Abstract
The cysteine desulfurase, IscS, provides sulfur for Fe-S cluster synthesis in vitro, but a role for IscS in in vivo Fe-S cluster formation has yet to be established. To study the in vivo function of IscS in Escherichia coli, a strain lacking IscS was constructed and characterized. Using this iscS deletion strain, we have observed decreased specific activities for proteins containing [4Fe-4S] clusters from soluble (aconitase B, 6-phosphogluconate dehydratase, glutamate synthase, fumarase A, and FNR) and membrane-bound proteins (NADH dehydrogenase I and succinate dehydrogenase). A specific role for IscS in in vivo Fe-S cluster assembly was demonstrated by showing that an Fe-S cluster independent mutant of FNR is unaffected by the lack of IscS. These data support the conclusion that, via its cysteine desulfurase activity, IscS provides the sulfur that subsequently becomes incorporated during in vivo Fe-S cluster synthesis. We also have characterized a growth phenotype associated with the loss of IscS. Under aerobic conditions the deletion of IscS caused an auxotrophy for thiamine and nicotinic acid, whereas under anaerobic conditions, only nicotinic acid was required. The lack of IscS also had a general effect on the growth of E. coli because the iscS deletion strain grew at half the rate of wild type in many types of media even when the auxotrophies were satisfied.
Keywords: NifS, FNR, oxidative stress repair, sulfur metabolism
Iron–sulfur proteins are ubiquitous in nature and participate in many vital cellular functions such as DNA repair, transcriptional regulation, nucleotide and amino acid biosynthesis, and energy metabolism. Although many studies have focused on the activities of Fe-S proteins, little is known about how Fe-S clusters are assembled and inserted into these proteins in vivo. Under reducing conditions in vitro, such clusters are slowly assembled into proteins when only Fe2+ and S2- are provided (1). However, studies of the N2-fixation (nif) gene cluster from Azotobacter vinelandii suggest that cells contain activities that increase the efficiency of Fe-S cluster assembly (2). In particular, the characterization of NifS led to the discovery that this protein is a pyridoxal 5′-phosphate-containing cysteine desulfurase that transfers the sulfur from cysteine to an active site cysteinyl residue of NifS, forming an enzyme-bound persulfide (3). Upon reduction and addition of an iron source, the S0 can be released and efficiently incorporated into the cluster of the Fe-S protein of nitrogenase (4).
Proteins similar to NifS have been discovered in non-N2-fixing bacteria (5), eukaryotes and archaea (blast, National Center for Biotechnology Information), indicating that such cysteine desulfurases might serve a general role in Fe-S cluster formation. In support of this notion, a homolog of NifS, called Nfs1, was identified in the yeast Saccharomyces cerevisiae, where it appears to play a role in mitochondrial Fe-S cluster assembly (6–8). In addition, cysteine desulfurases homologous to NifS have been purified from A. vinelandii and Escherichia coli (9, 10). These proteins have been named IscS for iron sulfur cluster to distinguish them from their counterparts encoded within the N2-fixation gene clusters of N2-fixing bacteria (5). Although IscS from E. coli and A. vinelandii have been shown to facilitate Fe-S cluster assembly in vitro, their role in in vivo Fe-S cluster formation has not yet been established. In addition, Flint et al. (11) also showed by using E. coli cell extracts that other proteins can function to donate sulfur for Fe-S cluster assembly, leaving open the question as to which protein is the physiologically relevant sulfur donor. Thus, the goal of this study was to investigate the role of IscS in donating sulfur for in vivo Fe-S cluster assembly.
Fe-S cluster assembly is important for the de novo synthesis of a broad spectrum of proteins and also the repair of certain Fe-S proteins that are damaged during oxidative stress. A family of dehydratases, including the major fumarase and aconitase isozymes, use [4Fe-4S]2+ clusters as Lewis acids during catalysis. These solvent-exposed clusters can be rapidly oxidized by superoxide, and the oxidized clusters are unstable, losing one and perhaps more iron atoms (12). The inactivation of these enzymes is responsible for many of the characteristic deficiencies of superoxide-stressed cells. Work in several labs has shown that these clusters are reassembled in vivo, using iron drawn from ferritin and a presumptive electron donor that has not been identified (13, 14). In fact, the fully demetallated form of one such enzyme, dihydroxyacid dehydratase, was used as a substrate during the purification of the E. coli IscS protein (10). However, it is not clear whether cluster disintegration in vivo progresses to the point where sulfur atoms are lost, so that the role of IscS in cluster repair is unclear.
An attempt to delete iscS from A. vinelandii was unsuccessful, apparently because the gene is essential (5). In addition, studies in yeast have demonstrated that the IscS homologue (Nfs1) is also essential in this organism (6). In this study, we report the successful disruption of iscS from E. coli, allowing us to analyze the function of IscS in vivo. We have found that the lack of IscS leads to an overall decrease in the activity of a number of Fe-S proteins and results in a general growth defect.
Materials and Methods
Growth Experiments.
Cultures were grown aerobically overnight at 37°C in Luria broth (15), pelleted, washed, resuspended in glucose minimal medium (15), and used as an inoculum at a 1:1,000 dilution. Growth was monitored by measuring the OD600. When indicated, the growth medium was supplemented with yeast extract (5 mg/ml), thiamine (2 μg/ml), nicotinic acid (12.5 μg/ml), chloramphenicol (20 μg/ml), ampicillin (50 μg/ml), tetracycline (10 μg/ml), and kanamycin (Kn, 40 μg/ml).
Plasmid Construction.
