Abstract
To test the ability of triple helix-forming oligonucleotides (TFOs) to promote recombination within chromosomal sites in mammalian cells, a mouse LTK− cell line was established carrying two mutant copies of the herpes simplex virus thymidine kinase (TK) gene as direct repeats in a single chromosomal locus. Recombination between these repeats can produce a functional TK gene and occurs at a spontaneous frequency of 4 × 10−6 under standard culture conditions. When cells were microinjected with TFOs designed to bind to a 30-bp polypurine site situated between the two TK genes, recombination was observed at frequencies in the range of 1%, 2,500-fold above the background. Recombination was induced efficiently by injection of both psoralen-conjugated TFOs (followed by long-wave UVA light; 1.2%) and unconjugated TFOs alone (1.0%). Control oligomers of scrambled sequence but identical base composition were ineffective, and no TFO-induced recombination was seen in a control LTK− cell line carrying an otherwise identical dual TK gene construct lacking the 30-bp polypurine target site. TFOs transfected with cationic lipids also induced recombinants in a highly sequence-specific manner but were less effective, with induced recombination frequencies of 6- to 7-fold over background. Examination of the TFO-induced recombinants by genomic Southern blotting revealed gene conversion events in which both TK genes were retained, but either the upstream (57%) or the downstream gene (43%) was corrected to wild type. These results suggest that, with efficient intracellular delivery, TFOs may be effective tools to promote site-specific recombination and targeted modification of chromosomal loci.
Keywords: gene targeting, recombination, thymidine kinase
Genetic manipulation of mammalian cells has been a major research tool. Gene transfer methods coupled with techniques to select for cells in culture that have undergone site-specific homologous recombination have facilitated specific gene replacement and enabled the development of genetically altered “knockout” mice (1). These selection methods improve the apparent efficiency of gene targeting but do so by eliminating clones arising from nonhomologous events. They do not alter the absolute frequency of homologous recombination, which is typically low in mammalian cells in gene transfer experiments (2). This low frequency of homologous recombination limits the extension of this technology to gene therapy, and therefore efforts have been made to improve the efficiency of gene targeting.
A series of studies have focused on modification of the recipient chromosomal site to create a substrate prone to homologous recombination. The site-specific endonuclease I-SceI can induce double-strand breaks (DSBs) within both extrachromosomal and chromosomal DNA loci engineered to carry the rare 18-bp recognition site (3). Such targeted DSBs have been shown to boost substantially the frequency of intramolecular and intermolecular recombination in mammalian cells and also in Xenopus oocytes (4–8). However, this approach requires the prior introduction of the recognition site within the genome.
In addition to DSBs, DNA damage from UV light, alkylating agents, and photoreactive molecules such as psoralen has been shown to be recombinagenic (9–11) but in a non-site-specific way. However, in previous work, we found that site-specific DNA damage could be introduced in mammalian cells by taking advantage of the sequence specificity of oligonucleotide-mediated triple helix formation (12–15). Triplex DNA can be formed when oligonucleotides bind in the major groove of the double helix in a sequence-dependent manner at polypurine/polypyrimidine stretches in duplex DNA (reviewed in ref. 16). The specificity arises from the base triplets formed by either Hoogsteen or reverse-Hoogsteen hydrogen bonding between the third strand and the purine strand of the duplex. In previous studies, we demonstrated that psoralen-conjugated triplex-forming oligonucleotides (TFOs) could mediate the introduction of base pair-specific psoralen adducts (and consequently mutations) in mammalian cells (12).
Using this strategy, we found that triple helix-targeted psoralen photoadducts could induce recombination within a simian virus 40-based shuttle vector carrying two mutant copies of the supF reporter gene (17). In addition, prompted by data showing that intermolecular triple helix formation, even in the absence of covalent DNA damage, could provoke DNA repair (13), we also tested the ability of non-psoralen-conjugated TFOs to induce recombination in the episomal simian virus 40 target. We found that third strands capable of high-affinity binding to the target DNA were able to stimulate recombination in a pathway that depended on nucleotide excision repair (NER; ref. 18).
These results raised the possibility that high-affinity TFOs, with or without an associated DNA reactive conjugate, might serve as tools to sensitize a chromosomal site to recombination. To investigate the feasibility of such an approach, we established a mouse LTK− cell line carrying two mutant copies of the herpes simplex virus thymidine kinase (TK) gene as direct repeats in a single chromosomal locus. In this construct, recombination can be detected by reconstruction of a functional TK gene. Using a series of oligonucleotides, we performed experiments showing that high-affinity TFOs, with or without psoralen, are capable of stimulating recombination between the tandem TK genes. Experiments in which oligonucleotides were transfected into cells by cationic lipids demonstrated that the induced recombination depended on the specificity of the TFO for the target locus but yielded modest levels of induction. In contrast, intranuclear delivery of TFOs by direct microinjection produced recombinants at frequencies greater than 1%, 2,500- to 3,000-fold over background and in the range of the best results in model systems employing I-SceI for DSB generation. Analysis of the TFO-induced recombinant clones was consistent with a pathway of homology-directed gene conversion. The results suggest that, with effective delivery, TFOs can be potent agents for promoting homologous recombination at targeted chromosomal sites.
