Abstract
Toward the goal of understanding system properties of biological networks, we investigate the global and local regulation of gene expression in the Saccharomyces cerevisiae metabolic network. Our results demonstrate predominance of local gene regulation in metabolism. Metabolic genes display significant coexpression on distances smaller than the average network distance, a behavior supported by the distribution of transcription factor binding sites in the metabolic network and genome context associations. Positive gene coexpression decreases monotonically with distance in the network, while negative coexpression is strongest at intermediate network distances. We show that basic topological motifs of the metabolic network exhibit statistically significant differences in coexpression behavior.
Keywords: expression, genome context, metabolism, motifs, network
Introduction
Recent studies of general topological organization of metabolic networks have provided valuable insights into the functional properties of these systems (Jeong et al, 2000; Ravasz et al, 2002). The topological characteristics, however, provide only a static description of the biological networks. Functions of cellular networks are highly regulated in a temporal fashion at a number of levels, including transcriptional regulation. The expression dynamics of protein–protein and protein–DNA interaction networks have been previously investigated and small, but statistically significant, expression correlations were found between network neighbors (Ge et al, 2001). Earlier studies have analyzed coexpression in individual metabolic pathways (Gerstein and Jansen, 2000; Hughes et al, 2000; Karp et al, 2002; Pavlidis et al, 2002; Ihmels et al, 2003), identified highly coexpressed modules (Hanisch et al, 2002; Ihmels et al, 2003; Patil and Nielsen, 2005) and have characterized coexpression properties of metabolic junctions (Ihmels et al, 2003). In the present study, we demonstrate several novel aspects of the expression dynamics in yeast metabolic network. We also show that the observed patterns of the mRNA coexpression are similar to the patterns of other context-based associations between genes: phylogenetic profiles and chromosomal distance.
In our analysis we represent metabolism as a graphical model, with nodes of the graph corresponding to genes encoding metabolic enzymes, and edges to metabolic connections between corresponding enzymes (see Materials and methods). We investigate how positive and negative correlation of mRNA expression profiles depends on the metabolic network distance, and determine the maximum distance at which genes display statistically significant coexpression. At the level of individual biochemical reactions, we examine coexpression and functional association patterns of local topological motifs of the metabolic network (Shen-Orr et al, 2002). Our results show that different motifs exhibit distinct coexpression patterns, which may elucidate dynamic design principles of metabolic networks.
Results and discussion
Global properties of gene coexpression
Our analysis uses Saccharomyces cerevisiae metabolic network recently reconstructed by Forster et al (2003). In contrast to currently available protein–protein interaction data, yeast metabolic network has a substantially smaller error rate compared to commonly available physical interaction data (Mering et al, 2002; Spinzak et al, 2003). Using the established metabolic connectivity between S. cerevisiae enzyme-encoding genes, we investigate how the degree of gene coexpression changes with the network distance. A similar question was posed recently for adjacent genes in the protein–protein physical interaction network (Ge et al, 2001; Jansen et al, 2002).
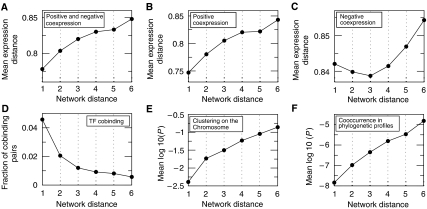
An intuitive expectation is that genes close in the metabolic network would also be coexpressed. The dependency of mean expression distance (distance=1−correlation coefficient, see Materials and methods) on metabolic network distance is shown in Figure 1A. Expression distance increases monotonically with network distance, demonstrating that genes closer to each other in metabolic network tend to have, on average, higher level of coexpression. The extent of local coexpression in the metabolic network can be characterized by calculating the maximal network distance, at which one finds genes that are, on average, significantly more coexpressed than the rest of the metabolic genes. We find that mean expression distance at network distances 1–3 is significantly (Wilcoxon test P<10−3) lower than that of all other pairs, making the radius of significant coexpression equal to 3 (Supplementary Figure 1), which is just below the network diameter (3.7).
Figure 1.
