Abstract
Biological systems are orchestrated by heterogeneous regulatory programs that control complex processes and adapt to a dynamic environment. Recent advances in high-throughput experimental methods provide genome-wide perspectives on such regulatory programs. A considerable amount of data on the behavior of model systems in a variety of conditions is rapidly accumulating. Still, the dominant paradigm is to analyze new genome-wide experiments separately from any other extant data, for example, by clustering the new data alone. Here we introduce a new methodology for analyzing the results of a new functional genomic study vis-à-vis a large compendium of previously published results from heterogeneous experimental techniques. We demonstrate our methodology on Saccharomyces cerevisiae, using a compendium of some 2000 experiments from 60 different publications. Most importantly, we show how the integrated analysis reveals unexpected connections among biological processes, and differentiates between novel and known effects in the analyzed experiments. Such characterization is impossible when new data sets are studied in isolation. Our results exemplify the power of the integrative approach in the analysis of genomic scale data sets and call for a paradigm shift in their study.
Keywords: data integration, gene expression analysis, biclustering
Introduction
Over the last several years, a variety of methods have been used to monitor biological processes on a genomic scale. In a typical study, the researchers define a target cellular response (e.g., a signaling pathway, response to certain environment or disease), select a set of conditions (genetic perturbations, environmental stimulations, a new drug, etc.) and perform high-throughput experiments in these settings. A rapidly increasing pool of techniques allows measurements of gene expression (DeRisi et al, 1997), transcription factor binding (Ren et al, 2000; Iyer et al, 2001), selection from a mutant collection (Birrell et al, 2002), two-hybrid analysis (Schwikowski et al, 2000), synthetic lethality (Tong et al, 2004) and more. The typical computational analysis, following Eisen et al (1998), clusters the data obtained and then tries to characterize each cluster's gene set using known gene functions and promoter analysis. Usually, a laborious expert scrutiny of specific genes and their behavior is needed in order to reach meaningful biological conclusions. This methodology has proven very effective in identifying primary trends in the experimental results. Following the publication of many dozens of high-throughput studies, a very rich resource is now available, containing thousands of different molecular snapshots of wild-type and mutant cells under different conditions. The current analysis paradigm does not directly take full advantage of this vast resource. In analogy, the current method for analyzing microarray profiles is similar to trying to find structural motifs in a small number of new cDNA clones without using homology searches in appropriate sequence databases.
In this work, we introduce a new methodology that takes advantage of a large data compendium in the analysis of novel high-throughput experiments. Previously, we have shown how data from different sources can be integrated using the SAMBA biclustering algorithm (Tanay et al, 2004b). Here we develop a method to characterize the response of biological pathways to various stimuli at the system level, capturing not only the dominant primary responses but also finer and less-easily tractable processes. Previous approaches to integrated analysis of functional genomics data focused on predicting single gene functions (Kemmeren et al, 2002; Wu et al, 2002; Troyanskaya et al, 2003), studied the global organization of molecular networks and transcriptional programs (Ihmels et al, 2002; Beer and Tavazoie, 2004; Segal et al, 2004; Tanay et al, 2004b) or combined several experimental approaches to construct and test networks for specific systems (Ideker et al, 2001; Prinz et al, 2004). The approach introduced here is aimed at the analysis of a data set from one new study in the context of a large compendium of data from many diverse and heterogeneous prior studies. We combine the broad perspective of global analysis, with a focused and easy-to-use dissection of a single experimental data set, very much like standard clustering-based analysis. Our approach thus allows for rapid interpretation of novel data sets in terms of the activity of known and novel biological modules.
To illustrate the use of the new methodology, we reanalyzed publicly available data on the yeast Saccharomyces cerevisiae. We assembled a comprehensive collection of data from 60 different studies and close to 2000 different experiments. We show that using this compendium and our algorithms, it is possible to greatly extend the understanding of complex regulatory mechanisms, beyond what can be done using single studies. Our tools, data compendium and comprehensive results are available through a new web interface (www.cs.tau.ac.il/~rshamir/simba/).
Results
An integrated compendium of yeast functional data
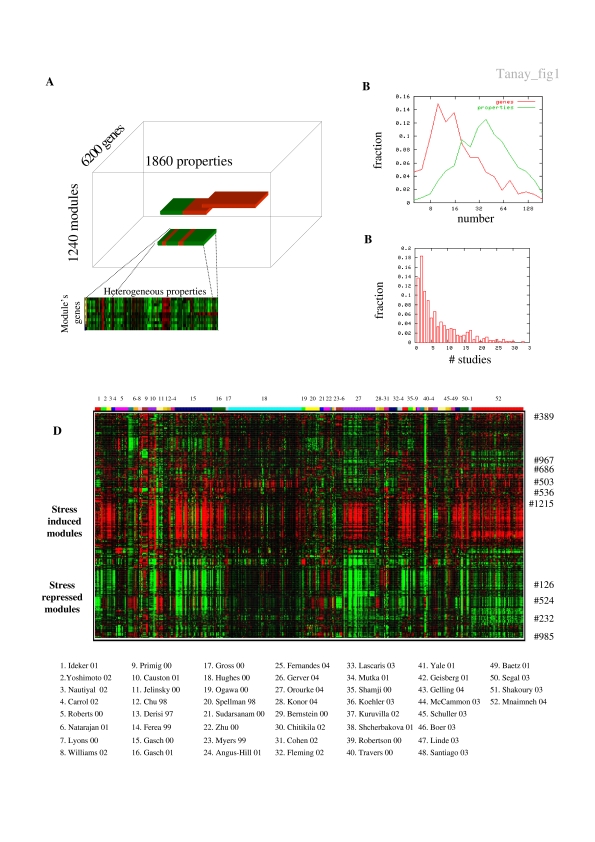
We have built a compendium of yeast functional data including profiles from 52 gene expression studies, five transcription factor location studies, three synthetic lethality studies and data on protein interactions from the GRID database (http://biodata.mshri.on.ca/yeast_grid/servlet/SearchPage). The complete list of references for all data sources is available on Supplementary website (www.cs.tau.ac.il/~rshamir/simba/). Our algorithmic framework (Tanay et al, 2004b) transforms all sources of information into generalized conditions and analyzes them together (Materials and methods). We applied biclustering to the combined data set and derived a set of ∼1200 statistically significant modules. A module consists of a set of genes and a set of conditions, such that the genes have significant and correlated values over the set of conditions. For example, a bicluster may be defined by a set of genes that are (1) coexpressed in several conditions (2) are targeted by the same specific transcription factors and (3) their protein products are likely to interact with a certain protein. To understand the biology behind specific modules, we automatically associated them with known processes and regulatory mechanisms. We assigned modules to biological processes using functional enrichment tests based on the SGD GO annotation (Materials and methods). We searched for known and novel enriched cis-elements in the promoters of the genes in each module and manually annotated the discovered motifs (Materials and methods). When discussing modules, we use the module number, the primary biological process associated with it (when available) and the module's number of genes and properties, for example, module #524 (RNA processing, 76 × 211). A single biological process may be represented by several modules of varying sizes and specificities, but our algorithm guarantees that no two modules are similar.