Materials and Methods
Oligonucleotides.
Unconjugated and psoralen-linked oligonucleotides were synthesized by the Keck Facility at Yale University by using standard phosphoramidite chemistry and materials from Glen Research (Sterling, VA). All oligomers contained phosphodiester backbones and were synthesized to contain a 3′ propylamine group to minimize susceptibility to degradation by 3′ exonucleases (19). The oligomers were purified by either gel electrophoresis or HPLC, followed by Centricon-3 filtration in distilled water (Amicon). The psoralen was incorporated into the oligonucleotide synthesis as a psoralen phosphoramidite, resulting in an oligonucleotide linked at its 5′ end via a six-carbon linker arm to 4′-hydroxymethyl-4,5′,8-trimethyl psoralen. The oligonucleotides used in this study were pso-AG30, (5′ psoralen-AGGAAGGGGGGGGTGGTGGGGGAGGGGGAG-3′), AG30 (same as pso-AG30 but without 5′ psoralen), pso-SCR30 (5′ psoralen-GGAGGAGTGGAGGGGAGTGAGGGGGGGGGG-3′), and SCR30.
Plasmids.
Plasmid pJS-3, containing two mutant copies of the herpes simplex virus TK gene along with the neomycin-resistance gene, was obtained from M. Liskay (Oregon Health Sciences University, Portland, OR; ref. 20). The TK genes in pJS-3 contain XhoI linker insertion mutations at positions 735 (TK26) and 1,220 (TK8). They are present as 2.0-kilobase (kb) and 2.5-kb fragments in direct repeat orientation in the HindIII and BamHI sites in the vector, respectively (Fig. 1). A 200-bp fragment carrying the supFG1 gene from the plasmid pSupFG1 (12) was amplified by PCR with primers incorporating ClaI recognition sites and was inserted into the unique ClaI site in pJS-3 situated in the middle of the 1.3-kb stretch between the TK genes, yielding the plasmid pTK2supF (Fig. 1). The supFG1 gene contains a 30-bp G-rich polypurine site that affords high-affinity triple helix formation in the antiparallel purine motif by the TFO AG30.
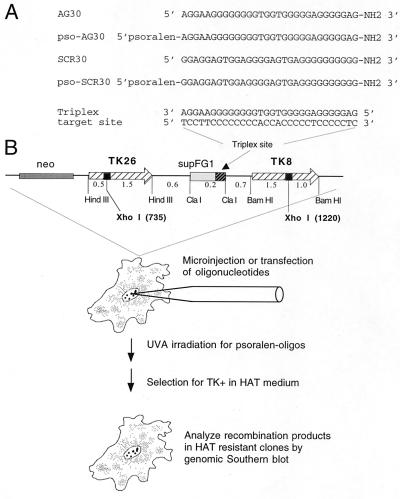
Figure 1.
Experimental scheme to investigate induction of intrachromosomal recombination by TFOs. LTK− cells carrying, at a single chromosomal locus, two mutant copies of the TK gene as direct repeats flanking a polypurine third-strand binding site were used to test the ability of transfected or microinjected TFOs to promote recombination. A purine-rich oligonucleotide of length 30 (AG30) was designed to form a triple helix in the antiparallel triplex motif at the G-rich target site, as shown. As a control, Scr30, containing the same base composition but a scrambled sequence, was used. In some experiments, the AG30 and the SCR30 oligonucleotides were conjugated at their 5′ ends to 4′-hydroxymethyl-4,5′,8-trimethylpsoralen via the 4′-hydroxymethyl position. In this case, by formation of the triple helix, psoralen intercalation and photoaddition is targeted to the thymidines at the predicted duplex-triplex junction. Potential recombinants are identified as TK+ clones growing in selective HAT (1 × 10−4 M hypoxanthine/2 × 10−6 M aminopterin/1.6 × 10−5 M thymidine)-containing medium.
Cells.
Mouse LTK− cells were obtained from the American Type Culture Collection and were grown in DMEM supplemented with 10% (vol/vol) FBS. The LTK− cells were transfected with 20 ng of plasmid pTK2supF linearized at the HpaII site by using Lipofectamine (Life Technologies, Bethesda, MD) as directed by the manufacturer. Selection for transfectants was carried out in 400 μg/ml G418 in DMEM. Transfectants were analyzed for integrated plasmid structure and copy number by Southern analysis of genomic DNA as described (20). The clone designated FL-10, determined to contain a single copy of the pTK2supF construct (data not shown), was chosen for further study. The LTK−-derivative cell line pJS-3-10, containing a single copy of the original pJS-3 vector (lacking the supFG1 polypurine target site), was obtained from M. Liskay (21) and was used as a control.
Microinjection of Oligonucleotides and Recombination Assay.