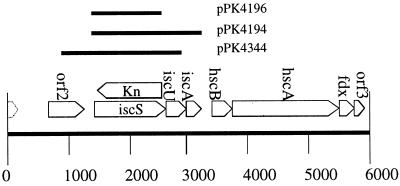
Plasmids pPK4071 and pPK4083 were made by digesting λ430 (16) with NruI and cloning this 5-kb fragment containing iscRSUAhscB (17) into the SmaI site of pUC19 (18) and pAlter-1 (Promega), respectively. Plasmids pPK4196 and pPK4194 were generated by cloning PCR products containing only the ORFs for iscS and iscSUA into pET11a (19), respectively (Fig. 1). Plasmid pPK4344 was generated by cloning into pGem2 (Promega), a PCR product containing sequence beginning within the iscR ORF and ending within the iscA ORF (Fig. 1). For DNA amplification by PCR from chromosomal or plasmid DNA, either Pfu DNA polymerase (Stratagene) or Taq DNA polymerase (Promega) was used. The sequences of oligonucleotides (Operon Technologies, Alameda, CA) used in this work are available on request.
Figure 1.
Genomic organization of the isc region. Plasmids used in this study are indicated. The genotype of the iscS mutant is indicated with the iscS coding region having been replaced by the KnR gene.
Strain Construction.
Codons 7 and 399 of iscS in pPK4083 were altered by site-directed mutagenesis (Promega Altered Sites), creating XhoI restriction endonuclease sites and generating plasmid pPK4119. The iscS coding region was deleted by digestion of pPK4119 with XhoI, the ends made blunt, and a blunt-ended KnR cartridge from Tn903 (20) was inserted by ligation to generate pPK4155 (Fig. 1). A DNA fragment containing approximately 1 kb of flanking sequence on either side of the KnR gene was PCR-amplified from pPK4155 and then cloned into pK03 (chloramphenicol, sacB, pSC101ori), which is temperature-sensitive for replication (21). The resulting plasmid, pPK4258, was transformed into PK4304, a strain containing a chromosomal Tn10 insertion in phe (located approximately 2 min from iscS) and F′142 that covers both the isc and phe regions. Strain PK4304 was made by mating CAG18469 (phe∷Tn10) with KL709 (F′142) (22). Strains containing pPK4258 integrated into the chromosome of PK4304 were selected at the nonpermissive temperature (42°C) with a frequency of 10−3-10−4. Cells containing the integrated plasmid were plated on 5% sucrose to select for those that had resolved and subsequently lost the plasmid. Sucrose-Kn-resistant colonies arose at a frequency of 10−3, which was the source of PK4326. P1 phage were generated from PK4326, and the ΔiscS∷KnR marker was transduced into RZ4500, PK3292, or PK3293 (Table 1) by selecting on Kn, generating strains PK4331, PK4357, and PK4359, respectively.
Table 1.
E. coli strains
| Strain | Genotype | Reference |
|---|---|---|
| CAG18469 | MG1655 phe∷Tn10 | (56) |
| CAG18481 | MG1655 zff-208∷Tn10 | (56) |
| RZ4500 | MG1655 lacZΔ145 | (57) |
| PK3292 | RZ4500, fnr+, dmsA′-lacZ | This lab |
| PK3293 | RZ4500, fnr−, dmsA′-lacZ | This lab |
| PK4304 | CAG18469, F′142 | This study |
| PK4326 | CAG18469, ΔiscS∷KnR, F′142 | This study |
| PK4331 | RZ4500 ΔiscS∷KnR | This study |
| PK4357 | PK3292 ΔiscS∷KnR | This study |
| PK4359 | PK3293 ΔiscS∷KnR | This study |
Fe-S Protein Assays.
Isocitrate and potassium cyanide were from Aldrich; all other reagents were from Sigma. Cultures for assays of enzymatic activities were grown to 0.4–0.7 OD in media that would ensure consistent, substantial synthesis of the relevant enzymes. In all cases except for that of fumarase, the media were chosen to avoid a dependence on Fe-S enzymes for central metabolism. NADH DH1 (Ndh1) and succinate dehydrogenase (SDH) were assayed by using inverted membrane vesicles prepared from cells grown in minimal A salts (15) containing 1% casein hydrolysate, 0.5% yeast extract, and 0.5% NaCl (pH 7), and NADH DH2 (Ndh2) was assayed in vesicles derived from cells grown in LB medium containing 0.2% glucose. For aconitase assays cells were grown in LB medium; for 6-phosphogluconate dehydratase assays, in minimal A salts containing 0.2% gluconate and 0.2% casein hydrolysate; for glutamate synthase, in minimal A salts containing 0.2% glucose; and for fumarase and isocitrate dehydrogenase, in minimal A salts containing 40 mM fumarate.
To stabilize Ndh1 for assay, vesicles were prepared by sonication in 50 mM MES, pH 6. Ndh1 activity was measured by the oxidation of deaminoNADH (23), which was prepared by the enzymatic reduction of deaminoNAD+ using the protocol for NADH preparation (24). Ndh2 was assayed by the oxidation of NADH after Ndh1 had been inactivated by the overnight incubation of the vesicles in 50 mM KPi, pH 7.8, on ice. Succinate oxidase was measured by monitoring O2 consumption with an OROBOROS oxygraph (Paar, Gratz, Austria) during incubation of inverted vesicles with 0.1 M succinate in 50 mM KPi, pH 7.8, at 37°C; succinate/quinone oxidoreductase activity was determined by plumbagin-dependent cytochrome c reduction (25).
Because anaerobiosis is essential for maintenance of the full activity of aconitase (S. M. Varghese and J.A.I., unpublished results), cell extracts were prepared by sonication in anaerobic 50 mM Tris⋅HCl, pH 7.8, at 1% the culture volume, in the anaerobic chamber. Aliquots were immediately diluted into 100 mM isocitrate in the Tris buffer and sealed inside air-tight cuvettes, and activity was quantitated by the production of cis-aconitate (26). Because some aconitase activity is from the stable isozyme aconitase A, this latter activity was determined by the inactivation of aconitase B by treatment with 1 mM EDTA for 1 h before assay (S. M. Varghese and J.A.I., unpublished results) and subtracted to indicate the aconitase B activity.