Synergism between different sources of data
We first asked how much synergism exists among the experimental data from different studies. The distribution of module dimensions (Figure 1B and Supplementary Figure 1) indicates that the comprehensive compendium gives rise to highly specific modules, with 10–50 genes supported by 20–100 conditions. The distribution of the number of studies contributing properties to each module (Figure 1C) demonstrates a high level of synergism in the multistudy data compiled. A total of 86% of the modules used data from more than one study and 68% used data from three studies or more, showing that indeed, information was extracted from multiple data sets and is not biased by one predominant source. A global representation of the compendium and its dissection into modules is obtained by clustering the mean module expression across all experimental conditions. Since the same gene may be part of several modules, such clustering allows the ‘unfolding' of the function of pleiotropic genes and differs substantially from a standard gene-by-condition clustering. The resulting representation (Figure 1D) shows how two opposite environmental stress responses (ESRs; Gasch et al, 2000) dominate the entire compendium. This response to stress is so strong and widespread that other, condition-specific regulatory programs are hard to detect without the combination of multiple studies and the application of sensitive algorithms. As we shall see below, separating the general stress response into specific modules and comparing their activities in different conditions provides further insights into the complex regulation of this biological process.
Figure 1. Integrating yeast functional data.
(A) Bicluster analysis. Our SAMBA biclustering algorithm analyzes an integrated data set to discover an extensive collection of modules. Each module consists of a set of genes and is supported by a set of functional properties. We analyze novel data sets by testing their effect on the modules derived from the entire compendium. (B) Modules' dimensions. The distribution of the number of genes and properties in each module indicates that modules are characterized by specific sets of genes (10–50) and a large number of different experiments (20–100). (C) Synergism among studies. The graph shows the distribution of the number of studies contributing to each module. A total of 86% of the modules use data from more than one study. (D) The module–condition view. To obtain a global view of the behavior of our modules across all conditions, we clustered the module mean values across all conditions. Rows represent modules and columns represent conditions, with numbered colored bars indicating the study reporting each condition (the full references of the studies are available on the website). We show low means in green and high means in red. The global view reveals that the massive repression and induction of genes in stressful conditions dominates the compendium. Using our integrative analysis, we can dissect this response into components and study their specific regulation. The numbers on the right refer to modules addressed in this study.
The cytokinesis transcriptional module
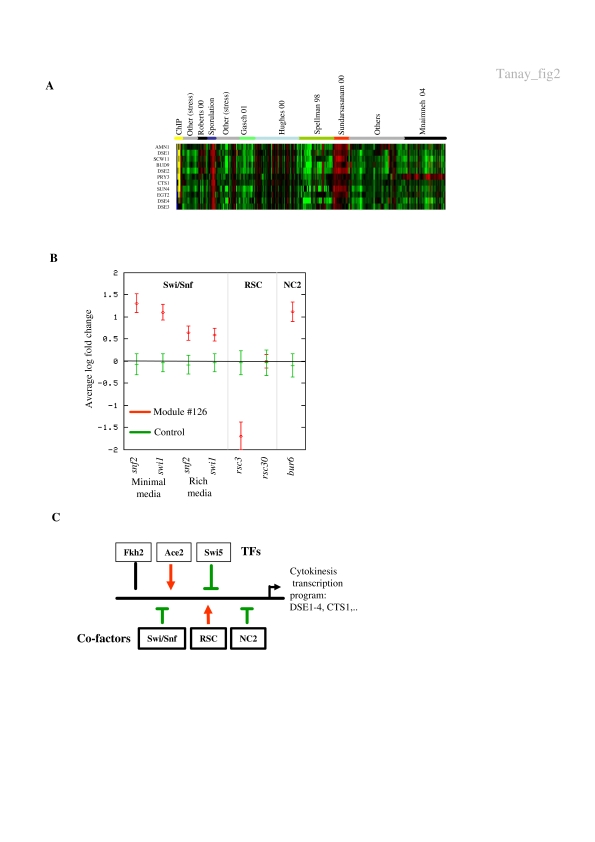
Defined by data from many different experiments, modules can characterize highly specific biological phenomena. Module #126 (Figure 2A and Supplementary website) consists of 11 genes related to cytokinesis and daughter-specific expression. Of these genes, DSE1-4, SCW11, CTS1, EGT2, AMN1 and BUD9 are known to be localized to the daughter cell during late mitosis, and are associated with cell wall separation and exit from mitosis (Colman-Lerner et al, 2001). SUN4 is also known to be involved in cell septation (Velours et al, 2002) and PRY3 encodes a cell wall-specific protein of unknown function. The association of these genes into a single module was based on gene expression data from 261 conditions taken from 30 different studies, and the transcription factor location profiles of the cell cycle regulators Ace2, Swi5 and Fkh2. Indeed, Ace2 and Swi5 are known to have positive and negative effects, respectively, on the transcription of some of the genes in this module (Doolin et al, 2001). Fkh2 is known to regulate genes required for the G2/M transition and has been implicated (together with Ndd1 and Mcm1) in the regulation of the SWI5 and ACE2 genes (Simon et al, 2001), but its direct association to cytokinesis genes, to the best of our knowledge, was not noted before. This possible role for Fkh2 is supported by evidence for its involvement in the regulation of pseudohyphal growth (Zhu et al, 2000) and by its synthetic lethality with CLA4 (Goehring et al, 2003), a gene involved in polarization and budding, which functions in a cascade regulating exit from mitosis (Hofken and Schiebel, 2002). The association of Fkh2 with cytokinesis genes may reflect the need to inhibit the function of these genes until mitosis is completed or during transition to pseudohyphal growth.