A day before microinjection, selected LTK−-derived cell lines (either FL-10 or pJS-3-10) were seeded at a density of 375 cells per cm2 in 30-mm dishes on which grids consisting of 25 squares were drawn. One cell at the top left of each square was microinjected. The injection needles were pulled with micropipette puller model P-87 (Sutter Instruments, Novato, CA) from 1.2/0.94-mm (outside diameter/inside diameter) borosilicate capillaries with filaments (World Precision Instruments, Sarasota, FL). The cells were microinjected with an Eppendorf 5170 micromanipulator and an Eppendorf 5242 microinjector equipped with Zeiss Axiovert 135 microscope. Solutions of selected oligonucleotides at 8 μM were injected in volumes of ≈15 fl directly into cell nuclei to deliver an estimated 72,000 oligonucleotides per injection, yielding an intranuclear concentration in the range of 2 × 10−7 to 2 × 10−6 M, depending on the precise number of molecules injected and the actual nuclear volume.
In the case of the psoralen-linked oligomers, 1 h after injection, the cells were exposed to 1.8 J/cm2 of UVA irradiation as described (15). The cells were incubated for an additional 24 h in nonselective medium, after which the medium was changed to DMEM supplemented with HAT (1 × 10−4 M hypoxanthine/2 × 10−6 M aminopterin/1.6 × 10−5 M thymidine) to select for potential recombinants expressing wild-type TK.
Transfection with Cationic Lipids.
Cells at a density of 1.67 × 104 per cm2 (1 × 106 in 100-mm dishes) were transfected with 10 μg of oligonucleotide DNA per dish mixed with 66 μl of GenePorter and diluted into a total of 2 ml of serum-free medium, as directed by the manufacturer (Gene Therapy Systems, San Diego). In the case of the psoralen-conjugated oligomers, UVA irradiation was given 5 h after transfection, after which the cells were placed in full growth medium supplemented with 10% (vol/vol) FBS. Medium was changed to HAT selection 24 h later.
Southern Blot Analyses.
HAT-resistant colonies representing candidate TK+ recombinant clones were identified after 2 weeks. The clones were expanded and genomic DNA was isolated by using Genzyme genomic DNA purification kits as described by the supplier. Genomic DNA was subject to restriction by either BamHI and HindIII or BamHI, HindIII, and XhoI (New England Biolabs). Southern analysis was performed by using standard methods by gel electrophoresis through 0.9% agarose and transfer to Hybond-N membranes (Amersham Pharmacia), with the 32P-labeled 2.5-kb BamHI TK fragment from pTK2supF used as a probe.
Sequence Analysis of Genomic DNA.
A 200-bp region flanking the polypurine triplex target site was amplified from the genomic DNA of selected HAT-resistant clones by PCR. Primers used were JS3-P6 (5′-CATGACATTAACCTATAAAAATAGGCG-3′) and JS3-P7 (5′-GGTTAAGTCCTCATTTAAATTAGGCA-3′). An internal primer, JS3-P11 (5′-TTAAATTAGGCAAAGGAATTC-3′) was used for DNA sequence analysis via automated methods (22).
Results
Experimental Strategy.
To investigate TFO-induced recombination at a chromosomal site, we established a mouse LTK− cell line subclone (FL-10) carrying a pair of mutant TK genes in a single locus as direct repeats separated by 1.3-kb (Fig. 1). In this construct, the region between the TK genes was engineered to contain a 30-bp G-rich polypurine sequence (present in the supFG1 gene insert in the ClaI site) that previous work had shown to be a site amenable to high-affinity third-strand binding in the antiparallel triplex motif (23) by the TFO designated AG30 (ref. 12; Fig. 1). The TK genes contain inactivating XhoI linker insertion mutations at different sites (positions 735 in TK 26 and 1,220 in TK8). The TK26 and TK8 genes are present as 2.0-kb and 2.5-kb fragments within HindIII and BamHI sites, respectively. The stable integration of this construct into the LTK− cells and its presence in a single copy was confirmed by Southern analysis of genomic DNA from the FL-10 subclone (data not shown).
In the assay, recombination between the two TK genes has the potential to produce a functional gene. Because the parental LTK− cells lack the cellular TK, cells in which the mutant TK genes have recombined to generate a wild-type TK can be selected by growth in the presence of HAT medium. Induction of recombination by selected oligonucleotides is quantified by enumerating the HAT-resistant colonies as a proportion of the total number of cells treated.
For comparison, a similar LTK− cell line obtained from M. Liskay was also used (21). This line, pJS-3-10, contains a single chromosomally integrated copy of a dual TK construct similar to that in FL-10 but lacks the supFG1 gene insert. Hence, in the pJS-3-10 cells, the recombination substrate lacks the 30-bp polypurine target site for AG30. Also, in FL-10 cells, TK26 is upstream of TK8, whereas in pJS-3-10 cells, TK8 is upstream of TK26, with polarity defined with respect to the direction of TK gene transcription. Previous work by Liskay and colleagues (21) showed that the TK8 and TK26 genes individually revert to wild type at frequencies in the range of 10−8 or lower and thus generation of a functional TK gene at a measurable frequency requires information transfer between the two genes. In the pJS-3-10 cells, recombination between the TK genes occurs at spontaneous frequencies in the range of 10−6 (21).
TFO-Induced Intrachromosomal Recombination.
