Abstract
Interactions between short modified peptide motifs and modular protein domains are central events in cell signal-transduction. We determined interaction partners to all cytosolic tyrosine residues of the four members of the ErbB-receptor family in an unbiased fashion by quantitative proteomics using pull-down experiments with pairs of phosphorylated and nonphosphorylated synthetic peptides. Each receptor had characteristic preferences for interacting proteins and most interaction partners had multiple binding sites on each receptor. EGFR and ErbB4 had several docking sites for Grb2, while ErbB3 was characterized by six binding sites for PI3K. We identified STAT5 as a direct binding partner to EGFR and ErbB4 and discovered new recognition motifs for Shc and STAT5. The overall pattern of interaction partners of EGFR and ErbB4 suggests similar roles during signaling through their respective ligands. Phosphorylation kinetics of several tyrosine resides was measured by mass spectrometry and correlated with interaction partner preference. Our results demonstrate that system-wide mapping of peptide-protein interactions sites is possible, and suggest shared and unique roles of ErbB-receptor family members in downstream signaling.
Keywords: phosphorylation, post-translational modification, protein-protein interaction, receptor tyrosine kinase signaling, SILAC
Introduction
Modification-dependent protein-protein interactions between domains and characteristic peptide motifs are key organizing principles of signaling events in all organisms. For example, specific, extended peptide sequences containing phosphotyrosine interact with cognate SH2 or PTB domains (Pawson and Nash, 2003; Schlessinger and Lemmon, 2003; Pawson, 2004). Consensus peptide sequences involved in recognition by domains have been well studied using oriented peptide libraries (Songyang et al, 1994), peptide arrays (Landgraf et al, 2004) or phage display (Cesareni et al, 2002). Individual interactions between activated tyrosine kinase receptors and specific downstream signaling molecules have been investigated using immunoprecipitation and detection of selected proteins by Western blotting (see Olayioye et al, 2000 and references therein). However, most of these methods lack specificity for modification-dependent interactions.
In the past, there have been several studies for large-scale protein-protein interaction mapping in yeast (Ito et al, 2000, 2001; Uetz et al, 2000; Gavin et al, 2002; Ho et al, 2002) and in higher organisms (Bouwmeester et al, 2004; Goehler et al, 2004) using the yeast two-hybrid system or tagging of full-length proteins, followed by immunoprecipitation and mass spectrometric identification of bound partners. This work provided important insights into protein interaction networks but did not specifically address signal-dependent interactions. However, to serve as a basis for systems biology, static and dynamic, signal-dependent interactions need to be determined in a comprehensive manner. Since protein-protein interactions during signaling events occur in a controlled and ordered way, proteins serving as nodes in interaction networks need to provide distinct binding sites for the variety of interaction partners, in response to different stimuli. In order to improve our understanding of how interaction specificity of different pathways is achieved, it is important to obtain an overview of the intramolecular distribution of binding sites. Therefore, there is a great need for methods that can determine interactions at the resolution of single modification sites and with high-throughput. This would help in systematizing our increasing knowledge of protein interaction networks and signaling pathways.
Members of the ErbB-receptor family are expressed in many different tissues and play a crucial role in cell proliferation and differentiation. They are activated by ligand binding, which leads to homo- or heterodimerization followed by transphosphorylation of characteristic tyrosines. This triggers downstream signaling cascades by recruitment of specific substrate proteins. EGFR (ErbB1) and ErbB4 are fully functional receptor tyrosine kinases, whereas ErbB2 has no endogenous ligand and ErbB3 has no functional kinase domain. Changes in expression and aberrant activation, especially of EGFR and ErbB2, have been shown to be associated with a variety of cancers (Blume-Jensen and Hunter, 2001). The EGFR and its signaling pathway has been well studied with respect to protein-protein interactions, and for the main players in the pathway, their interaction sites have been mapped. The three other members of the ErbB family are less well studied (Olayioye et al, 2000), although interaction partners have also been identified for a number of phosphotyrosine residues in ErbB2, ErbB3 and ErbB4. A general model of how this receptor family integrates different signals from different stimuli through unique and redundant binding sites will be crucial to our understanding of the diverse biological roles of this receptor family. Therefore, systematic mapping of protein interaction sites of whole protein families and motif classes combined with unbiased protein detection is necessary.
Using a recently developed quantitative proteomics technology for determining protein-protein interactions (Schulze and Mann, 2004), we here present results of a first systematic profiling of phosphotyrosine-dependent interaction sites on the ErbB-receptor family. Our screen resulted in novel and global insights into specificity and distribution of protein interaction sites by giving a broad picture of the direct interaction partners of all phosphotyrosine residues of the ErbB family members. The peptide-protein interaction screen turned out to be extremely specific in the detection of primary interaction partners, and revealed that each of the receptors has distinct patterns of binding partners, indicating distinct roles in signaling events. Strikingly, EGFR and ErbB4 show high similarities in their phosphotyrosine interaction partners, suggesting integrative roles in signaling pathways of their respective ligands.
Results
Detection of protein-protein interactions has previously been a balance between specificity (background reduction) and affinity (detection of weak interactions). Recently, the introduction of stable isotope labeling, to distinguish specific from unspecific interaction partners (Blagoev et al, 2003; Ranish et al, 2003), has enabled detection of weak binders in the presence of high levels of background proteins (see below). Based on this principle, we have developed a proteomic screen for peptide motif-based interactions (Schulze and Mann, 2004). Using synthetic peptide pairs in phosphorylated and unphosphorylated form, pull-down experiments are performed to enrich specific binding partners to the phosphorylated bait peptides. These proteins are subsequently identified and quantified by mass spectrometry. In this study, by further method development, we greatly increased the throughput of the analysis allowing us to profile the entire phosphotyrosine interactome of the ErbB-receptor family.