Total fumarase activity was assayed by the conversion of 50 mM malate to fumarate. To distinguish the oxidatively labile fumarase A isozyme from fumarase C, fumarase A was inactivated by exposure of the extract to xanthine oxidase and xanthine for 15 min at room temperature before reassay (27). The residual activity was caused by fumarase C. Isocitrate DH was assayed by the production of NADPH (28). To test the responsiveness of the SoxRS (superoxide) regulon to superoxide stress, the cells were cultured in the presence or absence of 10 μM paraquat for more than three generations before harvesting for fumarase C determinations.
The activity of 6-phosphogluconate dehydratase was assayed by the two-step procedure for the determination of pyruvate (29). Glutamate synthase activity was determined by the oxidation of NADPH (30, 31). Protein was measured by the Coomassie dye-binding assay (Pierce).
The activity of FNR (for fumarate nitrate reduction) in whole cells was determined in strains containing lacZ under the control of an FNR-activated promoter, PdmsA (DMSO reductase). Cells were grown aerobically or anaerobically to an OD600 of 0.1–0.2 or 0.3–0.7, respectively, in glucose minimal medium supplemented with yeast extract (5 g/liter). β-galactosidase activity was measured as described (15).
Results
Generation of a iscS Null Strain.
To address the function of IscS in E. coli, a gene disruption was created by replacing the iscS coding region with a gene encoding KnR. The genotype of the ΔiscS∷KnR strain was verified by PCR using gene-specific primers (data not shown). Genetic mapping using P1 generalized transduction and a Tn10 (zff-208, 57.4 min) tightly linked to iscS (57.3 min) also showed that the KnR marker was at the expected location in the E. coli chromosome.
IscS Is Not Essential in E. coli.
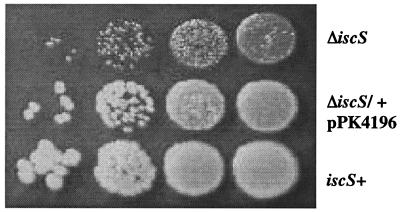
The iscS mutant (PK4331) had a small colony phenotype on LB plates (Fig. 2), with colony formation taking twice as long for the mutant as for the iscS+ wild-type (WT) parent cells (RZ4500). P1 transduction demonstrated that the small colony phenotype was always associated with the KnR marker (data not shown), and when the WT iscS locus was cotransduced with a nearby Tn10 marker (zff-208) into the KnR mutant, all of the Kn-sensitive (iscS+) transductants grew as well as WT. In addition, the phenotype was corrected by the addition of iscS in trans on plasmid pPK4196 (Fig. 2). Because the iscS mutant took 2 days to form colonies, and because similar attempts to create a iscS null strain in A. vinelandii were unsuccessful (5), we wanted to address the possibility that the iscS mutant strain might have acquired a secondary mutation for viability. This appears not to be the case, because upon selection of the ΔiscS∷KnR marker or an unlinked phe∷Tn10 marker transduced independently from the same strain similar numbers of transductants were obtained. If a suppressor mutation was required for viability, then fewer ΔiscS∷KnR transductants would be expected because of the additional event that would be needed for growth. Therefore, in contrast to the situation in A. vinelandii and yeast (5–7), iscS is not essential for viability in E. coli, allowing us to analyze the function of IscS in vivo.
Figure 2.
The iscS mutant has a small colony phenotype. Colony phenotype of the iscS mutant (PK4331; Top) after 48 h of growth on LB plates at 37°C compared with the isogenic IscS+ strain (RZ4500; Bottom). Each spot represents a 10-fold serial dilution. The iscS mutant complemented with iscS in trans on plasmid pPK4196 (Middle).
The iscS Mutant Has a Slow Growth Phenotype in Rich Media and Is Auxotrophic for Thiamine and Nicotinic Acid Under Aerobic Growth Conditions.
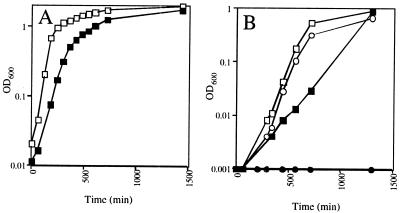
In addition to producing small colonies on LB solid medium, the iscS mutant also exhibited a slow growth phenotype in LB liquid medium with a doubling time of 62 min for the iscS mutant compared with 31 min for WT (Fig. 3A). The iscS mutant did not grow at all in glucose minimal medium (Fig. 3B), even when casamino acids were present (data not shown). However, the addition of yeast extract to glucose minimal medium restored growth to the rate observed in LB medium (data not shown). Subsequent experiments determined that the addition of thiamine and nicotinic acid (Fig. 3B) to glucose minimal medium was sufficient for growth of the iscS mutant under aerobic conditions, with doubling times of 67 min for WT and 122 min for the iscS mutant. However, under anaerobic growth conditions the growth requirement for thiamine was eliminated (data not shown). To verify that the growth phenotypes resulted from the iscS deletion, we added iscS in trans (pPK4196), which relieved the thiamine and nicotinic acid auxotrophies and restored the growth rate to that of WT (data not shown). The iscS mutant also grew more slowly than WT in fumarate minimal medium, with doubling times of 230 min and 110 min, respectively. Therefore, under all conditions where the iscS mutant grew, the growth rate was consistently half the rate of WT growth, suggesting that this effect on growth rate is not related to nutritional requirements.
Figure 3.