Coexpression and functional associations on the scale of the whole metabolic network. Mean expression distance is plotted as a function of the metabolic network distance separating the metabolic gene pairs for (A) all metabolic gene pairs; (B) positively coexpressed pairs and (C) negatively coexpressed pairs. (D) Fraction of metabolic gene pairs that share at least one transcription factor binding site in their promoter region as a function of metabolic network distance. (E) Mean chromosome clustering gene pair association score dependency. (F) Mean phylogenetic profile cooccurrence association score dependency.
Local coregulation can also be illustrated by examining distribution of transcription factor binding in the metabolic network. Because genes sharing DNA binding transcription factors are, generally, expected to be coregulated, the distribution of transcription factor binding cooccurrences in the metabolic network should confirm the pattern observed for coexpression. Indeed, we find (Figure 1D) that the fraction of gene pairs binding a common transcription factor is highest in the metabolically adjacent genes, and decreases with network distance. Similar results were recently observed for E. coli metabolic network (Spirin et al, 2003). It is also worthwhile to note that both cobinding fraction and coexpression distance fall off is somewhere between linear and exponential, as evidenced by the semi-log scale plots (Supplementary Figures 2 and 3).
The expression distance dependency is shown separately for gene pairs with positive and negative correlation of expression profiles (Figure 1B and C). While expression distance of positively coexpressed gene pairs monotonically increases with metabolic network distance, there is no such simple relationship in the case of negative coexpression. Negative coexpression is strongest at intermediate distances (around 3) and is relatively weak at short and long network distances. In all cases, behavior of mean coexpression magnitude is matched by the relative abundance of highly coexpressed pairs at each network distance (Supplementary Figure 1). The relationships between network distance and gene coexpression observed in the combined expression data set (see Materials and methods) agree with the relationships in individual data sets: Rosetta (Hughes et al, 2000), Brown (Gasch et al, 2000) and Young (Causton et al, 2001) (see Supplementary Figures 4–6).
Both coexpression and transcription factor binding site cooccurrence reflect the degree of functional association between metabolic genes. To generalize our results, we have examined other functional association evidence based on the genome context: physical clustering of genes on the chromosome (Overbeek et al, 1999) and gene cooccurrence in phylogenetic profiles (Pellegrini et al, 1999) (see Materials and methods). We find that associations based on both cooccurrence in phylogenetic profiles (Figure 1E) and physical clustering of genes on the chromosome (Figure 1F) also exhibit the local property observed for gene coexpression, with the same radius of significant association.
Coexpression in the metabolic network depends not only on the network distance between genes, but also on the characteristics of connecting metabolites. Analyzing the dependency between the total number of enzyme pairs connected by a given metabolite (metabolite enzyme pair number) and mean expression of the connected pairs (see Supplementary Figure 1), we find that increasing enzyme pair number corresponds, on average, to weaker positive coexpression (Spearman rank r=0.21, P=4.7 × 10−5) and stronger negative coexpression (r=−0.14, P=2.7 × 10−2). The details of this analysis and of similar genome context association dependencies are given in Supplementary information.
It should be noted that our analysis examines coexpression across a large number of conditions. As it has been recently demonstrated (Patil and Nielsen, 2005), individual environmental perturbations may affect sets of metabolic genes connected by some of the common cofactors. Nevertheless, our study shows that the overall coexpression is stronger for genes connected by metabolites used in a small number of reactions. In general, a decrease in the reaction number of connecting metabolite increases linearity of the pathways going through this node. Consequently, our results suggest that positive coexpression and strong functional associations dominate in linear parts of the network, while negative coexpression is stronger in highly branched pathways.
Gene coexpression in metabolic motifs
To understand patterns of local regulation in the metabolic network, we compare coexpression properties of elementary topological motifs formed by adjacent enzymes in the metabolite graph. Several studies have recently investigated local regulatory motifs in bacteria and yeast (Lee et al, 2002; Shen-Orr et al, 2002). These studies identified elementary topological motifs that are significantly more abundant in real biological networks than expected by chance. In contrast, we analyze coexpression of all possible two-gene motifs and a majority of three-gene substructures of the metabolite graph (see Materials and methods). Mean expression distances of these motifs are given in Supplementary Table 1. The same analysis was repeated on each individual expression data set, and the results are shown in Supplementary Tables 2–4. Below, we examine behavior established by positive coexpression as being predominant in magnitude and consistent between different expression data sets (considering both positive and negative coexpression results in the same motif ordering).