The FL-10 and pJS-3-10 cell lines were used to test the ability of TFOs to stimulate intrachromosomal recombination in a site-specific manner. The cells were transfected with a series of oligonucleotides by two different methods (either comixture with cationic lipids or direct intranuclear microinjection), and the production of HAT-resistant colonies expressing wild-type TK was determined.
The oligonucleotides used are shown in Fig. 1 and included both psoralen-conjugated oligomers as well as unconjugated ones (with the only modification in the latter case being 3′ end protection with a 3′ propylamine group to provide nuclease resistance). The TFOs specific for the polypurine site between the two TK genes, AG30 and pso-AG30, were determined in previous work to bind with high-affinity to the duplex target site, with equilibrium dissociation constants of 1 × 10−8 M and 3 × 10−9 M, respectively (12, 13). The control oligomers, Scr30 and pso-Scr30, have the same G-rich base composition as the AG30 TFOs but in a scrambled sequence (creating 14 mismatches of 30 in the third-strand binding code for the polypurine target site versus only 2 mismatches in the case of AG30). These oligomers show no detectable binding to the 30-bp target site even at concentrations up to 10−5 M in gel mobility-shift assays (unpublished results). In the case of the psoralen-conjugated oligomers, UVA irradiation (1.8 J/cm2) was given to the cells after the transfections to photoactivate the psoralen for potential photoadduct formation at the target site. The timing of the UVA irradiation differed depending on the transfection method. The microinjected cells were irradiated 1 h after injection, whereas the cells transfected with cationic lipids were irradiated 5 h after addition of the oligonucleotide/lipid mixtures.
The results (Table 1) show that only AG30 and pso-AG30 were effective in inducing HAT-resistant clones in the FL-10 cells at frequencies substantially above the background level of 4 × 10−6 in the untreated cells. However, the efficiency of induction by the specific TFOs varied significantly depending on the method of TFO delivery. Transfection of the FL-10 cells with AG30 and pso-AG30 (plus UVA light) by using cationic lipids yielded recombinants at frequencies of 25 × 10−6 and 30 × 10−6, respectively, values 6- and 7-fold above the background in untreated cells. Remarkably, microinjection of AG30 and pso-AG30 produced HAT-resistant colonies at frequencies of 1.0% and 1.2%, 2,500- and 3,000-fold above background. These values were calculated as the ratio of the number of HAT-resistant colonies produced divided by the total number of cells injected with the oligonucleotides. In the microinjection procedure, a volume of 15 fl of a solution containing 8 μM oligonucleotide was injected into the nuclei, yielding approximately 72,000 oligonucleotide molecules per nucleus. Cell viability after microinjection was estimated to be ≈70%, based on experiments that used the same protocol to inject a green fluorescent protein expression vector instead of the TFOs (data not shown). However, this viability estimate was not used to correct the induced recombination frequencies, because we do not know exactly the postinjection viability in the oligonucleotide experiments.
Table 1.
Intrachromosomal recombination induced by TFOs
| Cells | Oligonucleotide | Transfection method | UVA | TK+ clones/total cells | Frequency, % |
|---|---|---|---|---|---|
| FL-10 | None | None | 4/1 × 106 | 0.0004 | |
| None | Cationic lipids | + | 40/6 × 106 | 0.0007 | |
| pso-Scr30 | Cationic lipids | + | 44/6 × 106 | 0.0007 | |
| AG30 | Cationic lipids | 149/6 × 106 | 0.0025 | ||
| pso-AG30 | Cationic lipids | + | 178/6 × 106 | 0.0030 | |
| pso-Scr30 | Microinjection | + | 0/1,375 | ≤0.072 | |
| AG30 | Microinjection | 14/1,375 | 1.0 | ||
| pso-AG30 | Microinjection | + | 16/1,375 | 1.2 | |
| pJS-3-10 | None | None | 12/2 × 106 | 0.0006 | |
| pso-AG30 | Microinjection | + | 0/650 | ≤0.15 |
In comparison, microinjection of pso-Scr30 into the FL-10 cells, followed by UVA irradiation, yielded no HAT-resistant clones (Table 1). As an additional control, pso-AG30 was also microinjected into the pJS-3-10 cells under the same conditions as with the FL-10 cells. The pJS-3-10 cells, like the FL-10 cells, are an LTK− subclone. They contain a dual TK construct similar to the one in the FL-10 cells, with the only differences being the absence of the polypurine target site and the order of the TK8 and TK26 alleles as they reside in the chromosome (21). As shown (Table 1), no recombinants were detected in 650 injected cells.
Analysis of HAT-Resistant Clones.
The HAT-resistant clones induced by microinjection of FL-10 cells with AG30 (n = 14) and pso-AG30 (n = 16) were isolated, recloned, and expanded. Genomic DNA was prepared for Southern analysis to examine the structure of the TK locus and to determine the nature of the recombination products.