Improvements and scale-up of the peptide-protein interaction screen
The ErbB family contains a total of 89 cytosolic tyrosine residues. In order to carry out a systematic analysis of binding partners to the corresponding phosphotyrosine motifs, the proteomic peptide-protein interaction screen had to be optimized and streamlined. In the process of scaling up the method, desthiobiotin instead of biotin was used as the tagging reagent during peptide synthesis. Desthiobiotinylated peptides were then immobilized on streptavidin-coated magnetic beads (Dynabeads MyOne Streptavidin), and after incubation with cell lysate, bait peptides and proteins bound to them were eluted with biotin. Bait peptides were removed from the reaction by precipitation of proteins. As a consequence, eluted proteins did not have to be separated over SDS-PAGE prior to analysis by mass spectrometry and were directly trypsin-digested in-solution. This eliminated the time-consuming in-gel digestion step and reduced the number of LC-MS/MS experiments from at least six to one run per pull-down experiment. Furthermore, in-solution digests are more readily automatable and can be performed at a scale of hundreds or thousands. Thus, the improved method allows interaction profiling at a system-wide scale.
In the later part of the study, we employed a linear ion trap Fourier transform mass spectrometer (LTQ-FT, ThermoFinnigan). This instrument has significantly higher sequencing speed and peptide mass accuracy, which resulted in significantly higher quality data.
Systematic analysis and general comparison
Here, we present results of the first modification-dependent protein interaction study involving systematic profiling of binding sites of a whole receptor subfamily. For many receptors, the physiological phosphorylation sites are not well documented, and algorithms for their prediction are still limited. The relatively high throughput of our screen allowed systematic study of all tyrosine sites, without making prior assumptions about the baits or their potential interaction partners. In particular, we wanted to demonstrate that the throughput of the method is sufficiently high, so that no information about the phosphorylation status (i.e. in vivo phosphorylated or not) needed to be taken into account. Therefore, we chose to study potential interaction partners to all cytosolic tyrosine residues of the ErbB-receptor family in their phosphorylated form in a systematic and unbiased approach. Altogether, we analyzed all 94 pairs of synthetic singly phospho-, nonphospho- and doubly phosphopeptides to a total of 89 cytosolic tyrosine residues in the four receptors EGFR, ErbB2, ErbB3 and ErbB4.
For a total of 25 phosphotyrosines, a single interaction partner was found, and for 13 additional tyrosine residues, we found more than one interacting protein (Table I). Not surprisingly, 49 out of the 89 investigated tyrosine residues did not have an interaction partner to their phosphorylated form. Most of the tyrosine residues without interaction partners are located in and around the kinase domain, whereas the residues that interacted with specific partners accumulated at the C-terminal regions of the receptors. Strikingly, all binding partners with a significant ratio in our assay had either an SH2 or a PTB domain.
Table 1.
The number of cytosolic tyrosine residues of each receptor analyzed and the number of residues to which none, one or more than one interacting protein was found
| EGFR | ErbB2 | ErbB3 | ErbB4 | |
|---|---|---|---|---|
| Total number of cytosolic tyrosines | 20 | 19 | 23 | 27 |
| No interactions | 8 (40%) | 11 (57%) | 14 (60%) | 16 (59%) |
| One interaction partner | 6 | 7 | 8 | 7 |
| More than one interaction partner | 6 (30%) | 1 (5%) | 1 (4%) | 4 (14%) |
| Number of different binding partners | 9 | 4 | 4 | 8 |
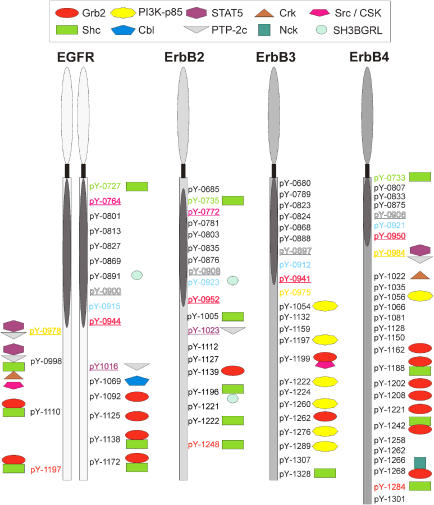
An overview of the distribution of the interaction partners of the different members of the ErbB-receptor family shows clear differences between individual receptors, but also a large overlap (Figure 1, Supplementary Table I). EGFR is the family member with most interaction partners and the highest percentage of tyrosine residues with more than one binding partner. ErbB3 is characterized by a large number of binding sites for phosphatidylinositol-3-kinase (PI3K), while ErbB2 has only few interaction partners with Shc as the most frequent one. ErbB4 and EGFR have a variety of phosphotyrosines that bind Grb2, or Grb2 and Shc. EGFR and ErbB4 have a greater diversity of interaction partners than ErbB2 and ErbB3. All interactions found in this study have been submitted to the public Molecular INTeraction database (Zanzoni et al, 2002).
Figure 1.
Summary of systematic interaction profiling of the ErbB-receptor tyrosine family. All cytosolic residues of the ErbB-receptor family were used in pull-down assays. The interaction partners found are indicated by symbols. The kinase domain of the receptors is designated as an oval. Underlined and colored tyrosine residues mark identical sequence regions between different receptors; regions around colored residues show strong homology between receptors. Most interaction partners to tyrosine residues are found at the C-terminal end outside the kinase domain. The EGFR has multiple interaction partners, and several binding sites for Grb2. ErbB2 has few interaction partners; of them Shc is the most common. ErbB3 interacts mainly with P13-Kinase subunit p85, and ErbB4 again shows a diversity of interaction partners, also with several binding sites for Grb2. Receptors are not drawn to scale. The dimerization is indicated by EGFR dimer. Residues are labeled according to the full-length sequence of each of the receptors.