Figure 2. Using a massive functional compendium—the cytokinesis module.
(A) Module #126 is defined by data from 30 different studies and contains a highly coherent set of 11 genes, all but two of which are known to be involved in late mitosis and in cell septation. (B) Regulation by coactivators and corepressors. We plot the average module expression (red) and the background genome-wide mean and standard deviation for a random set containing 11 genes (green) under conditions in which the Swi/Snf complex are not expressed in minimal and rich media (Sudarsanam et al, 2000), in conditions blocking components of the RSC complex (Angus-Hill et al, 2001) and in a strain lacking Bur6, a component of the NC2 cofactor (Geisberg et al, 2001). There is significant induction in all Swi/Snf conditions and in the NC2 experiment, indicating a possible negative role for these cofactors in regulating the module. There is also a significant repression in the rsc3 strain (but not in the rsc30 strain), indicating that RSC may have a positive role in the regulation of the module. (C) Extended regulatory model for the cytokinesis module. See text for details.
The wealth of functional information used to construct the module enabled us to explore the behavior of this important transcriptional program across many different experimental conditions. In particular, we analyzed the behavior of the module genes in experiments perturbing different transcriptional coactivators and corepressors (Sudarsanam et al, 2000; Angus-Hill et al, 2001; Geisberg et al, 2001) to try and refine our understanding of the mechanisms of transcriptional regulation used in timing the mitotic events. The module exhibits a statistically significant response in several such experiments (Figure 2B). Strong induction is observed upon perturbation of the SWI/SNF chromatin remodeling complex (t-test, P<0.0001 in minimal media, P<0.001 in rich media, for both mutants). Strong repression was observed in an experiment that inactivated the RSC factor Rsc3 (P<0.0004), but no effect was detected when the RSC factor Rsc30 was inactivated (P<0.89). In addition, a strain knocked out for NC2 activity (BUR6 deletion) exhibited strong increase in the expression of this module (P<0.0002). Interestingly, the behavior of module #126 in the SWI/SNF, RSC and NC2 experiments is unique among all the modules (Supplementary Table 1), suggesting that the particular combination of cofactors uncovered may define the particular regulatory behavior of this module. Taken together, our analysis suggests that the module is controlled by an extended regulatory program that includes the well-known Ace2/Swi5 and Fkh2 transcription factors and a unique combination of coactivators and corepressors (Figure 2C). The cytokinesis module thus exemplifies the power of our methodology to unravel the complex regulation network of a group of coordinated genes.
Regulation of the galactose system
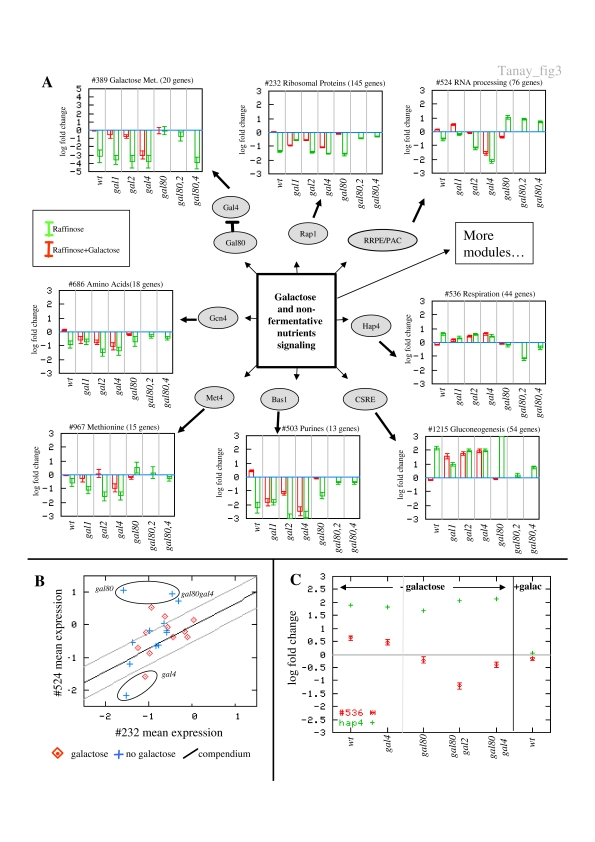
We next turned to the analysis of a single high-throughput data set versus the entire compendium. The yeast galactose utilization pathway is among the best-characterized biological systems. In a systematic set of experiments, Ideker et al (2001) measured the transcriptional response of yeast strains knocked out for a set of enzymes and regulators involved in galactose metabolism. The data were then clustered and analyzed in light of the known Gal4–Gal80–Gal3 regulatory circuit. We used the galactose data set as a test case for our methodology. Instead of clustering yeast genes given their expression in the galactose data set only, we screened our complete set of modules, which are based on almost 2000 experiments, for modules that are responsive in at least one of the conditions analyzed by Ideker et al. Since the data defining our modules are relevant to many different aspects of the yeast regulatory network, we were able to interpret galactose-related conditions from a broad perspective. We depict the effect of galactose-related conditions on several central modules in Figure 3A (interactive visualization of all modules is available on the website). As expected, the strongest effects are well known and were easily observed using clustering of the galactose data set alone. For example, module #389 (Galactose metabolism, 20 × 160), the classical Gal4 regulon, consists mainly of enzymes required for the utilization of galactose (GAL1,2,7,10) and is strongly repressed when galactose is lacking from the medium or when knockouts in the GAL pathway compromise its yield. The response of other modules, however, is less predictable and reveals novel regulatory relations between different processes.