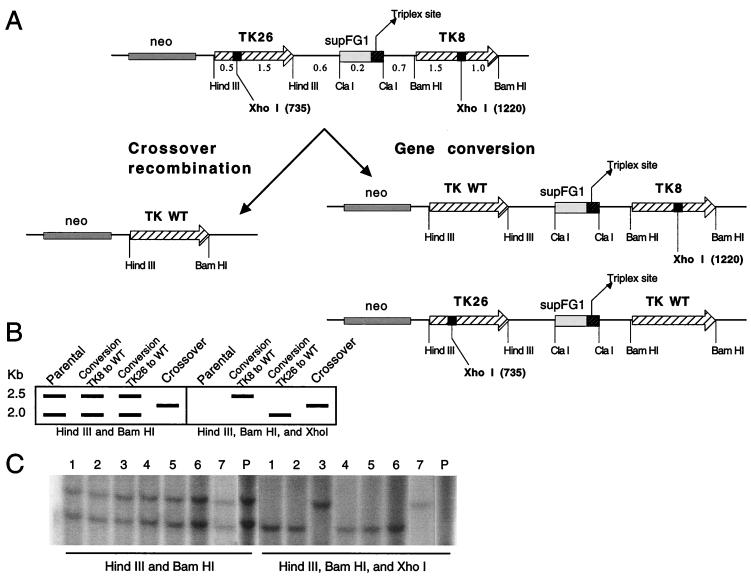
Possible recombination pathways that can generate a functional TK gene are illustrated in Fig. 2A. A nonconservative event involving crossover recombination can produce a single copy of a wild-type TK gene. However, because of the orientation of the inactivating mutations in TK26 and TK8, a single crossover would generate a doubly mutant TK gene retained in the chromosome and would pop out the reconstructed wild-type TK gene. Multiple crossovers would be required for a nonconservative event to yield a wild-type allele in the chromosome. Alternatively, there can be information transfer from one TK gene to the other to correct the XhoI linker insertion mutation, as diagrammed, in a conservative process of gene conversion that retains both TK gene fragments, one now wild-type and one still mutant.
Figure 2.
Analysis of TFO-induced recombinant clones expressing wild-type (WT) TK. (A) Possible pathways to generate a wild-type TK gene from the tandem mutant TK genes in FL-10 cells. The diagram illustrates crossover recombination, yielding (via multiple crossovers) a single wild-type TK gene in a nonconservative event, and gene conversion, in which there is information transfer from one TK gene to the other to correct the XhoI linker insertion mutation, with retention of both TK genes in a conservative event. (B) Expected pattern of bands on Southern analysis of parental and recombinant HAT-resistant clones, depending on the nature of the recombination event. The hypothetical band pattern is based on the indicated restriction digestion of genomic DNA and hybridization with a TK gene fragment as a probe. (C) Southern blot analysis of genomic DNA from the parental FL-10 cells (P) and seven HAT-resistant clones produced by TFO microinjection (lanes 1–7). The samples were restricted with the indicated enzymes, and the Southern hybridization was performed with the 2.5-kb TK gene BamHI fragment as a probe.
The expected pattern of bands on Southern analysis from each of these possible pathways is illustrated schematically in Fig. 2B. In the parental HAT-sensitive FL-10 cells, genomic DNA restricted with BamHI and HindIII and probed with a TK gene fragment should yield two bands of 2.0 and 2.5 kb. These bands represent the 2.0-kb TK26 HindIII fragment and the 2.5-kb TK8 BamHI fragment. Both of these bands will be eliminated by the addition of XhoI to the restriction digest, because both mutant TK fragments contain the XhoI linker insertions.
In the case of a nonconservative crossover recombinant, only a single band should be visualized in the BamHI and HindIII lane; however, this band should persist when XhoI is added, because the recombinant fragment would contain the wild-type TK sequence without either of the XhoI sites.
The gene conversion events can involve either correction of TK8 to wild type or correction of TK26 to wild type. In the former case, the 2.0-kb and 2.5-kb bands will be present on double digestion, but the 2.0-kb band will be lost in the triple digestion (because the XhoI linker insertion remains in the TK26 gene). The 2.5-kb band will remain, because the gene conversion event would eliminate the XhoI site in the TK8 gene. With conversion of TK26 to wild type, again both bands will be present in the BamHI and HindIII double digest lane, but in this case, the 2.5-kb band will be lost with addition of XhoI, and the 2.0-kb band will be resistant to XhoI and thus will persist in the three-enzyme sample.
An example of this analysis performed on one series of recombinant clones induced by pso-AG30 is shown in Fig. 2C. All seven clones analyzed in this blot showed two bands on BamHI and HindIII digestion. With the addition of XhoI, clones 1, 2, 4, 5, and 6 showed loss of the 2.5-kb band and retention of the 2.0-kb one, whereas, conversely, clones 3 and 7 showed loss of the 2.0-kb band and persistence of the 2.5-kb one. These results are consistent with all seven clones having arisen from gene conversion events, with 1, 2, 4, 5, and 6 being convertants from TK26 to wild type and 3 and 7 resulting from gene conversion from TK8 to wild type.
Table 2 presents a summary of the results of the Southern analysis. All 14 HAT-resistant clones induced by AG30 and all 16 induced by pso-AG30 were found to have arisen by gene conversion. In both groups, the distribution of gene conversion events was almost identical, with 43% TK26 converting to wild type and 57% TK8 converting to wild type for the AG30-induced events and 44% and 56% for pso-AG30-induced events, respectively.