Growth phenotype of the iscS mutant. (A) Growth curves of the iscS mutant (PK4331; ■) and the isogenic IscS+ parent strain (RZ4500; □) grown aerobically in LB medium. (B) Growth curves of the iscS mutant (●) and the parent strain (○) grown aerobically in glucose minimal medium, and the iscS mutant (■) and the parent strain (□) grown in glucose minimal medium supplemented with thiamine and nicotinic acid.
IscS Is Required for Full Activity of Many Fe-S Proteins.
The cysteine desulfurase IscS previously has been shown to participate in in vitro Fe-S cluster assembly. Therefore, the activity of a number of Fe-S proteins was measured in the iscS mutant to assess the role of IscS in vivo. The lack of IscS resulted in a 2- to 50-fold decrease in the specific activities of a number of proteins containing [4Fe-4S] clusters (Table 2 and Fig. 4). IscS appeared to be important for the activity of both soluble and membrane-bound Fe-S enzymes, because proteins from both classes had reduced activities in the iscS mutant. Enzymes that did not contain Fe-S clusters, such as isocitrate DH (Table 2) and the respiratory Ndh2 (0.99 unit/mg in the mutant compared with 0.94 unit/mg in the parent), were unaffected by the mutation. Ndh1 activity was decreased to a greater extent (50-fold) than aconitase B activity (2-fold) even when both were assayed from the same cell culture, demonstrating that the distinct growth conditions necessary to assay certain enzyme activities (e.g., fumarase) did not cause the variable effect the iscS mutation had on different enzymes.
Table 2.
Effect of the iscS mutation upon enzyme activities
| + | − | −/S | −/PsSU | |
|---|---|---|---|---|
| NADH DH I | 100 (0.47) | 1.4 | 6.3 | 28 |
| SDH (succinate oxidase) | 100 (0.033) | 17 | 22 | 73 |
| (quinone reduction) | 100 (0.047) | 12 | 23 | 70 |
| Glutamate synthase | 100 (0.42) | 34 | 57 | 51 |
| Aconitase B | 100 (0.021) | 52 | 60 | 90 |
| 6-phosphogluconate dehydratase | 100 (0.098) | 11 | 43 | 52 |
| Fumarase A | 100 (1.3) | 49 | 59 | ND |
| Isocitrate DH | 100 (1.04) | 107 | 113 | 103 |
Averages are shown from 2–3 assays with <5% variation and are normalized to those of the parent iscS+ strain. The values in parentheses are the specific activities (units/mg) of the iscS+ strain. 1 unit = 1 μmol/min of product, except for succinate oxidase, where it represents 1 μmol per min O2 consumed. ND, not determined. +, the iscS+ strain, −, the iscS− strain, −/S, the iscS− strain containing plasmid pPK4196, and −/PsSU, the iscS− strain containing plasmid pPK4344.
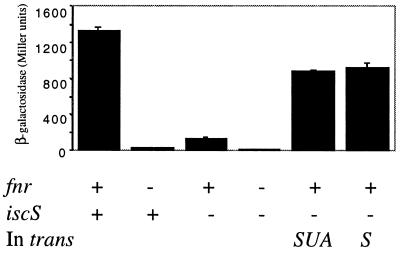
Figure 4.
The iscS mutant has reduced FNR activity that can be complemented by the addition of iscS in trans. FNR activity was measured by assaying the amount of β-galactosidase (Miller units) produced in strains grown under anaerobic conditions containing the FNR-dependent dmsA′∷lacZ promoter fusion. The genotypes are indicated below the figure. Complemented strains contained either pPK4196 (iscS) or pPK4194 (iscSUA) and as with complemented strains in Figs. 2 and 3, expression of the isc gene(s) depended on fortuitous promoter elements from the vector (pET11a). The strains are all derivatives of either PK3292 (FNR+) or PK3293 (FNR-; Table 1).
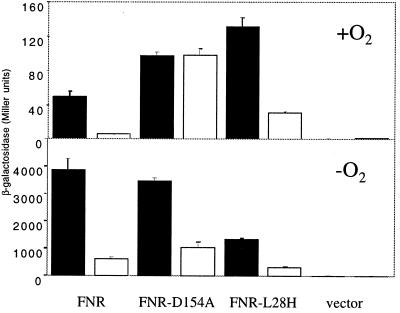
To demonstrate a direct role for IscS in in vivo Fe-S cluster formation, the anaerobic global regulator, FNR, was analyzed to take advantage of FNR mutants that have altered Fe-S cluster requirements. FNR is normally active only under anaerobic conditions because of the O2 lability of its [4Fe-4S] cluster. FNR activity was monitored by assaying β-galactosidase activity from strains containing an FNR-dependent dmsA′∷lacZ reporter fusion construct. As was the case with other Fe-S proteins analyzed, FNR activity was significantly decreased in the iscS mutant (PK4357) to 10% of the activity observed in the iscS+ strain (PK3292) grown under anaerobic conditions (Fig. 4). This decrease in FNR activity cannot be explained by a decrease in FNR protein levels because Western blot analysis showed that FNR protein levels were equivalent in both the WT and the iscS mutant strains (data not shown). To demonstrate that the decrease in FNR activity was directly caused by a defect in Fe-S cluster synthesis, we measured the activity of a FNR mutant (D154A), which unlike WT FNR, has measurable activity under aerobic conditions when the protein is devoid of a Fe-S cluster (32, 33). The aerobic activity of FNR-D154A was not altered by the deletion of iscS (Fig. 5 Upper), consistent with the fact that FNR-D154A activity is independent of a Fe-S cluster under aerobic growth conditions. A decrease in the activity of FNR-D154A is observed in the IscS- strain under anaerobic conditions as is found with WT FNR (Fig. 5 Lower), consistent with previous studies showing that the majority of FNR-D154A activity under anaerobic conditions depends on the [4Fe-4S] cluster (33, 34). Thus these results show that the loss of IscS only reduces the activity of those forms of FNR that depend on an Fe-S cluster for activity.