Interactions mediated by specific peptide motifs
ErbB family members show high homology in the kinase domain (59-81% identity), whereas the C-terminal domains are more divergent (11-25% identity). ErbB3 was the receptor showing the weakest homology to the other three receptors. Nevertheless, characteristic sequences around two tyrosine residues are identical in all four receptors: tyrosines Y900 of EGFR, Y908 of ErbB2, Y897 of ErbB3 and Y906 of ErbB4, all are embedded in the sequence SDVWSYGVTVW; and tyrosine Y944 of EGFR, Y952 of ErbB2, Y941 of ErbB3 and Y950 of ErbB4, all have the identical sequence CTIDVYMIMVK (underlined and color coded in Figure 1). Tyrosine 944 of the EGF-receptor is phosphorylated by Src-kinase (Stover et al, 1995). To both of these shared tyrosine-containing sequences no interaction partners were found, suggesting that these sequences are conserved for structural functions or that they mediate other types of effects. Furthermore, two pairs of phosphotyrosine-containing sequences are identical in EGFR and ErbB2. One of these pairs (pY764 in EGFR and pY772 in ErbB2) has no interaction partner and the other (pY10l6 in EGFR and pY1023 in ErbB2) mediates interaction with PTP-2c. In addition, the protein sequence around pY978 in EGFR and pY984 in ErbB4 is identical, and in our experiments, binds STAT5 and PTP-2c when phosphorylated.
Besides these identical sequences, the ErbB-receptor family also has a highly conserved region around Y915 (EGFR), Y923 (ErbB2), Y912 (ErbB3) and Y921 (ErbB4), to which no interaction partner was found. Similar to Y944, Y915 was also shown to be a Src-kinase phosphorylation site in EGFR (Stover et al, 1995). Two other conserved regions are around Y1197 (EGFR), Y1248 (ErbB2) and Y1284 (ErbB4), as well as around Y727 (EGFR), Y735 (ErbB2) and Y733 (ErbB4). All six corresponding phosphopeptides showed specific interactions to Shc. It is interesting to note that ErbB3 clearly differs from the other three receptors with respect to its interaction partners and conserved regions around tyrosine residues.
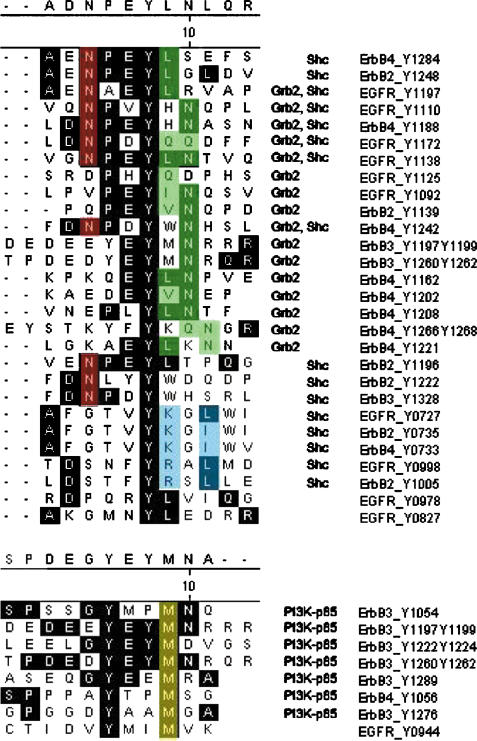
Interaction of phosphotyrosine peptides with Grb2, Shc or PI3K were dependent on specific consensus sequences that have been well documented (Songyang et al, 1994; Yan et al, 2002; Pawson, 2004). The protein Shc, which contains a PTB-domain and an SH2-domain, was found to bind to two different consensus sequences. The majority of Shc-binding sites conformed to the typical N-P-X-pY recognition motif for the PTB domains (Gustafson et al, 1995). However, an alignment of the sequences around five Shc-binding sites revealed no conserved N-terminal motif. Instead, we found a consensus sequence pY-[KR]-X-[LI] involving a basic residue neighboring the phosphotyrosine and a hydrophobic residue at position +3 (Figure 2). Since PTB domains have recognition sequences N-terminal to pY and SH2 domains have C-terminal binding motifs (Yaffe, 2002), Shc is likely to bind these bait peptides through its SH2 domain. A consensus binding motif for the Shc SH2-domain of pY-φ-X-[LI], with strongest selection on the position +3 has been described in pool-sequencing experiments (Songyang et al, 1994). However, our results indicate that the strong selection of position +3 alone comprising the motif pY-X-X-[LI] is not sufficient to specify binding of endogenous Shc, as Shc did not bind to pY915, pY978 of EGFR, pY8O3, pY923 of ErbB2, pY912, pY975, pY1307 of EbB3 or pY921, pY984 of ErbB4, which have a pY-X-X-[LI] sequence. Supporting evidence for our motif also comes from experiments that demonstrated binding of recombinant SH2 domain of Shc to phosphopeptides containing the pY-[KR]-X-[LI] sequence around pY727 and pY998 of EGFR (Ward et al, 1996).
Figure 2.
Binding motifs of the major interaction partners of the ErbB-receptor family. Consensus motifs for binding of Grb2, Shc and P13-Kinase are presented. Sequences shaded in black are common to the majority of all aligned sequences. Residues shaded in red denote essential residues for PTB-domain binding of Shc, while green shades indicate Grb2 binding sites. Residues shaded in blue indicate a motif for SH2-domain binding of Shc. Yellow residues mark essential amino acids in the binding motif for the SH2 domain of P13K. The binding motif for the PTB domain of Shc is N-P-X-pY. The residue N at position −3 is essential, whereas the residue P at position −2 can also be replaced. Besides the known Grb2-binding motif pY-X-N, five binding sites were found for Grb2, which did not contain an N in position +2 after the phosphotyrosine. A binding target for the SH2 domain of Shc was found to contain pY-[KR]-X-φ. The binding motif for P13-Kinase subunit p85 is pY-X-X-M.