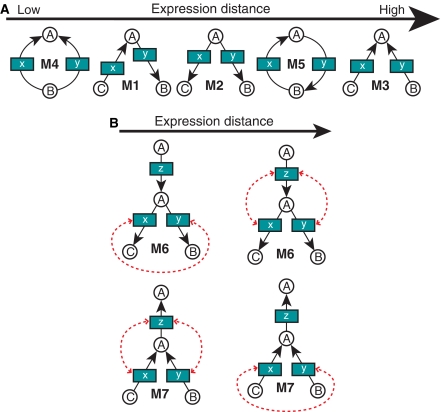
The ordering of irreversible two-gene motifs established by positive coexpression is shown in Figure 2A. Functionally, these motifs represent serial (M1), divergent (M2), convergent (M3), parallel (M4) and cyclic (M5) topologies. Some of the motif coexpression patterns are intuitive: for example, high level of coexpression in the M1 motif representing sequential genes in a pathway, or low level of coexpression in the M5 motif representing a futile cycle. A highly coexpressed M4 motif includes homologous enzymes frequently resulting from gene duplication, which tend to be correlated in their expression patterns (Wagner, 2002). Indeed, exclusion of homologous gene pairs (see Materials and methods) affects only the mean coexpression level of the M4 motif, reducing it below the level of the M2 motif (see Supplementary Figure 7 and Supplementary Table 5).
Figure 2.
Coexpression in motifs of metabolite graph. (A) All irreversible metabolite graph motifs consisting of two genes (x and y) are analyzed using only positively coexpressed gene pairs. The motifs are ordered according to the mean expression distance of gene pairs belonging to each motif. (B) Two three-gene motifs (M6 and M7). Two distinct types of gene pairs within each motif are connected by dashed arrows. The pair types are ordered (from left to right) in the order of increasing mean expression distance.
The M2 and M3 motifs represent pairs of divergent and convergent metabolic reactions correspondingly. The mean level of positive coexpression of genes within the M2 motif is significantly higher than within the M3 motif (Wilcoxon test P=3.4 × 10−3), implying that coregulation of divergent metabolic pathways is generally stronger compared to convergent pathways. This suggests that regulation of the metabolic network emphasizes reactions in which one metabolic precursor, such as a carbon source, is simultaneously used to synthesize a variety of compounds required for biomass growth.
In analyzing three-gene motifs, we compare coexpression among different types of gene pairs within each motif. The M6 and M7 motifs (Figure 2B) represent three-gene extensions of the two-gene M2 and M3 motifs, and their behavior reflects the coexpression patterns established by the two-gene motifs. For example, positive coexpression of the convergent (type 2) pair in the M7 motif is significantly lower than that of the serial (type 1) pair (P=1.0 × 10−3), which can be explained by the corresponding behavior of the M1 and M3 motifs. This result supports earlier findings by Ihmels et al (2003).
In contrast to the M7 motif, the pattern of positive coexpression for the three-gene M6 motif differs from the behavior observed in two-gene motifs. Here, enzymes consuming the same metabolite exhibit, on average, stronger coexpression with each other than with the enzyme producing the metabolite. Although the observed pattern for the M6 motif coexpression differs from the conclusions of the divergent junction analysis reported by Ihmels et al (2003), no statistical significance has been established in either case (Wilcoxon test P=0.33 for positively coexpressed pairs, P=0.54 for positive and negative coexpression, no P-value was reported by Ihmels et al). Overall, coexpression patterns of both M6 and M7 motifs support predominance of coregulation in divergent versus convergent branches demonstrated in the analysis of the two-gene motifs.
The coexpression behavior of local topological motifs is matched by the strength of genome context associations between genes. Genome context association of the metabolite graph motifs (clustering of genes on the chromosome, and gene cooccurrence in phylogenetic profiles) is given in Supplementary Tables 6 and 7. Since genome context scores rely on ortholog identities, these analyses exclude motifs formed by homologous gene pairs (see Materials and methods).