Figure 3. Revisiting the galactose system.
(A) Response of selected modules to disruptions in the GAL system. We plot the mean and standard deviation of the expression of several key modules that our algorithm associated with conditions from the galactose data set (Ideker et al, 2001). For each module, we plot the behavior in four galactose-related mutants and two double mutants grown with (red) and without (green) galactose. Module #389 (Galactose metabolism) is strongly repressed when galactose is lacking or when the GAL pathway yield is compromised. Modules #232 (Ribosomal proteins) and #524 (RNA processing) are repressed when growth is slower. Interestingly, module #524 is particularly repressed when gal4 is knocked out, and is induced when gal80 is knocked out and galactose is not available (right-most bars). Modules #536 (Respiration) and #1215 (Gluconeogenesis) are induced when galactose is not available or not processed. Here again, the gal80 mutants exhibit altered behavior (module #536 is repressed). Modules #503 (Purines), #686 (Amino-acid biosynthesis) and #967 (Methionine metabolism) are repressed when growth is slower. (B) Disrupted coupling of two stress-related modules. The plot shows the mean expression of modules #232 (Ribosomal proteins) and #524 (RNA processing) in the galactose pathway experiments, together with the linear regression line of the dependency between the mean expression levels of the two modules over the entire compendium (Materials and methods). Broken lines indicate ±1 standard deviation. The gal80 mutants exhibit increased expression, while the gal4 mutants (excluding gal80gal4) exhibit decreased expression relative to the compendium trend, supporting a possible involvement of the gal80–gal4 circuit in the regulation of the modules. (C) Hap4-independent repression of module #536 in gal80 strains. We plot the mean expression of module #536 (Respiration) and the expression of the gene coding for its direct regulator Hap4 in selected conditions. When galactose is not available, module #536 is induced via increased expression of HAP4. Similar effect is observed in several other conditions, for example in the gal4 strain. In gal80 strains, we observe repression (or lack of induction) of the module, although the HAP4 gene is expressed at high levels.
A first surprising effect revealed by our analysis is the repression of module #524 (RNA processing, 76 × 211) in gal4 strains, in both galactose-containing (paired t-test, gal4+galactose/wt+galactose, P<10−21) and galactose-free media (gal4−galactose/wt−galactose, P<10−22). The repression of this module in mutants lacking structural enzymes is much weaker, and so is the response of the wild-type strain to lack of galactose (gal4+galactose/wt−galactose, P<10−10). Moreover, in three strains knocked out for Gal80 (the Gal4 inhibitor), grown in medium lacking galactose, we observe induction of module #524 (gal80/wt, P<10−17; gal80gal2/wt, P<10−25; gal80gal4/wt, P<10−20). This result includes the double mutant gal4gal80, implying that the effect is Gal4-independent. The induction of module #524 is particularly interesting given the slow growth and transcriptional repression of module #232 (Ribosomal proteins, 145 × 269) in the gal80 strains. Across the entire compendium, the expression of modules #524 and #232 is tightly coupled, as both are strongly repressed under general stress conditions (Gasch et al, 2000). The correlation between the mean expressions of the two modules across 1500 gene expression conditions is indeed very high (Pearson=0.73; Supplementary Figure 2). The marked difference between the expression of the two modules in the gal80 and gal4 experiments (Figure 3B) represents a regulatory discrepancy whose mechanistic causes are still unclear. Module #524 is regulated by the two highly enriched cis-elements PAC (GCGATGAG) and RRPE (GAAAATTTT) (Hughes et al, 2000), but it is still not known which factors bind these sites. Module #232 is regulated by Rap1 and possibly by additional factors (Marion et al, 2004). Some interaction between these factors, their coactivators/repressors and the Gal4/Gal80 circuit may account for the mutants altered response.
Mutations in genes of the galactose pathway and changes in the carbon source have an extensive effect on the yeast metabolism as a whole. The transcriptional regulation of nonfermentative metabolism involves a complex network of transcriptional regulators, coactivators and corepressors (Schuller, 2003). Many of the modules that were associated with the galactose data set are linked to different metabolic activities. Using data from different studies, we can dissect the general metabolic response into basic building blocks, thereby shedding light on the regulatory interactions that gave rise to it (Figure 3). Overall, we observe two types of behavior. Modules #1215 (Gluconeogenesis, 54 × 86) and #536 (Respiration, 44 × 156) are generally induced in conditions in which the yield of the galactose pathway is compromised. Modules #503 (Purine metabolism, 13 × 198), #686 (Amino-acid biosynthesis, 18 × 150) and #967 (Methionine metabolism, 15 × 156) are repressed under these conditions. This general trend fits well with our understanding of the yeast regulatory program. Yeast cells respond to the lack of galactose-based energy by increasing the activity of the respiratory pathway and adapt to slower growth by reducing biomass production. Given these general, well-documented trends, the behavior of the gal80 strains again remains unexplained. Module #536 (Respiration), for example, is repressed in gal80, gal2gal80 and gal4gal80 strains in the absence of galactose (Figure 3C), although there is no yield from the galactose pathway under these conditions. The repression cannot be explained by constitutive expression of GAL genes, given that expression is reduced also in the gal4gal80 double mutant. Module #536 is regulated by the Hap2–5 complex, and HAP4 is itself part of the module (Schuller, 2003). There is a strong correlation between Hap4 expression and expression of module #536 across the entire compendium (Pearson=0.65; Supplementary Figure 3). Nevertheless, in the three conditions in which GAL80 is inactivated, Hap4 is strongly induced while its module exhibits significant repression, suggesting the involvement of other factors in the repression of the respiratory genes. Other modules show different deviant responses to the gal80 knockout. For example, a Met4/31 module (#967) is induced in the gal80 strain, in contrast to its general repression in other conditions with reduced energy flux. Given the involvement of Gal80 in the repression of SAGA recruitment to Gal4-binding sites (Carrozza et al, 2002) and the similar acetylation patterns found in the Gal4-, Hap4- and Met4-activation sites (Deckert and Struhl, 2001), we hypothesize that in media without galactose addition, Gal80 is capable of affecting the recruitment of coactivators or corepressors for factors other than Gal4. Overall, our results provide an explanation of the slow growth phenotype of the gal80 strain, suggesting that deletion of this central regulator has far reaching implications, most notably breaking of the coupling between Ribosomal proteins and RNA processing modules, and the blocking of Hap4-dependent activation of the Respiration module.