Figure 5.
The effect of the iscS mutation on the activity of FNR mutants that exhibit altered Fe-S cluster requirements. β-galactosidase activity produced from the dmsA′∷lacZ promoter fusion was assayed from strains (IscS+; PK3293, filled columns, IscS-; PK4359, open columns) grown under aerobic conditions (Upper) and anaerobic conditions (Lower). WT FNR, FNR-D154A, and FNR-L28H were present on the multicopy plasmid pACYC184 (58).
The IscS Defects Resulted from a Block in de Novo Fe-S Cluster Synthesis.
Two lines of evidence suggest that the decrease in Fe-S cluster protein activity in the iscS deletion strain was caused by a decrease in de novo cluster synthesis rather than by an inability to repair oxidized clusters. First, using another well-characterized FNR mutant, L28H, we show that the decreased activity observed in the iscS mutant is independent of O2. The [4Fe-4S] cluster in FNR-L28H has been shown to be less O2 labile than the WT protein, resulting in increased activity of this protein under aerobic conditions (35). Nevertheless, when cells are grown aerobically the [4Fe-4S] cluster of FNR-L28H is likely to be more susceptible to oxidative damage than when cells are grown anaerobically. Thus, if IscS were functioning in the repair of oxidatively damaged clusters, then the aerobic activity of FNR-L28H should show a greater dependence on IscS. However, the iscS mutant shows the same 4-fold decrease in activity under both aerobic and anaerobic growth conditions (Fig. 5), suggesting that IscS does not preferentially function in a cluster repair pathway; rather, IscS appears to function primarily in de novo cluster synthesis.
Additional evidence supporting the role of IscS in de novo cluster synthesis is that neither 6-phosphogluconate dehydratase nor aconitase B could be reconstituted by treatment of the iscS mutant extracts with DTT and Fe2+ (data not shown). In contrast, this protocol restored the activities of these enzymes when they were oxidatively damaged in vitro or in WT cells (36, 37) (data not shown). It is likely that cluster oxidation to the [4Fe-4S]3+ state triggers demetallation to an inactive [3Fe-4S]1+ form that can be converted back to the active form without a sulfur source. Evidently the proteins in the iscS mutant accumulate incomplete clusters that are deficient in sulfur atoms, which differ from protein clusters after oxidation by superoxide, suggesting that the Fe-S protein defects observed in the iscS deletion strain reflect deficiencies in de novo Fe-S cluster assembly.
Complementation of the iscS Mutant.
In addition to pPK4196 (iscS+) complementing the growth phenotype of the iscS mutant, the level of FNR activity restored by the presence of the same plasmid was 80% of the levels observed in the iscS+ parent (Fig. 4). Although this represents a significant restoration in FNR activity, we were surprised that the levels were not the same as that observed in the iscS+ strains. Because the iscS mutant strain contained a KnR gene in place of iscS, we considered the possibility that the KnR insertion also might be affecting expression of the downstream genes iscUA. iscUA (Fig. 1) show similarity to two other A. vinelandii nif genes, nifU and orf6, that also have been implicated in Fe-S cluster assembly (5). However, the amount of FNR activity observed in the strain containing iscSUA in trans (pPK4194) was no different from that observed in the strain containing just iscS in trans (Fig. 4), suggesting that a decrease in the amount of the iscUA gene products is not contributing to the mutant phenotype. In contrast to the results obtained with FNR, the activity of several of the other Fe-S proteins was only minimally restored in strains containing either pPK4196 (iscS; Table 2) or pPK4194 (iscSUA; data not shown). Because all of the strains in Table 1 lack T7 RNA polymerase, expression of the isc genes from these plasmids depends on fortuitous vector promoter sequences, and thus IscS levels are likely to be low. Therefore, we introduced a plasmid containing the native iscS promoter along with iscSU (pPK4344). Although strains containing pPK4344 did not show any increase in FNR activity (data not shown), this plasmid led to increased activity for many of the Fe-S proteins (Table 2). Taken together, these data suggest that the degree of complementation may be related to the level of iscS expression. If this is indeed the case, then the decreases in Fe-S protein activities observed in the ΔiscS∷KnR strain are most easily explained by the lack of iscS.
Discussion
The Role of IscS and Other Possible Cysteine Desulfurases in in Vivo Fe-S Cluster Formation.
Previous studies of IscS have indicated that the cysteine desulfurase activity of this enzyme is used for in vitro Fe-S cluster formation (10). In this study, we observed that all examined Fe-S proteins had decreased levels of activity in the strain lacking IscS, indicating that IscS is also a major donor of sulfur atoms for the construction of Fe-S clusters in vivo. The fact that all Fe-S enzymes exhibited some activity demonstrates that a modest amount of cluster assembly persists in the iscS mutant. The efficiency of the residual cluster building pathway(s) varied among the enzymes that were tested, being particularly inadequate for Ndh1, SDH, and 6-phosphogluconate dehydratase. Although Ndh1 and SDH share the property of requiring multiple clusters for activity and these clusters are buried inside the holoenzyme, neither of these properties holds true for 6-phosphogluconate dehydratase, indicating no clear trends with this data set.
The E. coli genome contains three ORFs that encode proteins homologous to A. vinelandii NifS. IscS, the focus of this study, is 40% identical to NifS, and this homology exists through the entire protein. The two remaining NifS homologs, CSD and CsdB, represent a separate class in that they are both 24% identical to NifS with the highest homology in the N-terminal half of the proteins. Although both CSD and CsdB have cysteine desulfurase activity, selenocysteine is the preferred substrate (38, 39). In addition to CSD and CsdB, several other enzymes from E. coli were able to desulfurylate cysteine in vitro, stimulating Fe-S cluster assembly in apoenzymes (11). However, the enzymes that were shown to carry out this activity have other, well-established roles in cysteine metabolism, so their cluster assembly activity in vitro could be fortuitous or they could represent minor reactions for these enzymes. Nevertheless, these activities may account for the residual cluster assembly that occurs in the iscS mutant.