The ordering of irreversible two-gene motifs (M1–M5) established by the genome context associations is identical to the order established by positive coexpression, with the exception of M1–M2 switch in the phylogenetic profile association ordering (Supplementary Figure 7). Significantly larger association of genes in divergent (M2) compared to convergent (M3) motif is also supported by the difference in the mean chromosome clustering scores (Wilcoxon test P=4.6 × 10−19) and phylogenetic profile cooccurrence (Wilcoxon test P=3.8 × 10−30). Similarly, the relative coexpression behavior of the three-gene M6 and M7 motifs is matched by the relationship of genome context associations.
Conclusion
The presented results reveal interesting and statistically significant patterns of coregulation in the metabolic network. We find that regulation of metabolic genes is local and extends, generally, to distances smaller than the mean network distance. Such regulation implies that genes close in the metabolic network are usually coexpressed together, possibly to optimize local metabolic fluxes (Zaslaver et al, 2004). Positive coexpression is strongest among adjacent genes and decreases monotonically with network distance. In contrast, negative coexpression is most prominent at intermediate distances. Functional associations based on the genome context analysis exhibit the same local property observed in the case of positive coexpression. These results suggest that regulation of the metabolic network establishes a number of local, positively coexpressed regions that may exhibit some degree of negative coexpression between each other. Furthermore, we find that positive coexpression and functional associations are strongest in the linear parts of metabolism, while negative coexpression is more pronounced in highly branched regions.
Our analysis of the elementary topological motifs illustrates that coexpression in divergent branches is significantly stronger than that observed in convergent branches. This pattern suggests emphasis on coregulation of biomass synthesis or degradation from common metabolic precursors.
Good agreement between the mRNA coexpression and genome context associations suggests that the observed patterns of metabolic regulation are reflected in genome evolution and affect the location of genes on the chromosomes. In future studies, it will be important to confirm our findings using the metabolic networks of other organisms. It would also be interesting to perform similar analysis of other cellular networks, for example signaling and protein–protein interaction networks.
Materials and methods
Metabolic dependency graph and separation between genes
Metabolism was represented in a form of a connectivity graph. The nodes of the graph correspond to metabolic genes, and edges correspond to connections established by metabolic reactions. Metabolic genes X and Y are considered connected if and only if there exists a metabolite that is present among the list of either reactants or products of reactions catalyzed by enzymes encoded by both X and Y.
The metabolic connectivity graph is used to calculate network distance (or metabolic separation) between genes. We define a pair of directly connected metabolic genes as being separated by distance 1. In general, we define network distance between the genes X and Y as the length of the shortest path from X to Y on the metabolic connectivity graph. A hand-curated metabolic network model of S. cerevisiae (Forster et al, 2003) was used to construct a comprehensive metabolic connectivity graph. While any metabolite can be used to deduce gene connectivity, the relationships established by the common cofactors, such as ATP, are not likely to connect genes with similar metabolic functions. In compiling a global metabolic connectivity graph, we consider a subset of metabolites, which excludes most highly connected metabolic species. An exclusion threshold was determined based on the connectivity of the resulting gene dependency graph (Supplementary Figure 1). A total of 14 most highly connected metabolites (ATP, ADP, AMP, CO2, CoA, glutamate, H, NAD, NADH, NADP, NADPH, NH3, orthophosphate, pyrophosphate) and their mitochondrial and external analogs were excluded. The general trends described in the paper are not sensitive to the precise choice of the metabolite set; however, the actual values change when more or less metabolites are considered. For detailed analysis, see Supplementary information. Genes encoding enzymes that are part of known complexes, according to MIPS complex database (http://mips.gsf.de) and SGD (http://www.yeastgenome.org/), were masked as unassigned enzymes, so that their expression profiles would not be included in any of the analysis (36 enzyme-encoding genes in total).