Response to hyperosmotic stress
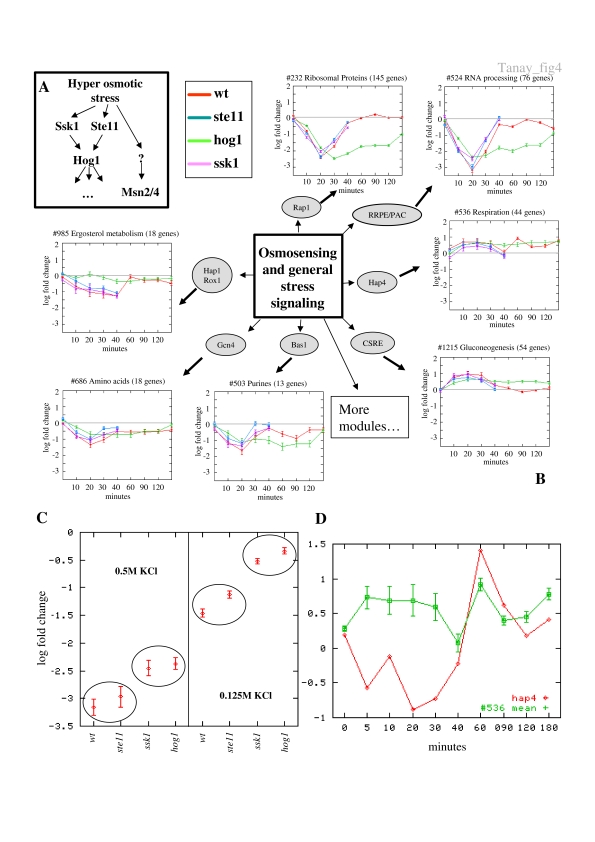
In response to hyperosmotic stress, yeast cells activate a combination of signaling pathways and transcriptional programs (Hohmann, 2002). We applied our analysis framework to a set of 129 expression profiles obtained in experiments that tested the response of S. cerevisiae to varying levels of osmotic stress in strains knocked out for Hog1, Ssk1 and Ste11, three important proteins in the HOG pathway (O'Rourke and Herskowitz, 2004). The response to high levels of osmotic stress is widespread and involves at least one-fifth of the yeast genome. We found that this massive response can be dissected into finer transcriptional programs that govern specific modules (Figure 4A). For example, modules #232 (Ribosomal proteins, 145 × 269) and #524 (RNA processing, 76 × 211) are strongly repressed in 0.5 M KCl. In the wild type, repression peaks at 20 min and is alleviated in a HOG1-dependent manner after 40 min. This joint effect was noted before, based on standard clustering analysis. Using the compendium, we uncover a refined regulatory program. In module #524, the hog1 and ssk1 strains exhibit reduced repression in the presence of 0.5 M KCl (paired t-test; hog1/wt, P<10−20; ssk1/wt, P<10−14; Supplementary Figure 4A), but no reduction is observed for ste11 (ste11/wt, P<0.14). Derepression by a hog1/ssk1 knockout is also noticeable in a medium containing 0.125 M KCl, (hog1/wt, P<10−28; ssk1/wt, P<10−20; Supplementary Figure 4B), and the effect is almost identical for the two knockouts (hog1/ssk1, P<0.002). Our analysis thus suggests that in medium/low osmotic shock, an Ssk1/Hog1-transmitted signal represses the RNA processing module activity, whereas during high osmotic shock, a Hog1-independent pathway is repressing the module additively to the Ssk1/Hog1-mediated effect (Figure 4B and C).
Figure 4. Revisiting the response to hyperosmotic stress.
(A) Outline of the hyperosmotic stress signaling pathway. Two Hog-dependent (Ssk1, Ste11) and one Hog-independent (Msn2/4) pathways mediate the hyperosmotic stress signal. (B) Response of selected modules to osmotic stress. We plot the average expression of several modules that our algorithm associated with osmotic stress conditions, in several strains knocked out for key players in the HOG pathway. The graphs show modules' mean expression time courses after treatment with 0.5 M KCl. In general, modules #232 (Ribosomal proteins) and #524 (RNA processing), #686 (Amino-acid biosynthesis), #503 (Purines) and #985 (Ergosterol biosynthesis) are repressed as part of the ESR, with peak response observed at 20 min and re-establishment of normal transcription after 40–60 min. Modules #536 (Respiration) and #1215 (Gluconeogenesis) are induced with similar kinetics. Specific modules show particular deviation from these two general trends. (C) Multiple signals additively regulate module #524. We plot the mean expression of module #524 and its standard deviations in four strains (wt, hog1, ste11, ssk1) under two levels of hyperosmotic shock (0.5 and 0.125 M KCl). There is marked difference between the ssk1 and hog1 strains and the wt, ste11 strains, suggesting the existence of two regulatory mechanisms. An osmotic stress-specific, Ssk1/Hog1-mediated signal represses the module in both low and high levels of osmotic shock. In high osmotic shock, a second, Hog1-independent signal (which is probably related to the general ESR) is active in parallel to the Hog1 signal and contributes additively to the repression of the module. (D) A two-phase regulatory program for module #536. We show the time courses of the mean expression of module #536 (Respiration) and its main regulator Hap4, when treated with 0.5 M KCl in the wt strain. The module exhibits weak and poorly correlated induction, which is Hap4 independent, during the primary phase of the osmoregulation program (0–40 min). A second phase is observed at 60–180 min, where a tightly correlated induction is facilitated by increase in HAP4 expression.