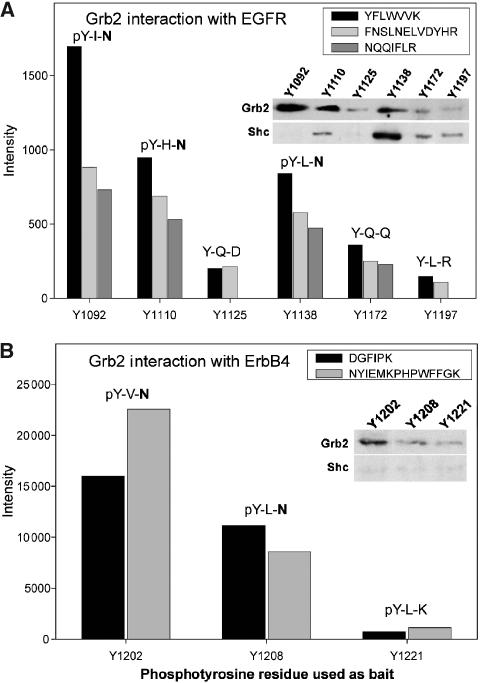
Grb2 was also found to bind sequences that deviate from the strict target sequence pY-X-N. Our experiments suggest the more generalized consensus motif pY-[φQ]-[NQFDK], where φ is a hydrophobic residue. Binding of Grb2 to motifs deviating from the pY-X-N sequence has previously been noted (Songyang et al, 1994; Ward et al, 1996). To describe Grb2 binding preferences, we compared ion intensities in extracted ion chromatograms of identical tryptic peptides of Grb2 from pull-down experiments with different phosphotyrosine residues (Figure 3). For the same mass spectrometric conditions, peptide ion signals from the same tryptic peptide in different experiments are indicative of the abundance of these peptides in different samples, a fact that is, for example, used in Protein Correlation Profiling (Andersen et al, 2003). Those bait peptides containing an asparagine residue at position +2 led to identification of Grb2 peptides with high ion intensities. This correlates with the strong binding of Grb2 to the pY-X-N motif. In contrast, bait peptides without asparagine at position +2 resulted in the identification of tryptic peptides with significantly lower ion intensities (Figure 3). Nevertheless, in all cases, Grb2 was identified as a specific binding partner to the bait peptide, and these results were confirmed by Western blots (insets in Figure 3). Furthermore, our findings are in agreement with in vitro studies of binding affinities of synthetic peptides to the recombinant SH2 domain of Grb2 (Ward et al, 1996).
Figure 3.
Interaction strength of Grb2 binding sites. Ion intensities of extracted ion chromatograms of identical tryptic peptides of Grb2, which were identified in pull-down experiments using different phosphotyrosine containing bait peptides. The two amino acids C-terminal to the bait phosphotyrosine are indicated. (A) Ion intensities of tryptic Grb2 peptides in pull-downs with phosphotyrosine baits of EGFR. (B) Ion intensities of tryptic Grb2 peptides in pull-downs with phosphotyrosine baits of ErbB4. Insets show abundance of interaction partner by Western blot.
In addition to direct binding of Grb2 to phosphotyrosine residues of receptor kinases, Grb2 can also be recruited to the receptor by binding to Shc when Shc is tyrosine phosphorylated as a result of receptor stimulation. Phosphorylation of Shc results in binding of Grb2 to the phosphorylated tyrosine of Shc via its SH2 domain. We found that five bait phosphopeptides had Grb2 and Shc as common binding partners. However, in all five cases, the bait peptide contained overlapping binding motifs for Grb2 and Shc (see also inset in Figure 3). Furthermore, the experiments were carried out in cells that were not especially stimulated with growth factor (even though they were grown in the presence of serum). Thus, most signal-dependent multi-protein complexes are not present and binding partners are not activated (phosphorylated). Therefore, the identification of Grb2 in our pull-down experiments is most likely due to direct binding and not indirect binding as an interaction partner of Shc. For the above reasons, we believe that our experiments generally yield the direct or primary phosphotyrosine interactome rather than protein complexes.
Y-E-Y motifs of ErbB3 and the role of double phosphorylation
ErbB3 contains three pairs of tyrosine residues that are separated by one glutamic acid residue comprising Y1197 and Y1199, Y1222 and Y1224, as well as Y1260 and Y1262. These Y-E-Y motifs were not found in any of the other ErbB-receptors, suggesting that these regions may perhaps have specific interaction partners also in the doubly phosphorylated state, or that double phosphorylation enhances or abolishes interaction of proteins in the singly phosphorylated peptides. To investigate this question, we used the Y-E-Y regions as bait peptides in unphosphorylated, singly phosphorylated and doubly phosphorylated states, with cell lysates that were metabolically labeled with either 12C6-arginine (Arg0), 13C6-arginine (Arg6) or 15N4 13C6-arginine (Arg10) and used in pull-down experiments with unphosphorylated, singly phosphorylated and doubly phosphorylated bait peptides.