IscS Appears to be Involved in de Novo Fe-S Cluster Synthesis.
Assembly of Fe-S clusters must occur in E. coli in at least two circumstances: during de novo cluster synthesis on the nascent apoenzyme and during the repair of oxidatively damaged clusters. Several lines of evidence indicate that the properties of the IscS mutant reported here resulted from inadequate de novo assembly rather than failed repair. The magnitude of the decrease in FNR-L28H activity was similar in cells grown both aerobically and anaerobically, whereas a defect in cluster repair would be expected to have a greater impact in air. The defects in FNR function and nicotinic acid biosynthesis persisted during anaerobiosis, when oxidative damage could not occur. Further, several proteins with oxidatively stable clusters (SDH, Ndh1, and glutamate synthase) were affected as much by the iscS mutation as were proteins with unstable clusters (aconitase B, fumarase A, 6-phosphogluconate dehydratase, and FNR). Finally, oxidative inactivation of even the labile dehydratase Fe-S clusters has been estimated to occur fairly slowly in routinely cultured aerobic cells (t1/2 = 40 min) (27). Because this rate is relatively low, the activity deficits that we measured cannot be ascribed solely to a defect in cluster repair. These results all indicate that the defects in Fe-S cluster assembly that were observed in this study occurred because IscS is necessary for efficient de novo cluster assembly in most or all Fe-S proteins. Whether IscS is additionally involved in some cluster repair processes remains to be determined.
The Role of IscS in [2Fe-2S] Cluster Synthesis.
All of the enzymes in Table 2 contain clusters of the [4Fe-4S] type, precluding any conclusion regarding the role of IscS in building [2Fe-2S] clusters. However, we found that treatment of the iscS mutant with paraquat led to a 10-fold induction of fumarase C, exactly as in the WT parent (data not shown). This result suggests that a substantial amount of SoxR protein, which mediates this response, contains its required [2Fe-2S] cluster (40). However, other work has shown that neither [4Fe-4S] nor [2Fe-2S] ferredoxins can be amply overproduced in E. coli unless the isc and hsc gene clusters are overexpressed as well (41). Additional work will be necessary to address the specificity of IscS in formation of [2Fe-2S] clusters and in the conversion of a [2Fe-2S] to a [4Fe-4S] cluster that occurs in some Fe-S proteins (42).
The Conserved Genomic Organization of the isc Genes and the hsc Genes Implies a Functional Relationship.
The grouping of iscU and iscA immediately downstream of iscS suggests that these two gene products are also likely to be involved in Fe-S cluster assembly, and this has recently been demonstrated in vitro for IscU from A. Vinelandii (43). Downstream from the isc genes are dnaJ and dnaK molecular chaperone homologs (hscBA, respectively) and a ferredoxin (fdx) that have been implicated in Fe-S cluster formation (5), and furthermore, this genomic organization is conserved in other prokaryotes (17, 44) (Fig. 1). Although hscBAfdx are located immediately downstream of iscSUA, a transcription start site was identified upstream of hscB (45), indicating that these genes are transcribed independently of iscSUA in E. coli. Thus it seems unlikely that the iscS gene disruption is affecting levels of the hscBAfdx gene products. Nevertheless, the hsc gene products appear to be playing a role in Fe-S cluster assembly, because FNR activity is reduced 4-fold in a hscA- strain (data not shown), and both the isc genes and the downstream hsc genes are needed for efficient overproduction of ferredoxins (41, 46). In addition, S. cerevisiae strains deficient in iscU homologs (isu1 and nfu1) or a hscA homolog (ssq1) exhibit a similar phenotype of decreased activity of Fe-S proteins (7, 47), indicating that these genes are functionally related in the process of Fe-S cluster assembly.
The Growth Phenotype of the iscS Mutant May Have Both Fe-S Cluster Protein-Dependent and -Independent Components.
The observation that the iscS mutant grew at a slower rate than WT was not unexpected because a number of metabolic enzymes contain Fe-S clusters. However, one cannot conclude at this time whether the slower growth rate is caused only by Fe-S protein defects or is related to other cellular functions requiring IscS as a sulfur donor, such as formation of thiouridine in tRNA (48).
In contrast, the requirement for nicotinic acid in the iscS mutant is most likely explained by a Fe-S cluster protein defect. In Bacillus subtilis, mutations in a nifS-like gene or the putative Fe-S protein NadA (quinolinate synthase A) in the NAD biosynthetic pathway lead to an auxotrophy for nicotinic acid, a precursor for NAD (49, 50). To explain the thiamine requirement is more complex because the thiazole ring itself contains sulfur and IscS could function as the sulfur donor. In support of this, ThiI, a protein required for both thiamine and tRNA modification (51, 52) recently has been shown to function in the transfer of sulfur from IscS to form thiouridine in tRNA (48). In addition, an enzyme in the thiamine biosynthetic pathway (ThiH) has been proposed to contain an Fe-S cluster (53), and the activity of this enzyme may be insufficient in the iscS mutant. Consistent with an Fe-S protein defect, oxidative stress, a condition known to damage Fe-S clusters, also induces a growth requirement for thiamine and nicotinic acid (54). Interestingly, similar growth requirements have been observed in a Salmonella typhimurium strain with an insertion into iscR, the gene upstream of iscS (59). It remains an open question as to why the thiamine requirement was alleviated under anaerobic conditions. If thiamine production is reduced in the IscS- strain, a simple explanation is that the level of thiamine needed to support anaerobic growth is less than that needed under aerobic conditions. This would not be surprising because expression of two major thiamine requiring enzymes, pyruvate dehydrogenase and αketoglutarate dehydrogenase, is repressed under anaerobic conditions (55).