Similar decomposition of the general stress response into components is possible for the set of stress-induced genes. Two of the transcriptional modules that are activated in general stress conditions (and specifically in the 0.5 M KCl experiments) are module #536 (Respiration, regulated by Hap4) and module #1215 (Gluconeogenesis). Interestingly, while the response of both modules is remarkably similar in the early phases of the osmoregulation program (0–40 min), only the Respiration module shows a strong secondary induction after 60 min (Figure 4B). Examination of the expression of the HAP4 gene, which is generally coupled to the module's expression level (Supplementary Figure 3), also reveals an increase after 60 min (Figure 4D), supporting the hypothesis that module #536 undergoes two consecutive inductions, one via some common mechanism (which also affects module #1215) and a second that occurs later and is facilitated by the increased levels in Hap4 expression. This second wave of regulation is the adaptive response of yeast cells that have recovered from the osmotic shock, in preparation for further growth.
Analysis of the behavior of module #985 (Ergosterol biosynthesis, 18 × 69) provides another example for the power of the integrative approach. A clear Hog1-dependent repression is observed. This result is in sharp contrast to the general ESR pattern, in which only derepression is Hog1 dependent. Previous work has shown that ergosterol-related genes respond strongly to osmotic shock (Hohmann, 2002). Our analysis suggests that their repression directly depends on Hog1 through an unknown signaling pathway that does not involve Ssk1 or Ste11.
Discussion
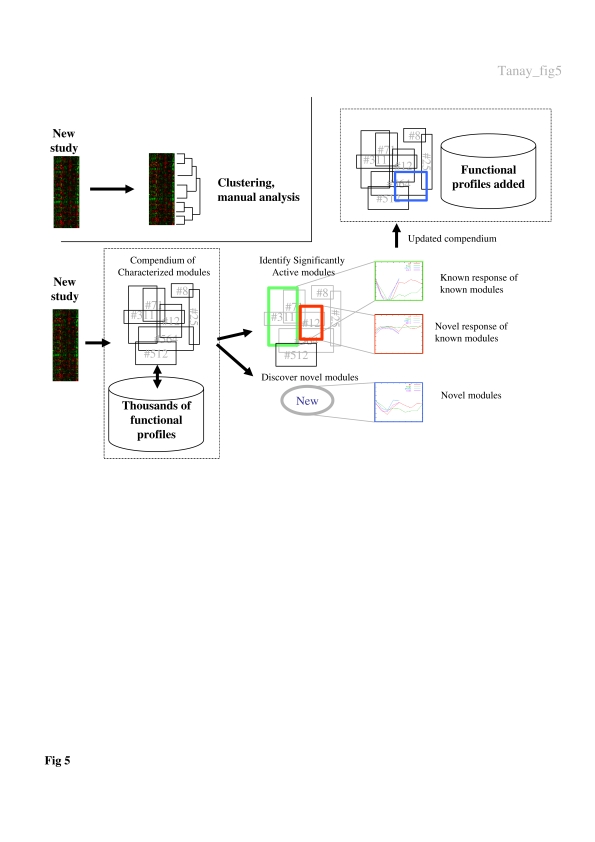
After almost a decade of microarray-based experiments, a revision of the paradigm for their computational analysis is appropriate. In this work, we have introduced a new method for the simultaneous analysis of new high-throughput data sets given a large compendium of diverse functional data. We have shown that the integrative approach greatly extends our understanding of the regulation of biological processes and allows the decomposition of seemingly global responses into characterized regulatory programs of specific biological modules. The methodology we envision (Figure 5) relies on a growing compendium of public data sets and on our robust algorithms for revealing biological correlations present within these data. Given the data of a new study, its integration with the large body of prior data allows us to recast the new experiments in terms of (a) the behavior of already characterized modules and (b) new modules that are discovered for the first time upon the addition of the new data. Using this approach, backed by appropriate community effort for modules nomenclature (e.g., based on Gene Ontologies), the results of high-throughput experiments will be easier to assess and share, as it will be clear what in the new experiments is new and what confirms previously published evidence. We are constructing an interactive web interface that will provide the infrastructure for this suggested methodology (www.cs.tau.ac.il/~rshamir/simba).
Figure 5. A new paradigm for analyzing functional genomic experiments.
According to the current prevalent paradigm (top part), novel data are analyzed in isolation, typically using clustering and expert manual analysis of specific clusters. We suggest a new approach (lower part) in which the community maintains the current publicly available data sets and the set of biological modules revealed by them. Modules may cover all aspects of biological processes and their regulation, as revealed, for example, by our biclustering algorithm. Using this resource, novel data sets can be represented in terms of the behavior of known and novel modules, providing an objective and transparent method for understanding, communicating and reusing high-throughput data.
In this paper, we have focused on the analysis of yeast data. Functional genomics resources are available for many other model systems, and are rapidly accumulating in repositories such as GEO (www.ncbi.nlm.nih.gov) and ArrayExpress (www.ebi.ac.uk). Recently, a set of literature-based and preprocessed gene sets were used to analyze a large cancer-related data compendium. The integration of data was shown to be synergistic, even across different cancer types (Segal et al, 2004). Our methodology allows integrated analysis together with the discovery of new modules, making it an effective approach for the routine analysis of new high-throughput data. Finally, recent studies have shown that transcriptional modules are sometimes highly conserved among species (Stuart et al, 2003; Bergmann et al, 2004; Tanay et al, 2004a). Having established deeper understanding of this conservation, it will be desirable to seek further integration of functional data across different species.
