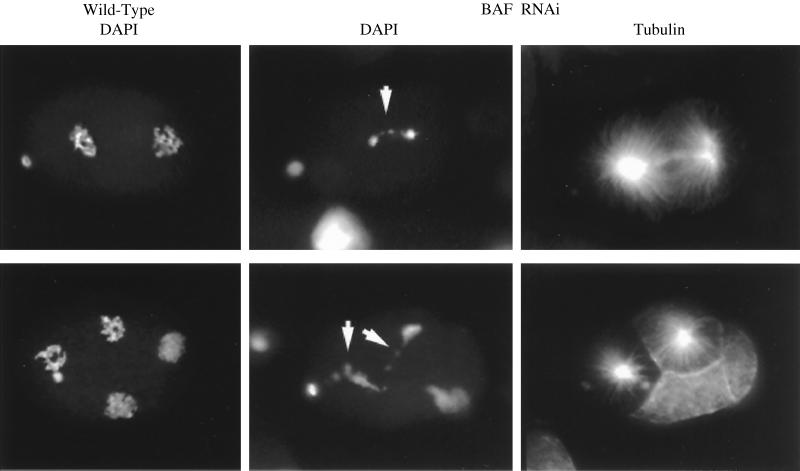
Double-stranded BAF RNA was injected into the gonads of C. elegans hermaphrodites, and the development of progeny embryos was followed by microscopy. Embryo development arrested at an early stage (<100 cells), with near 100% penetrance. Visualization of DNA with DAPI revealed abnormal segregation of chromosomes during mitosis, although spindle assembly appeared normal as judged by staining for tubulin (Fig. 6). Whereas condensed chromatin segregated as well defined clusters in wild-type embryos, those derived from C. elegans injected with the BAF RNAi exhibited trailing of chromatin between the separating clusters as if the daughter chromosomes were incompletely segregated. Although the phenotype reveals itself at the level of chromosome segregation during mitosis, identification of the primary defect and elucidation of the precise role of BAF require further investigation.
Figure 6.
Loss of BAF function causes a defect in chromatin segregation during mitosis. Injection of double-stranded C. elegans BAF RNA into the gonads of C. elegans resulted in an early arrest in embryo development. Two-cell (Upper) and four-cell (Lower) embryos for both wild-type and BAF RNAi are shown and stained with either DAPI or tubulin antibody as indicated. Arrested embryos invariably had a mitosis defect with a characteristic abnormal pattern of chromatin segregation, as revealed by DAPI staining. In contrast to the clean segregation of condensed chromatin in wild-type cells, a trail of chromatin linked the two sets of chromosomes at anaphase (white arrows). Staining for tubulin revealed a normal spindle. Embryo arrest likely is due to progressive aneuploidy after aberrant divisions.
Discussion
The DNA-bridging property of BAF is unique among known nonspecific DNA-binding proteins. Whereas BAF protein alone is a dimer in solution, the complex with DNA is formed from a dodecamer of BAF associated with five or six DNAs. We have not detected any association of DNA with the BAF dimer alone, suggesting that formation of the higher-order BAF multimer and DNA binding are coupled. Additional evidence for coupling of DNA binding and multimerization comes from studies with BAF mutant proteins that fail to bind DNA (data not shown). These mutant BAF proteins do not have a dominant-negative effect on the binding of wild-type BAF to DNA, suggesting that they also are unable to participate in the higher-order complex of BAF with DNA.
What role might BAF play for the cell? The bridging of DNA molecules provides an efficient means of compacting DNA, and an obvious possibility is that BAF is involved in organizing chromosomes. The ubiquitous expression of BAF mRNA among tissues (data not shown) suggests that it is essential for cell viability and serves such a housekeeping function rather than playing a regulatory role in cellular processes. Furthermore, the remarkable degree of conservation in the BAF amino acid sequence among species suggests that interactions with other macromolecules have constrained the divergence of BAF during evolution.
Chromosomes undergo major, dynamic structural changes during the cell cycle. Segregation of newly replicated chromosomes presents a daunting topological challenge that is, in part, solved by massive condensation of daughter chromatids before separation during mitosis. However, entanglement of DNA must be completely avoided during the condensation process, because even a single linkage would result in failure to segregate or a chromosomal break. Topoisomerases possess the necessary activity to unlink DNA, and they clearly play an important role in solving this problem. However, in the absence of a well defined pathway of condensation the task would appear insurmountable. In contrast to the extensive study of chromatin and its interaction with regulatory proteins at the nucleosome level, knowledge of the higher-order architecture of chromatin within the nucleus is still limited, although some proteins that play an important role in chromosome condensation have been identified (15). Based on the DNA-bridging activity of BAF, together with its association with metaphase chromosomes (9) and the RNAi phenotype in C. elegans, we propose that BAF plays a role in organizing chromosomes during mitosis.