The recent plethora of information about the isc genes and Fe-S cluster formation has led to a greater understanding of this essential cellular process. However, many questions remain, such as what other roles the IscS protein might perform in vivo, the source of the residual cluster assembly activity in the iscS mutant, whether [2Fe-2S] clusters are formed by the same assembly pathway as the [4Fe-4S] clusters, how the chaperone-like proteins are involved, and how these genes are regulated and this process is coordinated. Mutants in E. coli should provide a means by which these issues can be addressed at the molecular level.
Note Added in Proof.
Lauhon et al. (60) have recently made a similar observation that E. coli strains lacking Isc S require nicotinic acid and thiamin.
Acknowledgments
We thank N. Hintz for the deaminoNADH, V. Sutton for Western blots, L. Vickery for the hscA- strain, F. Blattner for unpublished E. coli genomic sequence, G. Church for pK03, and E. Skovran and D. Downs for sharing unpublished data. For reading this paper, we thank T. Donohue, C. Voisine, J. Newman, T. Isenbarger, J. Lewis, and the Kiley lab. This work was supported by National Institutes of Health Grants GM-45844 (to P.J.K.) and GM49640 (to J.A.I.).
Abbreviations
- Kn
kanamycin
- FNR
fumarate nitrate reduction
- WT
wild type
- SDH
succinate dehydrogenase
- DH
dehydrogenase
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.160261497.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.160261497
References
- 1.Beinert H, Holm R H, Münck E. Science. 1997;277:653–659. doi: 10.1126/science.277.5326.653. [DOI] [PubMed] [Google Scholar]
- 2.Dean D R, Bolin J T, Zheng L. J Bacteriol. 1993;175:6737–6744. doi: 10.1128/jb.175.21.6737-6744.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zheng L, White R H, Cash V L, Dean D R. Biochemistry. 1994;33:4714–4720. doi: 10.1021/bi00181a031. [DOI] [PubMed] [Google Scholar]
- 4.Zheng L, White R H, Cash V L, Jack R F, Dean D R. Proc Natl Acad Sci USA. 1993;90:2754–2758. doi: 10.1073/pnas.90.7.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng L, Cash V L, Flint D H, Dean D R. J Biol Chem. 1998;273:13264–13272. doi: 10.1074/jbc.273.21.13264. [DOI] [PubMed] [Google Scholar]
- 6.Li J, Kogan M, Knight S A B, Pain D, Dancis A. J Biol Chem. 1999;274:33025–33034. doi: 10.1074/jbc.274.46.33025. [DOI] [PubMed] [Google Scholar]
- 7.Strain J, Lorenz C R, Bode J, Garland S, Smolen G A, Ta D T, Vickery L E, Culotta V C. J Biol Chem. 1998;273:31138–31144. doi: 10.1074/jbc.273.47.31138. [DOI] [PubMed] [Google Scholar]
- 8.Kispal G, Csere P, Prohl C, Lill R. EMBO J. 1999;18:3981–3989. doi: 10.1093/emboj/18.14.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng L, Dean D R. J Biol Chem. 1994;269:18723–18726. [PubMed] [Google Scholar]
- 10.Flint D H. J Biol Chem. 1996;271:16068–16074. [PubMed] [Google Scholar]
- 11.Flint D H, Tuminello J F, Miller T J. J Biol Chem. 1996;271:16053–16067. doi: 10.1074/jbc.271.27.16053. [DOI] [PubMed] [Google Scholar]
- 12.Flint D, Smyk-Randall E, Tuminello J, Draczynska-Lusiak B, Brown O. J Biol Chem. 1993;268:25547–25552. [PubMed] [Google Scholar]
- 13.Kuo C F, Machino T, Fridovich I. J Biol Chem. 1987;262:4724–4727. [PubMed] [Google Scholar]
- 14.Keyer K, Imlay J A. J Biol Chem. 1997;272:27652–27659. doi: 10.1074/jbc.272.44.27652. [DOI] [PubMed] [Google Scholar]
- 15.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 16.Kohara Y, Akiyama K, Isono K. Cell. 1987;50:495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- 17.Blattner F, Plunkett G, Bloch C, Perna N, Burland V, Riley M, Colladovides J, Glasner J, Rode C, Mayhew G. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 18.Yanisch-Perron C, Vieira J, Messing J. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 19.Dubendorff J W, Studier F W. J Mol Biol. 1991;219:45–59. doi: 10.1016/0022-2836(91)90856-2. [DOI] [PubMed] [Google Scholar]
- 20.Harayama S, Leppik R A, Rekik M, Mermod N, Lehrbach P R, Reineke W, Timmis K N. J Bacteriol. 1986;167:455–461. doi: 10.1128/jb.167.2.455-461.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Link A J, Phillips D R, Church G M. J Bacteriol. 1997;179:6228–6237. doi: 10.1128/jb.179.20.6228-6237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Low K B. Bacteriol Rev. 1972;36:587–607. doi: 10.1128/br.36.4.587-607.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meinhardt S W, Matsushita K, Kaback R H, Ohnishi T. Biochemistry. 1989;28:2153–2160. doi: 10.1021/bi00431a029. [DOI] [PubMed] [Google Scholar]
- 24.Rafter G W, Colowick S P. Methods Enzymol. 1957;3:887–890. [Google Scholar]
- 25.Imlay J. J Biol Chem. 1995;270:19767–19777. [PubMed] [Google Scholar]