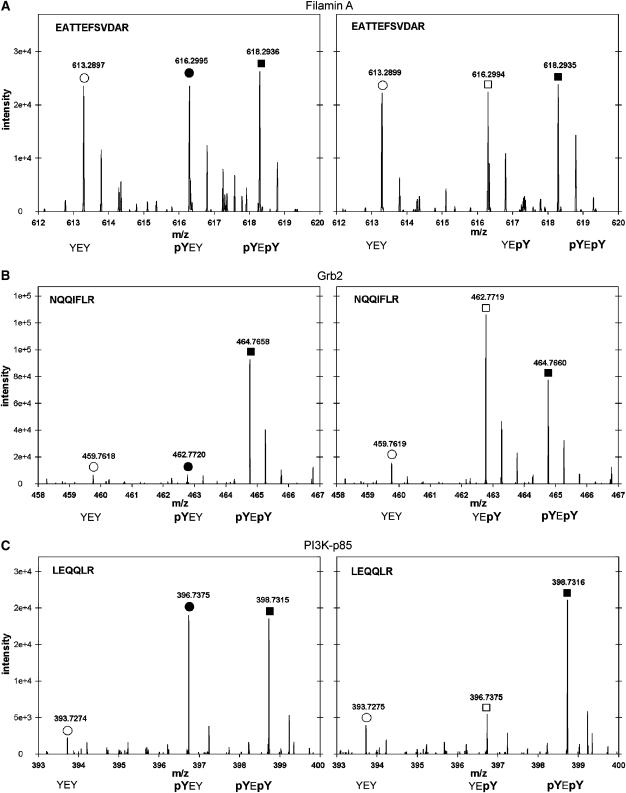
The results show that there are clear binding partners for each of the singly phosphorylated bait peptides of the Y-E-Y motifs (Figure 4). All three Y-E-Y motifs contain a binding site for PI3K at the first tyrosine residue (Y1197, Y1222 and Y1260; pY-E-Y), and two motifs contain a binding site for Grb2 at the second tyrosine residue (Y1199 and Y1268; Y-E-pY). The Y-E-Y motif with phosphorylated tyrosine residue Y1224 (Y-E-pY) did not result in specific binding of any protein. PI3K, as well as Grb2, also interacted with the doubly phosphorylated bait peptides. However, Grb2 interaction was weaker with the doubly phosphorylated bait peptides, compared to the interaction with the singly phosphorylated peptide, including the Grb2 binding site. Apparently, the doubly phosphorylated regions did not have characteristic specific interaction partners. Since both tyrosine residues are very close together, only one of the two possible interaction partners can bind at a time, and it remains open if the doubly phosphorylated form actually occurs in vivo.
Figure 4.
Interaction partners with the YEY motifs of ErbB3. Using metabolic labeling with Arg0, Arg6 and Arg10, interaction partners to Y-E-Y in all three phosphorylation combinations were determined. Results are shown for the two tyrosines around Yl 197 and Yl 199. (A) MS spectra of a tryptic peptide of Filamin A, an unspecific binding protein as indicated by the same ion intensity when precipitated by the nonphosphorylated, singly phosphorylated and doubly phosphorylated bait. (B) MS spectra of a tryptic peptide of Grb2. Grb2 binds to pY-E-pY and Y-E-pY. (C) MS spectra of a tryptic peptide of PI3-Kinase. PI3-Kinase subunit p85 binds to doubly phosphorylated peptide and pY-E-Y. No protein was found to bind specifically to the doubly phosphorylated peptides.
Direct interaction of EGFR and ErbB4 with STAT5
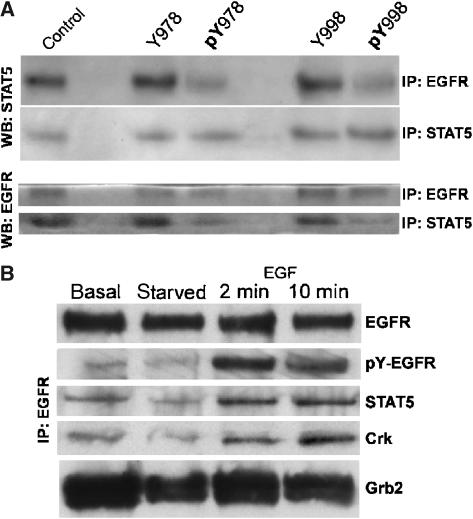
The systematic profiling of interaction partners not only allows a global view of interaction sites, but also reveals important detailed information that can be exploited further for the understanding of signaling processes, as will be illustrated with the example of STAT5 binding to EGFR. STAT5 is a transcription factor that is rapidly activated upon EGF stimulation. It then moves to the nucleus, where it is involved in the regulation of gene transcription. Experiments with truncated EGFR clones suggested that the region between tyrosine residue Y0978 and Y0998 was necessary for activation of STAT5 (Xia et al, 2002). However, it remained unclear whether this proposed interaction was direct or through a multiprotein complex. Systematic interaction profiling of all tyrosine residues of the ErbB-receptor family revealed STAT5 as a specific interaction partner to the phosphorylated bait peptides Y978 and Y998 of the EGF receptor and Y984 of ErbB4. We confirmed the direct interaction of STAT5 and EGFR by immunoprecipitation. When EGFR was immunoprecipitated from whole-cell lysates of cells under normal growth conditions (basal stimulation in 10% FBS), STAT5 was identified by Western blot and vice versa (Figure 5). To further confirm this finding, we added synthetic phosphorylated peptides pY978 and pY998 to the immunoprecipitation reaction, which resulted in reduced STAT5 detection after EGFR immunoprecipitation and in reduced EGFR detection after STAT5 immunoprecipitation. The unphosphorylated counterparts of these peptides had no influence on the immunoprecipitation reactions (Figure 5A). Figure 5B shows that the interaction of STAT5 depended on EGFR phosphorylation, which peaked after two minutes of EGF stimulation, and that normal growing HeLa cells also have a basal level of phosphorylated EGFR.
Figure 5.
Co-immunoprecipitation of STAT5 and EGFR. (A) In co-immunoprecipitation experiments, EGFR was detected by Western blot (WB) in immunoprecipitations (IP) with STAT5, and STAT5 was detected in immunoprecipitations of EGFR. Co-immunoprecipitation was reduced by addition of phosphorylated peptides pY978 and pY998 of EGFR. There was no effect when these peptides were added in their nonphosphorylated form as control (Y978 and Y998). In addition, equal amounts of EGFR and STAT5 were immunoprecipitated, as shown by the IP with STAT5, WB with STAT5 and the IP with EGFR, WB with EGFR, respectively. The experiments were performed on HeLa cells growing in 10% FBS. (B) Phospho-EGFR, STAT5 and Crk after immunoprecipitation of EGFR from normal growing HeLa cells (basal), serum-starved cells for 14 h (starved) and EGF-stimulated cells for 2 and 10 min. The antibody was used for IP and WB of EGFR against codons 998-1022, while the antibody against phospho-EGFR was directed against pYl 197 of EGFR. STAT5 antibody is directed against the C-terminal domain of Stat5b, but recognized isoforms STAT5a/b, and Crk antibody is directed against the N-terminus of Crk.