Materials and methods
Data preparation
We used data from 60 publications encompassing 1767 conditions. The complete list of publications and experiments is available on Supplementary website. Data were downloaded from papers' web supplements. For Affymetrix array experiments, we divided each condition's profile by a common reference condition (typically the zero time point of the experiment; see Supplementary website for more details). For other experiments, we used the normalization reported in the original papers.
In the SAMBA framework, each experiment defines one or several properties. For example, a gene expression experiment can be transformed into four properties, representing strong upregulation, weak upregulation, weak downregulation and strong downregulation in the tested condition. We assign genes with properties by applying translation functions that map experimental values to probabilities of having a property. For example, a gene with high gene expression readout in a condition X will be assigned with the property ‘strong upregulation in condition X' with high probability. The notion of property is very flexible and can accommodate diverse sources of data. For example, protein interaction data can be transformed into properties of the form ‘interacting with protein X' and phenotypes can be transformed to properties of the form ‘mutant is slowly growing on medium Y'. See Tanay et al (2004a, 2004b) for more details. We optimized the performance of SAMBA by testing the effect of changes in the parameters of translation functions, and selected parameters that were robust to addition of new data and are thus expected to provide good results as the compendium size increases. Note that for generating protein interaction properties, we used properties for proteins with at least 15 targets and discarded all others as they bias the statistical model. SAMBA detected statistically significant biclusters including half (133 out of 265) of the protein interaction properties. The other properties were either too distinct to be correlated with other properties or were too noisy for the statistical stringency of the model. All the parameters defining the translation functions we used are available on our website.
SAMBA biclustering
We applied the SAMBA 2.0 program to the entire compendium with standard parameters. The program searches the combined data set and outputs a set of modules, each of which is a set of genes that are correlated in a set of properties. Each gene may be part of several modules, allowing us to reveal multiple functions for it. Similarly, each property may belong to several modules, allowing us to associate it with different biological processes. The modules generated were then subjected to additional analysis. We associated biclusters with functional annotation terms using SGD GO associations and functional enrichment tests as previously described (Tanay et al, 2004b). We searched for enriched cis-elements in all bicluster gene sets using promoters including 600 bp upstream of each ORF, as described (Tanay and Shamir, 2004). Visualizations of modules and the effects of specific experiments on them are available on our prototype website (www.cs.tau.ac.il/~samba/). More information on the algorithms and their parameters is available on Supplementary website.
Module profiling
Given a target data set and the compendium, we derive the set of responding modules as those that contain at least one property from the target data set. SAMBA adds a property to a module, its genes having significant and correlated levels over the property; therefore, using this approach, we extract only modules that significantly respond in the analyzed experiments. We can then study the behavior of the responding modules in the entire analyzed data set.
Testing the significance of changes in modules' mean expression
To evaluate changes in the mean expression of a module between two conditions, we used a standard two-tailed paired t-test. To test the significance of an induction or repression of a module in a single condition, we performed two-tailed two-sample unpaired t-tests comparing the module's genes and the entire genome. To compute compendium trends of module #524 and #232, we used standard best linear fit and computed the standard deviation of the bias of samples from the linear curve. We also computed statistics using nonparametric tests with similar results. The above hypothesis testing procedures were used after employing SAMBA, as additional tests of claims on the regulation of specific modules.
URLs
More details on our results, including details of the expression compendium, algorithmic details and interactive module visualizations, can be found on our Supplementary website (www.cs.tau.ac.il/~rshamir/simba/).
Supplementary Material
Supplementary Material
Supplementary Figures
Acknowledgments
AT was supported by a Horvitz complexity fellowship. MK was supported in part by the ISF, the Recanati Fund and the Israeli Ministry of Health. RS holds the Raymond and Beverly Sackler Chair for Bioinformatics at Tel Aviv University, and was supported by the Israel Science Foundation (Grant 309/02).
References
- Angus-Hill ML, Schlichter A, Roberts D, Erdjument-Bromage H, Tempst P, Cairns BR (2001) A Rsc3/Rsc30 zinc cluster dimer reveals novel roles for the chromatin remodeler RSC in gene expression and cell cycle control. Mol Cell 7: 741–751 [DOI] [PubMed] [Google Scholar]
- Beer MA, Tavazoie S (2004) Predicting gene expression from sequence. Cell 117: 185–198 [DOI] [PubMed] [Google Scholar]
- Bergmann S, Ihmels J, Barkai N (2004) Similarities and differences in genome-wide expression data of six organisms. PLoS Biol 2: E9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birrell GW, Brown JA, Wu HI, Giaever G, Chu AM, Davis RW, Brown JM (2002) Transcriptional response of Saccharomyces cerevisiae to DNA-damaging agents does not identify the genes that protect against these agents. Proc Natl Acad Sci USA 99: 8778–8783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza MJ, John S, Sil AK, Hopper JE, Workman JL (2002) Gal80 confers specificity on HAT complex interactions with activators. J Biol Chem 277: 24648–24652 [DOI] [PubMed] [Google Scholar]
- Colman-Lerner A, Chin TE, Brent R (2001) Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell 107: 739–750 [DOI] [PubMed] [Google Scholar]