- 26.Henson C P, Cleland W W. J Biol Chem. 1967;242:3833–3838. [PubMed] [Google Scholar]
- 27.Gort A, Imlay J. J Bacteriol. 1998;180:1402–1410. doi: 10.1128/jb.180.6.1402-1410.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cribbs R, Englesberg E. Genetics. 1964;49:95–108. doi: 10.1093/genetics/49.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fraenkel D G, Horecker B L. J Biol Chem. 1964;239:2765–2771. [PubMed] [Google Scholar]
- 30.Meister A. Methods Enzymol. 1995;113:327–337. doi: 10.1016/s0076-6879(85)13045-4. [DOI] [PubMed] [Google Scholar]
- 31.Miller R, Stadman E. J Biol Chem. 1972;247:7407–7419. [PubMed] [Google Scholar]
- 32.Lazazzera B A, Beinert H, Khoroshilova N, Kennedy M C, Kiley P J. J Biol Chem. 1996;271:2762–2768. doi: 10.1074/jbc.271.5.2762. [DOI] [PubMed] [Google Scholar]
- 33.Lazazzera B A, Bates D M, Kiley P J. Genes Dev. 1993;7:1993–2005. doi: 10.1101/gad.7.10.1993. [DOI] [PubMed] [Google Scholar]
- 34.Bates D M, Lazazzera B A, Kiley P J. J Bacteriol. 1995;177:3972–3978. doi: 10.1128/jb.177.14.3972-3978.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bates D M, Popescu C V, Khoroshilova N, Vogt K, Beinert H, Münck E, Kiley P J. J Biol Chem. 2000;275:6234–6240. doi: 10.1074/jbc.275.9.6234. [DOI] [PubMed] [Google Scholar]
- 36.Gardner P R, Fridovich I. J Biol Chem. 1991;266:19328–19333. [PubMed] [Google Scholar]
- 37.Gardner P R, Fridovich I. J Biol Chem. 1991;266:1478–1483. [PubMed] [Google Scholar]
- 38.Mihara H, Kurihara T, Yoshimura T, Soda K, Esaki N. J Biol Chem. 1997;272:22417–22424. doi: 10.1074/jbc.272.36.22417. [DOI] [PubMed] [Google Scholar]
- 39.Mihara H, Maeda M, Fujii T, Kurihara T, Hata Y, Esaki N. J Biol Chem. 1999;274:14768–14772. doi: 10.1074/jbc.274.21.14768. [DOI] [PubMed] [Google Scholar]
- 40.Bradley T M, Hidalgo E, Leautaud V, Ding H, Demple B. Nucleic Acids Res. 1997;25:1469–1475. doi: 10.1093/nar/25.8.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takahashi Y, Nakamura M. J Biochem. 1999;126:917–926. doi: 10.1093/oxfordjournals.jbchem.a022535. [DOI] [PubMed] [Google Scholar]
- 42.Johnson M K. Curr Opin Chem Biol. 1998;2:173–181. doi: 10.1016/s1367-5931(98)80058-6. [DOI] [PubMed] [Google Scholar]
- 43.Agar J N, Zheng L, Cash V L, Dean D R, Johnson M K. J Am Chem Soc. 2000;122:2136–2137. [Google Scholar]
- 44.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J-F, Merrick J M, Dougherty B A. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 45.Lelivelt M J, Kawula T H. J Bacteriol. 1995;177:4900–4907. doi: 10.1128/jb.177.17.4900-4907.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakamura M, Saeki K, Takahashi Y. J Biochem. 1999;126:10–18. doi: 10.1093/oxfordjournals.jbchem.a022409. [DOI] [PubMed] [Google Scholar]
- 47.Schilke B, Voisine C, Beinert H, Craig E. Proc Natl Acad Sci USA. 1999;96:10206–10211. doi: 10.1073/pnas.96.18.10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kambampati R, Lauhon C T. Biochemistry. 1999;38:16561–16568. doi: 10.1021/bi991119r. [DOI] [PubMed] [Google Scholar]
- 49.Gardner P R, Fridovich I. Arch Biochem Biophys. 1991;284:106–111. doi: 10.1016/0003-9861(91)90270-s. [DOI] [PubMed] [Google Scholar]
- 50.Sun D, Setlow P. J Bacteriol. 1993;175:1423–1432. doi: 10.1128/jb.175.5.1423-1432.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Webb E, Class K, Downs D M. J Bacteriol. 1997;179:4399–4402. doi: 10.1128/jb.179.13.4399-4402.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mueller E G, Buck C J, Palenchar P M, Barnhart L E, Paulson J L. Nucleic Acid Res. 1998;26:2606–2610. doi: 10.1093/nar/26.11.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Begley T P, Downs D M, Ealick S E, McLafferty F W, Loon A P G M V, Taylor S, Campobasso N, Chiu H-J, Kinsland C, Reddick J J, Xi J. Arch Microbiol. 1999;171:293–300. doi: 10.1007/s002030050713. [DOI] [PubMed] [Google Scholar]
- 54.Brown O R, Seither R L. Fundam Appl Toxicol. 1983;3:209–214. doi: 10.1016/s0272-0590(83)80127-4. [DOI] [PubMed] [Google Scholar]
- 55.Guest J R. J Gen Microbiol. 1992;138:2253–2263. doi: 10.1099/00221287-138-11-2253. [DOI] [PubMed] [Google Scholar]
- 56.Singer M, Baker T A, Schnitzler G, Deischel S M, Goel M, Dove W, Jaacks K J, Grossman A D, Erickson J W, Gross C A. Microbiol Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Choe M, Reznikoff W S. J Bacteriol. 1991;173:6139–6146. doi: 10.1128/jb.173.19.6139-6146.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chang A C, Cohen S N. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Skovran E, Downs D. J Bacteriol. 2000;182:3896–3903. doi: 10.1128/jb.182.14.3896-3903.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lauhon C T, Kambampati R. J Biol Chem. 2000;275:20096–20103. doi: 10.1074/jbc.M002680200. [DOI] [PubMed] [Google Scholar]