Table 2.
Intrachromosomal recombination products induced by TFOs
| Oligonucleotide treatment | Frequency of TK+ colonies, % | Gene conversion
|
Crossover recombination | |
|---|---|---|---|---|
| TK8 to WT | TK26 to WT | |||
| pso-AG30 + UVA | 1.2 (16/1,375) | 9/16 (56%) | 7/16 (44%) | 0/16 (0%) |
| AG30 | 1.0 (14/1,375) | 8/14 (57%) | 6/14 (43%) | 0/14 (0%) |
WT, wild type.
Analysis of the Triple Helix Target Site.
In previous work, we had found that triple helix formation by TFOs can cause mutations at and around the target site (13). We therefore examined the putative third-strand binding site and surrounding sequences in randomly selected TFO-induced, HAT-resistant recombinants to determine whether the target region had undergone mutation at the same time that recombination had been stimulated. For each of 21 HAT-resistant clones tested, a 200-bp region of genomic DNA encompassing the polypurine/polypyrimidine site was amplified by PCR, and the DNA sequence of the region was determined. No mutations were seen in any of the clones. These results indicate that triplex-induced recombination is not necessarily accompanied by mutation of the third-strand binding site. However, a low frequency of TFO-induced mutagenesis cannot be ruled out. Also, the HAT-selection assay specifically identifies cells in which a functional TK gene has been generated; cells with other classes of nonparental products are not detected, and some of these might contain target site mutations.
Discussion
Results obtained with a mouse LTK− cell line carrying two mutant copies of the TK gene as direct repeats in a single chromosomal locus show that oligonucleotides designed to bind as third strands to a site between the genes can effectively stimulate intrachromosomal recombination. When the TFOs were introduced into the cells by intranuclear microinjection, the frequency of induced recombination was in the range of 1%, 2,500-fold greater than the background frequency in untreated cells. By Southern analysis of the genomic DNA from the nonparental TK+ colonies, the TFO-induced recombination products were found to have arisen from conservative gene conversion events in which one mutant gene was corrected via information transfer from the other. In addition, although triplex formation can induce mutations as well as recombination (13), no mutations were seen in or around the predicted TFO binding site in the triplex-induced recombinants.
Interestingly, the AG30 TFO itself, even without conjugation to a DNA-reactive molecule, was almost as effective as the psoralen-conjugated version, pso-AG30 (1.0% versus 1.2% when both were microinjected and 25 × 10−6 versus 30 × 10−6, respectively, when the TFOs were transfected with cationic lipids). These comparisons suggest two possibilities. (i) The yield of targeted photoadducts was low, possibly because the protocol for psoralen-TFO transfection and the timing of the UVA irradiation were not optimal. Hence, the results obtained with pso-AG30 may reflect primarily the effect of the triple helix formation alone. (ii) Targeted photoadducts were formed, but the TFO-mediated triplexes themselves are nearly as recombinagenic as the third-strand targeted psoralen adducts. Distinguishing between these possibilities will require further analyses, including direct measurements of third-strand-directed photoadduct formation at the target locus (24, 25). Nonetheless, the activity of the AG30 TFO by itself in promoting recombination demonstrates that TFOs capable of high-affinity, site-specific binding to DNA, even without the generation of covalent damage, can provoke DNA metabolism and thereby stimulate recombination.
The induced recombination frequencies achieved on microinjection of the TFOs (1% to 1.2%) are in the same range as results reported in experiments that used similar tandem repeat gene targets in which site-specific DSBs are produced within the target locus by expression of the I-SceI nuclease in the cells. For example, Taghian and Nickoloff (26) and Donoho et al. (27) observed I-SceI-induced recombination between neo gene repeats at frequencies of 1% and 3% in CHO and mouse embryonic stem cells, respectively. Although the recombination substrates in these studies were not exactly the same as in our work and although the I-SceI cleavage was targeted to sites within one of the duplicated genes rather than between them, this comparison nonetheless suggests that TFOs, with effective intranuclear delivery, can induce recombination at a chromosomal target with an efficiency rivaling that of a site-specific endonuclease.
In other experiments that provide a model for targeted gene replacement, Smih et al. (5) showed that, in cells with a single copy of the neo gene interrupted by an I-SceI recognition site insert, expression of the I-SceI nuclease could sensitize the target to recombination with a transfected neo gene fragment, at a frequency at least 50-fold above background. In a similar experimental design, Donoho et al. (27) also demonstrated the utility of I-SceI in promoting targeted recombination of a transfected DNA with a chromosomal locus containing an I-SceI site, observing targeting frequencies of up to 1%, 5,000-fold higher than in the absence of I-SceI expression. These experiments showed that induced strand breaks at a chromosomal locus can have a substantial effect on homologous recombination between the locus and an exogenously introduced DNA.
The experiments presented herein suggest that triplex formation can provoke sufficient DNA metabolism to stimulate intramolecular homologous recombination at a chromosomal locus at high efficiency. By analogy to the I-SceI data, we hypothesize that TFOs may also prove to be effective reagents for promoting intermolecular recombination between a transfected gene fragment and a chromosomal target site.