Dynamics of tyrosine phosphorylation after EGF stimulation
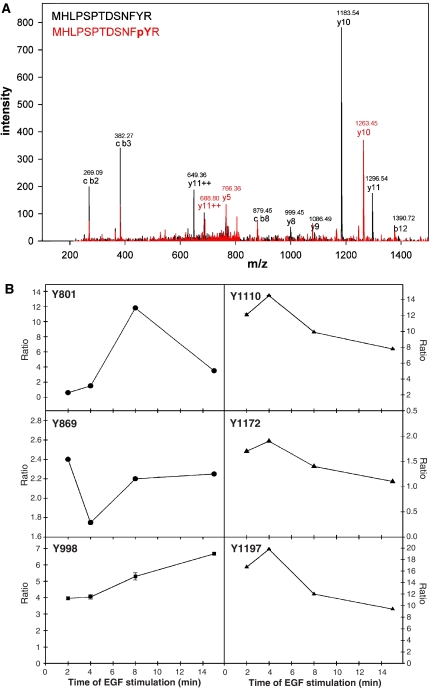
To correlate our findings with in vivo phosphorylation of the receptor, we directly analyzed the kinetics of phosphorylation of several sites upon EGF stimulation by mass spectrometry. Phosphopeptides of immunoprecipitated EGFR were analyzed by LC-MS/MS after different times of stimulation with EGF. For these experiments, different cell populations were completely SILAC labeled with either Arg0, Arg6 or Arg10. Arg0 populations were left unstimulated, Arg6 populations were stimulated for either 2 or 4 min, and Arg10 populations were stimulated for eight or 15 min. Ratios between tryptic phosphopeptides in their Arg0, Arg6 and Arg10 forms in the combined immunoprecipitations allowed one to monitor the abundance of these tryptic phosphopeptides as a function of time after EGF stimulation (Blagoev et al, 2004). In this experiment, we identified a novel EGFR phosphorylation site by the tryptic peptide containing the phosphorylated tyrosine residue Y998 (Figure 6A). In total, tryptic peptides to six phosphorylation sites were analyzed (Figure 6B). Peptides of phosphorylated Y1110, Y1172 and Y1197 showed highest ratios at 2-4 min of stimulation, and these ratios decreased with longer EGF stimulation. In contrast, Src-kinase phosphorylation site Y869 displayed a decrease in ratio at 4 min of stimulation, which was accompanied by an increase in ratio of the nonphosphorylated tryptic peptide. The novel phosphorylation site at Y998 displayed a gradual increase in ratio over 15 min of stimulation, and the phosphorylation site at Y801 was maximally activated as late as 8 min after start of EGF stimulation (Figure 6B). However, in interpreting the kinetics of such curves, it has to be kept in mind that the amount of EGFR immunoprecipitated by the phosphotyrosine antibody will depend on the total tyrosine phosphorylation of the receptor in addition to the phosphorylation of the site under investigation.
Figure 6.
Phosphorylation dynamics of EGFR. (A) MS-MS fragmentation spectrum of the tryptic peptide MHLPSPTDSNFYR and MHLPSPTDSNF[pY]R of EGFR showing phosphorylation of tyrosine 998. (B) Time course of phosphorylation at six different tyrosine residues of the EGFR upon EGF stimulation. Ratios of peak intensities after EGF stimulation versus no stimulation are shown. Tyrosine residues Y1110, Y1172 and Y1197 are autophosphorylation sites, tyrosine Y869 is phosphorylated by Src kinase, and tyrosine Y998 and Y801 are novel phosphorylation sites.
These results show that different phosphorylation sites show different phosphorylation dynamics after stimulation, reflecting distinct temporal roles in signaling. Interestingly, these differences in phosphorylation dynamics correlate with the interaction partners identified for each of these sites. The autophosphorylation sites at Y1110 and Y1197 display rapid phosphorylation with a maximum after 4 min and they have the same set of interaction partners (Grb2 and Shc). The novel phosphorylation site at Y998 is increasingly phosphorylated over 15 min, and interacts with STAT5, PTP-2c, Shc, Crk, and the phosphotyrosine kinase Src or Csk (Cole et al, 2003), which could not be distinguished based on the observed tryptic peptides. Crk was immunoprecipitated with EGFR in a phosphorylation-dependent manner (Figure 5B). In contrast, the phosphorylation sites at Y801 and Y869 showed distinct phosphorylation kinetics, but had no interaction partner in our assay.
Discussion
Specific protein-protein interactions are not only involved in the formation of stable multimeric complexes, but are important for a large number of rather short-term associations of proteins during signaling. Kinase and substrate need to interact, and temporary complexes are formed by means of scaffolding and adaptor proteins (Robinson and Bonifacino, 2001; Pawson, 2004). Knowing the binding partner to a phosphorylation site defines the role of that particular modification during signal transduction. Apart from resolving interactions to specific binding sites on different signaling proteins, an overall picture of the distribution of binding sites for interacting proteins may also lead to general conclusions of how signals are directed to different pathways. The approach taken in this study is thus complementary to other proteomic approaches in which whole signaling complexes are identified (see, e.g., Blagoev et al, 2003).