Distances between gene expression profiles
We used three data sets as sources of gene expression information. The Rosetta's ‘compendium' data set (Hughes et al, 2000) measures expression profiles of over 6200 S. cerevisiae open reading frames (ORFs) across 287 deletion strains and 13 chemical conditions. In addition, the data set contains 63 negative control measurements comparing two independent cultures of the same strain. These were used to establish individual error models for each ORF, providing not only the raw intensity and ratio measurement values for each experimental data point, but also a P-value gauging the significance of change in expression level. The ratio data were used for all analysis. We also used a data set from Brown's group, containing 173 environmental perturbations (Gasch et al, 2000), and a data set from Young's group with 34 conditions describing seven environmental perturbation time courses (Causton et al, 2001). Log 10 intensity ratios of each data set were normalized to have a mean of 0 and a variance of 1. Separate time courses contained in these data sets were first normalized individually and then combined. Supplementary Table 8 shows the relative variability of metabolic enzyme-encoding genes in each data set.
The expression distance measure between ORFs X and Y is then taken to be 1−S∣(px,py)∣, where px and py are expression profile vectors of X and Y, and S corresponds to Spearman rank correlation coefficient, calculated according to Press et al (2002). Combined expression profile vectors are formed by concatenation of log 10 ratio values from three data sets. Conditions missing experimental value for at least one of the genes were omitted in calculating the expression distance of a given gene pair. Genes that were missing expression values for more than 25% of conditions were not considered in the analysis.
Transcription factor binding
Information on transcription factor binding to the metabolic gene promoter sites was taken from Lee et al (2002). A P-value threshold of 0.001 was used to select transcription factor binding occurrences.
Clustering of genes on the chromosome
To assess genome context association based on the physical clustering of genes on the chromosome, we relied on gene order statistics. Chromosome clustering association score between genes x and y was calculated as S(x∣y)=∏g∈GP(dg(x,y)), where G is a set of genomes and P(dg(x, y)) is the probability of observing gene order distance dg(x, y) between genes x and y in a genome g, calculated based on the chromosome sizes of organism under the null hypothesis that genes are randomly ordered across the chromosomes. The scores were calculated using a set of 105 bacterial and three eukaryotic genomes. Orthology mapping was established using best bidirectional hits from KEGG SSDB (Itoh et al, 2004).
Cooccurrence in phylogenetic profiles
Phylogenetic profile cooccurrence association (Pellegrini et al, 1999) was assessed using hypergeometric probability, as described by Bowers et al (2004). The orthology data set was constructed based on best bidirectional BLASTP hits against NCBI NR protein data set. The calculation was limited to organisms containing orthologs for at least 1% of S. cerevisiae genes.
Metabolic motifs
The elementary topological motifs of the metabolite graph were classified in terms of the expression properties of the genes involved. The elementary node structures were enumerated, and sets of genes that were connected in the appropriate topology were extracted for each structure. It is possible for one gene to be included in multiple occurrences of the same motif; in other words, we counted any substructure with the correct topology as an occurrence. Motif instances formed around top 14 most connected metabolites, as well as their mitochondrial and external forms, were not included in the analysis. Mean expression distances of different types of gene pairs were compared using the Wilcoxon rank test (Wilcoxon, 1945).
In generating homolog-filtered data, a pair of metabolic genes was excluded from the analysis if the BLAST score comparing the two nucleotide sequences was below an E-value of 10−3.