However, the comparison to the I-SceI data does not necessarily imply that the TFOs mediate recombination by producing the same DSB type of damage as I-SceI. At this point, we have not performed experiments to examine directly the pattern of triplex-stimulated strand breaks. However, previous work has suggested that triple helix formation creates an altered DNA structure that is recognized by the NER complex in mammalian cells (13). It was found that triplexes formed on a plasmid substrate can stimulate DNA repair activity, as measured by DNA repair synthesis, in human cell extracts (13). Such repair activity is absent from extracts of cells deficient in the NER damage recognition factor XPA (unpublished results). The generation of repair synthesis tracts in plasmids to which the third stands have bound implies the generation of at least one single-strand break, which would be the minimum damage necessary to enable the incorporation of labeled nucleotides. Theoretically, if the canonical NER reaction (28) were carried out on the triplex-containing plasmid, a pattern of dual endonuclease incisions flanking the triple helix would be expected. The current data from the repair synthesis assay do not allow us to distinguish between the production of a single nick, followed by nick translation, and the generation of two nicks and a consequent gap, followed by gap-filling repair synthesis. However, if DSBs in the plasmids were produced at a significant frequency in the extracts, we would expect to see linearized DNA molecules, and no such species have been seen (ref. 13 and unpublished results).
At this point, we favor the possibility that one or more single-strand breaks are produced at or near the TFO-binding site via repair-directed endonuclease incision activity. However, the number, position, and strand orientation of the possible repair-mediated strand breaks remain to be determined. Also, the role of the NER pathway in the TFO-induced intrachromosomal gene conversion reported herein has not been tested explicitly. However, by extrapolation from our recent work demonstrating a requirement for NER in TFO-stimulated recombination in an episomal simian virus 40-based shuttle vector (18), we speculate that stimulation of recombination by triplex formation at a chromosomal site will also turn out to require NER activity.
The recombination products produced by the TFOs in the present work were all consistent with conservative gene conversion events. However, the manner in which the dual TK recombination substrate resides in the genome of the FL-10 cells favors the detection of gene conversion events. Detectable nonconservative events would require multiple crossovers and would therefore be rare. Single crossover events stimulated by triplex formation would yield doubly mutant TK genes and would not be detected in the assay. Hence, the results reported herein may underestimate the ability of TFOs to provoke intrachromatid recombination.
The ability of TFOs to stimulate recombination at a chromosomal locus demonstrates that chromatin is not an absolute barrier to third-strand binding to chromosomal DNA, consistent with recent studies that have detected TFO-directed mutagenesis at chromosomal sites (14, 15). However, in the mutagenesis studies, the TFOs were delivered into the cells by electroporation, passive fluid phase uptake, or cationic lipids, and the induced mutation frequencies, although above background, were in the 10−4 to 10−3 range. In the current work, a key finding is that microinjection of the TFOs yielded induced recombination frequencies several orders of magnitude greater than when the TFOs were introduced by comixture with cationic lipids. These results show that the biological obstacles to TFO-mediated genome modification include not only the chromatin structure of the target gene but also the cellular barriers that must be traversed by the TFOs to achieve an effective concentration in the appropriate intranuclear compartment for binding to the target site.
In our experiments, we estimate that intranuclear concentrations in the range of 2 × 10−7 to 2 × 10−6 M were achieved. These values are similar to those reported for oligomers transfected by other means, and thus the improved efficacy of the microinjection may be due to a more biologically effective intranuclear distribution of the TFOs. However, although microinjection is feasible as a research tool, its applicability for gene therapy is limited. Hence, facile techniques for effective intranuclear oligonucleotide delivery are needed to make this strategy more practical.
Nonetheless, the results reported herein support the utility of TFOs as tools to sensitize genomic sites to recombination. Although our experiments focused on recombination within a specially engineered chromosomal locus containing a tandem gene repeat, this strategy should be applicable to the stimulation of recombination between a chromosomal site and exogenously introduced DNA molecules. As such, this work may facilitate efforts directed at gene replacement or correction for research or clinical purposes. One current limitation is that triplex formation occurs predominately at polypurine/polypyrimidine sites. However, a considerable effort has been directed at extending the third-strand binding code (16), and in any case, polypurine sites are overrepresented in the genome. In addition, the present findings raise the possibility that not just TFOs but also other high-affinity DNA-binding molecules, such as peptide nucleic acids (29) and polyamides (30), may prove useful in strategies to promote site-specific recombination for the purpose of genome modification.
Acknowledgments
We thank M. Seidman and D. Carroll for helpful discussions and P. Chan, J. Yuan, S. Shiao, K. Vasquez, L. Narayanan, R. Franklin, S. J. Baserga, and L. Cabral for their help. This work was supported by National Institutes of Health Grant RO1 GM54731 and by a Scholar Award to P.M.G. from the Leukemia and Lymphoma Society. Z.L. was supported by National Institutes of Health Training Grant T32 CA09159. A.F.F. was supported in part by the Anna Fuller Foundation.