As signaling events are often characterized by phosphorylation of receptors and downstream interaction partners, the peptide-protein interaction screen (Schulze and Mann, 2004) was further developed for high sample throughput and used to map the primary binding partners of phosphorylated tyrosine residues of the ErbB-receptor family. Individual members of this family have in the past been studied with respect to interaction partners of selected residues using phosphopeptide competition analysis, receptor mutants and coimmunoprecipitation (Margolis et al, 1990; Birge et al, 1992; Fedi et al, 1994; Prigent and Gullick, 1994; Lombardo et al, 1995; Sorkin et al, 1996; Keilhack et al, 1998; Hellyer et al, 2001; Yarden and Sliwkowski, 2001; Moro et al, 2002; Blagoev et al, 2003). In this study, we take a new approach that allows comparison and mapping of the primary interaction partners and their binding sites in a single, comprehensive and unbiased system; that is, we do not need to make any assumptions about likely baits or interaction partners.
Specificity and sensitivity
The fact that all binding partners identified contained either SH2- or PTB-domains emphasizes the specificity of signal transduction modules (Pawson and Nash, 2003), combined with the observation that phosphotyrosines that were not expected to have binding partners did not show any proteins with significant ratios. This is in contrast to other protein-protein interaction screens, especially in large-scale format (von Mering et al, 2002), and we conclude that assay is extremely specific and capable of confirming known binding motifs and detecting new target sequences.
Our screen resulted in identification of the vast majority of expected direct interaction partners of the ErbB family that bind through phosphotyrosines. Among the few exceptions was PLC-γ, which we were unable to find as an interaction partner of Y1197 of the EGF receptor. This interaction has been described using immunoprecipitations and in vitro phosphopeptide competition and binding assays with cloned domains (Margolis et al, 1990; Rotin et al, 1992). The peptide-protein interaction assay as such is capable of identifying PLC-γ, since we previously described such an interaction using an IRS4 phosphopeptide as bait (Hinsby et al, 2004). We also could not find the interaction of Y1197 with SHIP1 (Keilhack et al, 1998), or of Y1248 of ErbB2 with Chk (Zrihan-Licht et al, 1998). Binding constants of SH2 domains to phosphotyrosines are in the range of 50-500 nM (Pawson, 2004), while binding constants of certain PTB domains to the respective bait peptides can be as low as 2 μM to 10 μM (Yan et al, 2002). The peptide-protein interaction screen was validated for binding constants as low as at least 5 μM (Schulze and Mann, 2004), but it is likely to work with much lower affinity interactions, too. Thus, we believe that the negative results are unlikely to be caused by low affinity of the interactions. However, some interactions may not only be motif-dependent, but also require cooperative binding or activation of the binding partner.
In this study, we identify Grb2 as a specific binding partner to tyrosines Y1199 and Y1268 of ErbB3. Other studies have identified Grb7 as a specific binding partner to those tyrosine residues using immunoprecipitation and Western blots as detection method, but assays with synthetic peptides indicated that Grb2 can also bind to Y1199 and Y1268, but with a lower affinity (Fiddes et al, 1998). We have not identified Grb7 at all in the peptide pull-downs using whole-cell lysate and could not detect Grb7 expression in HeLa cells by Western blotting (data not shown). Therefore, the differences in interaction partner may be due to the use of a different cell type. This example illustrates that the results of our assay depend on expression of the binding partner in vivo, and may be different for different cell types.
Finally, some proteins have previously been reported to interact with EGFR (van Delfi et al, 1997), such as EPS15 (Schumacher et al, 1995). In our study, we see EPS15 in pull-downs with a number of bait peptides, but always with a ratio of one to one, indicating no direct binding to the bait peptides. This would in any case be expected since EPS15 has no SH2 or PTB domain, and rather is recruited to EGFR in a larger protein complex.
Redundancy of binding sites
With very few exceptions, each of the proteins recruited to phosphotyrosines on the ErbB family had more than one binding site, although sometimes with different affinities. For example, EGFR has six binding sites for the adapter protein Grb2, and ErbB4 has five, each with different binding strength (see Figure 3 and Ward et al, 1996). PI3K is the sole binding partner to six tyrosines of ErbB3 and one in ErbB4. This redundancy allows combinatorial control of downstream effector pathways in several ways. Different upstream signals can induce different phosphorylation patterns on ErbB receptors–for example, in the case of EGF versus TGFα stimulation of EGFR (Guo et al, 2003). The combination of these different phosphorylation patterns with the different binding affinities allows a different cast of effector molecules to be assembled at the receptor. A second level of combinatorial control is the formation of different homo- or heterodimers, depending on the stimulatory condition (Olayioye et al, 1998; Hynes et al, 2001). Studies of cells with differential overexpression of ErbB family members in various combinations revealed that binding of ErbB2 to Shc, PI3K or Grb2 was dependent on the mode of activation, and that receptor phosphorylation also was dependent on the dimerization partner (Goehler et al, 2004). Thus, the interaction partners Grb2, Shc or PI3K may only interact with a specific phosphotyrosine under a given cellular condition, and with a different one under another stimulus or dimerization state. The redundancy in binding sites for specific adapter proteins thus may be essential for integration of different stimuli and the coordinated transduction of this information to downstream pathways and to ensure, in one way or another, the binding/activation of these very important signaling switches. However, it has to be kept in mind that some phosphorylation sites may be active only under very specific conditions in vivo, and some sites analyzed in our systematic screen are likely not be phosphorylated under any conditions.
Correlation to in vivo signaling events
In this study, we clearly identified STAT5 as a direct interaction partner to EGFR (Y978 and Y998) and ErbB4 (Y984). It was previously shown that Src-phosphorylated tyrosine residue Y869 is essential for activation of STAT5b, but does not mediate the interaction itself (Kloth et al, 2003). EGF stimulation and subsequent phosphorylation of EGFR at tyrosine Y978, Y998 and Y869 (see Figure 6B) would then lead to recruitment and activation of STAT5, which subsequently activates transcription. Our finding that ErbB4 also has a binding site for STAT5 is consistent with the observation that dimerization of ErbB2 and ErbB4 lead to activation of STAT5b (Olayioye et al, 1999) and that ErbB4 signaling is required for STAT5 activation during lactation (Jones et al, 1999). This is a further example showing that signaling through EGFR and ErbB4 can activate similar downstream processes in response to their different ligands.