- Deckert J, Struhl K (2001) Histone acetylation at promoters is differentially affected by specific activators and repressors. Mol Cell Biol 21: 2726–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO (1997) Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278: 680–686 [DOI] [PubMed] [Google Scholar]
- Doolin MT, Johnson AL, Johnston LH, Butler G (2001) Overlapping and distinct roles of the duplicated yeast transcription factors Ace2p and Swi5p. Mol Microbiol 40: 422–432 [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11: 4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisberg JV, Holstege FC, Young RA, Struhl K (2001) Yeast NC2 associates with the RNA polymerase II preinitiation complex and selectively affects transcription in vivo . Mol Cell Biol 21: 2736–2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehring AS, Mitchell DA, Tong AH, Keniry ME, Boone C, Sprague GF Jr (2003) Synthetic lethal analysis implicates Ste20p, a p21-activated potein kinase, in polarisome activation. Mol Biol Cell 14: 1501–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofken T, Schiebel E (2002) A role for cell polarity proteins in mitotic exit. EMBO J 21: 4851–4862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann S (2002) Osmotic stress signaling and osmoadaptation in yeasts. Microbiol Mol Biol Rev 66: 300–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JD, Estep PW, Tavazoie S, Church GM (2000) Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae . J Mol Biol 296: 1205–1214 [DOI] [PubMed] [Google Scholar]
- Ideker T, Thorsson V, Ranish JA, Christmas R, Buhler J, Eng JK, Bumgarner R, Goodlett DR, Aebersold R, Hood L (2001) Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science 292: 929–934 [DOI] [PubMed] [Google Scholar]
- Ihmels J, Friedlander G, Bergmann S, Sarig O, Ziv Y, Barkai N (2002) Revealing modular organization in the yeast transcriptional network. Nat Genet 31: 370–377 [DOI] [PubMed] [Google Scholar]
- Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO (2001) Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 409: 533–538 [DOI] [PubMed] [Google Scholar]
- Kemmeren P, van Berkum NL, Vilo J, Bijma T, Donders R, Brazma A, Holstege FC (2002) Protein interaction verification and functional annotation by integrated analysis of genome-scale data. Mol Cell 9: 1133–1143 [DOI] [PubMed] [Google Scholar]
- Marion RM, Regev A, Segal E, Barash Y, Koller D, Friedman N, O'Shea EK (2004) Inaugural article: Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc Natl Acad Sci USA 101: 14315–14322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke SM, Herskowitz I (2004) Unique and redundant roles for HOG MAPK pathway components as revealed by whole-genome expression analysis. Mol Biol Cell 15: 532–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz S, Avila-Campillo I, Aldridge C, Srinivasan A, Dimitrov K, Siegel AF, Galitski T (2004) Control of yeast filamentous-form growth by modules in an integrated molecular network. Genome Res 14: 380–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, Volkert TL, Wilson CJ, Bell SP, Young RA (2000) Genome-wide location and function of DNA binding proteins. Science 290: 2306–2309 [DOI] [PubMed] [Google Scholar]
- Schuller HJ (2003) Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae . Curr Genet 43: 139–160 [DOI] [PubMed] [Google Scholar]
- Schwikowski B, Uetz P, Fields S (2000) A network of protein–protein interactions in yeast. Nat Biotechnol 18: 1257–1261 [DOI] [PubMed] [Google Scholar]
- Segal E, Friedman N, Koller D, Regev A (2004) A module map showing conditional activity of expression modules in cancer. Nat Genet 36: 1090–1098 [DOI] [PubMed] [Google Scholar]
- Simon I, Barnett J, Hannett N, Harbison CT, Rinaldi NJ, Volkert TL, Wyrick JJ, Zeitlinger J, Gifford DK, Jaakkola TS, Young RA (2001) Serial regulation of transcriptional regulators in the yeast cell cycle. Cell 106: 697–708 [DOI] [PubMed] [Google Scholar]
- Stuart JM, Segal E, Koller D, Kim SK (2003) A gene-coexpression network for global discovery of conserved genetic modules. Science 302: 249–255 [DOI] [PubMed] [Google Scholar]
- Sudarsanam P, Iyer VR, Brown PO, Winston F (2000) Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae . Proc Natl Acad Sci USA 97: 3364–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanay A, Shamir R (2004) Multilevel modeling and inference of transcription regulation. J Comput Biol 11: 357–375 [DOI] [PubMed] [Google Scholar]
- Tanay A, Regev A, Shamir R (2004a) Evolutionary mechanisms underlying conservation and evolvability in regulatory networks (submitted)
- Tanay A, Sharan R, Kupiec M, Shamir R (2004b) Revealing modularity and organization in the yeast molecular network by integrated analysis of highly heterogeneous genomewide data. Proc Natl Acad Sci USA 101: 2981–2986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, Chen Y, Cheng X, Chua G, Friesen H, Goldberg DS, Haynes J, Humphries C, He G, Hussein S, Ke L, Krogan N, Li Z, Levinson JN, Lu H, Menard P, Munyana C, Parsons AB, Ryan O, Tonikian R, Roberts T, Sdicu AM, Shapiro J, Sheikh B, Suter B, Wong SL, Zhang LV, Zhu H, Burd CG, Munro S, Sander C, Rine J, Greenblatt J, Peter M, Bretscher A, Bell G, Roth FP, Brown GW, Andrews B, Bussey H, Boone C (2004) Global mapping of the yeast genetic interaction network. Science 303: 808–813 [DOI] [PubMed] [Google Scholar]
- Troyanskaya OG, Dolinski K, Owen AB, Altman RB, Botstein D (2003) A Bayesian framework for combining heterogeneous data sources for gene function prediction (in Saccharomyces cerevisiae). Proc Natl Acad Sci USA 100: 8348–8353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velours G, Boucheron C, Manon S, Camougrand N (2002) Dual cell wall/mitochondria localization of the ‘SUN' family proteins. FEMS Microbiol Lett 207: 165–172 [DOI] [PubMed] [Google Scholar]
- Wu LF, Hughes TR, Davierwala AP, Robinson MD, Stoughton R, Altschuler SJ (2002) Large-scale prediction of Saccharomyces cerevisiae gene function using overlapping transcriptional clusters. Nat Genet 31: 255–265 [DOI] [PubMed] [Google Scholar]
- Zhu G, Spellman PT, Volpe T, Brown PO, Botstein D, Davis TN, Futcher B (2000) Two yeast forkhead genes regulate the cell cycle and pseudohyphal growth. Nature 406: 90–94 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Supplementary Figures