Another important clue to the cellular role of BAF comes from the intriguing finding that it interacts with LAP2 (9, 16). LAP2, a protein associated with the nuclear lamina, can enhance DNA replication in Xenopus extracts (17) and may be involved in the expansion of reforming nuclei (17, 18). LAP2 protein includes distinct domains that are required for binding lamins and chromatin. The interaction of BAF with both LAP2 and DNA potentially provides a missing link in the chain of attachment of chromatin to the nuclear envelope.
Acknowledgments
We are indebted to Katherine Wilson for drawing our attention to the interaction of BAF with LAP2 and for stimulating discussions on the implications for nuclear assembly. We also thank Yuji Kohara for the expressed sequence tag clone of C. elegans BAF.
Abbreviations
- BAF
barrier-to-autointegration factor
- LAP2
lamina-associated protein 2
- RNAi
RNA interference
- DAPI
4′,6-diamidino-2-phenylindole
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.150240197.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.150240197
References
- 1.Adzuma K, Mizuuchi K. Cell. 1989;57:41–47. doi: 10.1016/0092-8674(89)90170-0. [DOI] [PubMed] [Google Scholar]
- 2.Craig N L. Annu Rev Biochem. 1997;66:437–474. doi: 10.1146/annurev.biochem.66.1.437. [DOI] [PubMed] [Google Scholar]
- 3.Brown P O. In: Retroviruses. Coffin J M, Hughes S H, Varmus H E, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 161–203. [Google Scholar]
- 4.Lee M S, Craigie R. Proc Natl Acad Sci USA. 1994;91:9823–9827. doi: 10.1073/pnas.91.21.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee M S, Craigie R. Proc Natl Acad Sci USA. 1998;95:1528–1533. doi: 10.1073/pnas.95.4.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen H M, Engelman A. Proc Natl Acad Sci USA. 1998;95:15270–15274. doi: 10.1073/pnas.95.26.15270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai M, Huang Y, Zheng R, Wei S Q, Ghirlando R, Lee M S, Craigie R, Gronenborn A M, Clore G M. Nat Struct Biol. 1998;5:903–909. doi: 10.1038/2345. [DOI] [PubMed] [Google Scholar]
- 8.Umland, T. C., Wei, S. Q., Craigie, R. & Davies, D. R. (2000) Biochemistry, in press. [DOI] [PubMed]
- 9.Furukawa K. J Cell Sci. 1999;112:2485–2492. doi: 10.1242/jcs.112.15.2485. [DOI] [PubMed] [Google Scholar]
- 10.Perkins S J. Eur J Biochem. 1986;157:169–180. doi: 10.1111/j.1432-1033.1986.tb09653.x. [DOI] [PubMed] [Google Scholar]
- 11.Fire A, Xu S Q, Montgomery M K, Kostas S A, Driver S E, Mello C C. Nature (London) 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 12.Miller D M, Shakes D C. In: Caenorhabditis elegans: Modern Biological Analysis of an Organism. Epstein H F, Shakes D C, editors. San Diego: Academic; 1995. pp. 365–394. [Google Scholar]
- 13.Ketting R F, Haverkamp T H A, van Luenen H G A M, Plasterk R H A. Cell. 1999;99:133–141. doi: 10.1016/s0092-8674(00)81645-1. [DOI] [PubMed] [Google Scholar]
- 14.Tabara H, Sarkissian M, Kelly W G, Fleenor J, Grishok A, Timmons L, Fire A, Mello C C. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- 15.Heck M M S. Cell. 1997;91:5–8. doi: 10.1016/s0092-8674(01)80002-7. [DOI] [PubMed] [Google Scholar]
- 16.Wilson K L. Trends Cell Biol. 2000;10:125–129. doi: 10.1016/s0962-8924(99)01708-0. [DOI] [PubMed] [Google Scholar]
- 17.Gant T M, Harris C A, Wilson K L. J Cell Biol. 1999;144:1083–1096. doi: 10.1083/jcb.144.6.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang L, Guan T L, Gerace L. J Cell Biol. 1997;139:1077–1087. doi: 10.1083/jcb.139.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]