Abbreviations
- TFO
triplex-forming oligonucleotide
- TK
thymidine kinase
- NER
nucleotide excision repair
- DSB
double-strand break
- kb
kilobase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.160004997.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.160004997
References
- 1.Capecchi M R. Science. 1989;244:1288–1292. doi: 10.1126/science.2660260. [DOI] [PubMed] [Google Scholar]
- 2.Hanson K D, Sedivy J M. Mol Cell Biol. 1995;15:45–51. doi: 10.1128/mcb.15.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rouet P, Smih F, Jasin M. Mol Cell Biol. 1994;14:8096–8106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lukacsovich T, Yang D, Waldman A S. Nucleic Acids Res. 1994;22:5649–5657. doi: 10.1093/nar/22.25.5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smih F, Rouet P, Romanienko P J, Jasin M. Nucleic Acids Res. 1995;23:5012–5019. doi: 10.1093/nar/23.24.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choulika A, Perrin A, Dujon B, Nicolas J F. Mol Cell Biol. 1995;15:1968–1973. doi: 10.1128/mcb.15.4.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brenneman M, Gimble F S, Wilson J H. Proc Natl Acad Sci USA. 1996;93:3608–3612. doi: 10.1073/pnas.93.8.3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Segal D J, Carroll D. Proc Natl Acad Sci USA. 1995;92:806–810. doi: 10.1073/pnas.92.3.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhattacharyya N P, Maher V M, McCormick J J. Mutat Res. 1989;211:205–214. doi: 10.1016/0027-5107(89)90003-1. [DOI] [PubMed] [Google Scholar]
- 10.Tsujimura T, Maher V M, Godwin A R, Liskay R M, McCormick J J. Proc Natl Acad Sci USA. 1990;87:1566–1570. doi: 10.1073/pnas.87.4.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vos J M, Hanawalt P C. Mol Cell Biol. 1989;9:2897–2905. doi: 10.1128/mcb.9.7.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang G, Levy D D, Seidman M M, Glazer P M. Mol Cell Biol. 1995;15:1759–1768. doi: 10.1128/mcb.15.3.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang G, Seidman M M, Glazer P M. Science. 1996;271:802–805. doi: 10.1126/science.271.5250.802. [DOI] [PubMed] [Google Scholar]
- 14.Majumdar A, Khorlin A, Dyatkina N, Lin F-L M, Powell J, Liu J, Fei Z, Khripine Y, Watanabe K A, George J, Glazer P M, Seidman M M. Nat Genet. 1998;20:212–214. doi: 10.1038/2530. [DOI] [PubMed] [Google Scholar]
- 15.Vasquez K M, Wang G, Havre P A, Glazer P M. Nucleic Acids Res. 1999;27:1176–1181. doi: 10.1093/nar/27.4.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan P P, Glazer P M. J Mol Med. 1997;75:267–282. doi: 10.1007/s001090050112. [DOI] [PubMed] [Google Scholar]
- 17.Faruqi A F, Seidman M M, Segal D J, Carroll D, Glazer P M. Mol Cell Biol. 1996;16:6820–6828. doi: 10.1128/mcb.16.12.6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faruqi A F, Datta H J, Carroll D, Seidman M M, Glazer P M. Mol Cell Biol. 2000;20:990–1000. doi: 10.1128/mcb.20.3.990-1000.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zendegui J G, Vasquez K M, Tinsley J H, Kessler D J, Hogan M E. Nucleic Acids Res. 1992;20:307–314. doi: 10.1093/nar/20.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liskay R M, Stachelek J L. Proc Natl Acad Sci USA. 1986;83:1802–1806. doi: 10.1073/pnas.83.6.1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Letsou A, Liskay R M. Genetics. 1987;117:759–769. doi: 10.1093/genetics/117.4.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Havre P A, Gunther E J, Gasparro F P, Glazer P M. Proc Natl Acad Sci USA. 1993;90:7879–7883. doi: 10.1073/pnas.90.16.7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beal P A, Dervan P B. Science. 1991;251:1360–1363. doi: 10.1126/science.2003222. [DOI] [PubMed] [Google Scholar]
- 24.Giovannangeli C, Diviacco S, Labrousse V, Gryaznov S, Charneau P, Helene C. Proc Natl Acad Sci USA. 1997;94:79–84. doi: 10.1073/pnas.94.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oh D H, Hanawalt P C. Nucleic Acids Res. 1999;27:4734–4742. doi: 10.1093/nar/27.24.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taghian D G, Nickoloff J A. Mol Cell Biol. 1997;17:6386–6393. doi: 10.1128/mcb.17.11.6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Donoho G, Jasin M, Berg P. Mol Cell Biol. 1998;18:4070–4078. doi: 10.1128/mcb.18.7.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sancar A. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 29.Faruqi A F, Egholm M, Glazer P M. Proc Natl Acad Sci USA. 1998;95:1398–1403. doi: 10.1073/pnas.95.4.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White S, Szewczyk J W, Turner J M, Baird E E, Dervan P B. Nature (London) 1998;391:468–471. doi: 10.1038/35106. [DOI] [PubMed] [Google Scholar]