Acknowledgments
We thank Patrik D'haeseleer, Philippe Marc, John Aach, Yonathan Grad, Leonid Mirny, Andrey Rzhetsky, Paul Pavlidis and Andrea Califano for useful discussions and critical reading of the manuscript. GMC was supported by the US Department of Energy, the Defense Advanced Research Projects Agency, and the PhRMA Foundation
References
- Bowers PM, Pellegrini M, Thompson MJ, Fierro J, Yeates TO, Eisenberg D (2004) Prolinks: a database of protein functional linkages derived from coevolution. Genome Biol 5: R35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causton HC, Ren B, Koh SS, Harbison CT, Kanin E, Jennings EG, Lee TI, True HL, Lander ES, Young RA (2001) Remodeling of yeast genome expression in response to environmental changes. Mol Biol Cell 12: 323–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster J, Famili I, Fu P, Palsson BO, Nielsen J (2003) Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Res 13: 244–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11: 4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge H, Liu Z, Church GM, Vidal M (2001) Correlation between transcriptome and interactome mapping data from Saccharomyces cerevisiae. Nat Genet 29: 482–486 [DOI] [PubMed] [Google Scholar]
- Gerstein M, Jansen R (2000) The current excitement in bioinformatics—analysis of whole-genome expression data: how does it relate to protein structure and function? Curr Opin Struct Biol 10: 574–584 [DOI] [PubMed] [Google Scholar]
- Hanisch D, Zien A, Zimmer R, Lengauer T (2002) Co-clustering of biological networks and gene expression data. Bioinformatics 18: S145–S154 [DOI] [PubMed] [Google Scholar]
- Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA (2000) Functional discovery via a compendium of expression profiles. Cell 102: 109–126 [DOI] [PubMed] [Google Scholar]
- Ihmels J, Levy R, Barkai N (2003) Principles of transcriptional control in the metabolic network of Saccharomyces cerevisiae. Nat Biotechnol 22: 86–92 [DOI] [PubMed] [Google Scholar]
- Itoh M, Akutsu T, Kanehisa M (2004) Clustering of database sequences for fast homology search using upper bounds on alignment score. Genome Inform Ser Workshop Genome Inform 15: 93–104 [PubMed] [Google Scholar]
- Jansen R, Greenbaum D, Gerstein M (2002) Relating whole-genome expression data with protein–protein interactions. Genome Res 12: 37–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H, Tombor B, Albert R, Oltavi ZN, Barabasi AL (2000) The large-scale organization of metabolic networks. Nature 407: 651–654 [DOI] [PubMed] [Google Scholar]
- Karp PD, Paley S, Romero P (2002) The Pathway Tools software. Bioinformatics 18: S225–S232 [DOI] [PubMed] [Google Scholar]
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, Zeitlinger J (2002) Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298: 799–804 [DOI] [PubMed] [Google Scholar]
- Mering C, Krause R, Snel B, Cornell M, Oliver SG, Fields S, Bork P (2002) Comparative assessment of large-scale data sets of protein–protein interactions. Nature 417: 399–403 [DOI] [PubMed] [Google Scholar]
- Overbeek R, Fonstein M, D'Souza M, Pusch GD, Maltsev N (1999) The use of gene clusters to infer functional coupling. Proc Natl Acad Sci USA 96: 2896–2901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil KR, Nielsen J (2005) Uncovering transcriptional regulation of metabolism by using metabolic network topology. Proc Natl Acad Sci USA 102: 2685–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlidis P, Lewis DP, Noble WS (2002) Exploring gene expression data with class scores. Pac Symp Biocomput 12: 474–485 [PubMed] [Google Scholar]
- Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO (1999) Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc Natl Acad Sci USA 96: 4285–4288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Press WH, Teukolsky SA, Vetterling WT, Flannery BP (2002) Numerical Recipes in C++: The Art of Scientific Computing. Cambridge University Press: Cambridge, UK [Google Scholar]
- Ravasz E, Somera AL, Mongru DA, Oltvai ZN, Barabasi AL (2002) Hierarchical organization of modularity in metabolic networks. Science 297: 1551–1555 [DOI] [PubMed] [Google Scholar]
- Shen-Orr SS, Milo R, Mangan S, Alon U (2002) Network motifs in the transcriptional regulation network of Escherichia coli. Nat Genet 31: 64–68 [DOI] [PubMed] [Google Scholar]
- Spinzak E, Sattah S, Margalit H (2003) How reliable are experimental protein–protein interaction data? J Mol Biol 327: 919–923 [DOI] [PubMed] [Google Scholar]
- Spirin V, Gelfand MS, Mirny LA (2003) Computational analysis of metabolic modules and pathways in E. coli metabolic network.ISCB 4: 239–240 [Google Scholar]
- Wagner A (2002) Asymmetric functional divergence of duplicate genes in yeast. Mol Biol Evol 19: 1760–1768 [DOI] [PubMed] [Google Scholar]
- Wilcoxon F (1945) Individual comparisons by ranking methods. Biometrics 1: 80–83 [Google Scholar]
- Zaslaver A, Mayo AE, Rosenberg R, Bashkin P, Sberro H, Tsalyuk M, Surette MG, Alon U (2004) Just-in-time transcription program in metabolic pathways. Nat Genet 36: 486–491 [DOI] [PubMed] [Google Scholar]