In a separate experiment, we have determined the activation profiles of several tyrosine phosphorylation sites in response to EGF. We found that activation kinetics can correlate with the type of interaction partner. While this experiment was limited in the number of sites analyzed, it clearly shows how integration of in vivo kinetics with a knowledge of binding partners can add more functional information of phosphorylation sites and interaction partners.
Implications for signaling roles of different ErbB family members
This study reveals striking similarities between EGFR and ErbB4 with respect to their interaction partners and the distribution of specific binding sites at the C-terminal end of the receptors. EGFR and ErbB4, the two fully functional receptor tyrosine kinases of the family, are also the two receptors that were found to have the greatest diversity of interaction partners. Both receptors contain redundant binding sites for Grb2, and both receptors were found to bind STAT5. However, EGFR was unique in having a binding site for Cbl, which did not interact with any of the other receptor bait peptides.
It is well known that EGFR functions as an integration site for different stimuli (Hackel et al, 1999; Zwick et al, 1999). Thus, redundancy of binding sites and a great diversity of different binding partners is not unexpected. Our data suggest that ErbB4 may serve a similar role during signaling through neuregulin-induced pathways. This idea is supported by findings of overexpression studies, in which EGFR dimerizes preferably with ErbB2 and ErbB3, while ErbB4 dimerizes with ErbB2 and ErbB3, but to a lesser extent with EGFR (Olayioye et al, 2000). In this respect, it should be kept in mind that ErbB2 has a nonfunctional extracellular ligand-binding domain and is a common dimerization partner of other members of the ErbB family (Graus-Porta et al, 1997). In contrast, ErbB3 has an inactive kinase domain; thus, transphosphorylation in different heterodimers may lead to phosphorylation of different sites in ErbB3.
An evaluation of existing microarray expression data (Affymetrix human U133A data set, Novartis Foundation) using hierarchical cluster analysis (Supplementary Figure 1) shows that EGFR and ErbB4 indeed are rarely coexpressed and that ErbB2 and ErbB3 are seldom expressed without either EGFR or ErbB4. Thus, our data are consistent with the idea that EGFR and ErbB4 function as 'control centers' and sites of integration for signaling processes, involving stimulation with their ligands, EGF and neuregulin, respectively. Furthermore, the striking difference in interaction partners of kinase-inactive ErbB3 compared to the other family members may indicate a specific role in signaling to the PI3K pathway as part of heterodimers with EGFR, ErbB4 or ErbB2 in different cell types or differentiation stages.
Perspective
The peptide-protein interaction screen integrates well with the broader goals of phosphoproteomics. While we have performed a systematic screen of all potential pY sites in this study, bioinformatic predictions of phosphorylation sites or experimentally verified sites could have been taken as a basis for bait selection. The identification of signal-dependent phosphorylation sites is thus a major experimental goal in the characterization of signaling networks. The peptide interaction screen then allows one to directly identify possible binding partners to these identified phosphopeptides with high confidence.
ErbB2 and EGFR are attractive targets for cancer therapy as they are often overexpressed or mutated in tumors. The drug HerceptinTM has been developed as an efficient treatment of cancer types that depend on overexpression of ErbB2, and small molecule kinase inhibitors have been directed against EGFR. Both strategies have the general goal of inhibiting aberrant signaling. However, many studies show that perturbation of signaling networks can have diverse and unanticipated effects. For example, a thorough analysis of Herceptin action revealed that it did not only affect ErbB2, but also the expression of EGFR and its downstream ligands (Smith et al, 2004). The understanding of receptor-signaling crosstalk and a detailed picture of different sites of interaction to downstream binding partners can be expected to contribute to description of signaling networks at the systems biology level. Systematic screens, such as the one performed here, can possibly enable entirely new ways of identifying interesting drug targets and potentially make important contributions to therapeutic research.
Our studies show that different receptors in the ErbB family clearly differ in their preferred interaction partners, indicating distinct roles in signaling. Furthermore, modification-dependent interaction does not occur at random, but rather requires characteristic motifs for specific classes of proteins to interact. Novel motifs were found for Shc and STAT5 binding. Importantly, we have shown that the peptide-protein interaction screen can be performed in a large-scale fashion and that it is indeed a suitable method to obtain a broad picture of shared and distinct interaction partners of a whole protein family. In the future, with further improvements in mass spectrometric performance, streamlining and automation, the screen can be applied system-wide to essentially all signaling proteins and their substrates.
Materials and methods
Cell culture
Human HeLa cells were grown in arginine-deficient Dulbecco's modified Eagle's medium (DMEM) with 10% dialyzed fetal bovine serum. One cell population was supplemented with normal isotopic abundance L-12 arginine (Sigma) and L-lysine, and the other with 99% isotopic abundance 13C6-arginine (Aldrich) and 13C6-lysine, as described (Ong et al, 2003). Thereby full labeling of all tryptic peptides was achieved. In some experiments, cell populations were metabolically labeled using D4-lysine instead of 13C6-lysine. For studies with singly and doubly phosphorylated peptides of the Y-E-Y motifs, three different cell populations were labeled with 12C6-arginine (Arg0), 13C6-arginine (Arg6), and 13C6 15N4-arginine (Arg10), resulting in mass differences of 6 and 10 Da to the Arg0 form, respectively. Each cell population was grown for at least five passages, encompassing a minimum of seven population doublings.
For experiments determining phosphorylation kinetics, three different cell populations were labeled with Arg0, Arg6 and Arg10, respectively. Prior to stimulation, cells were serum-starved for 18 h. Unlabeled cells were left unstimulated, while two cell populations labeled with Arg6 were stimulated for two minutes and four minutes, respectively, and two cell populations labeled with Arg10 were stimulated for 8 min and 15 min, respectively. EGF was added to a final concentration of 200 ng/ml. Lysates of cells after different times of EGF stimulation were combined as described (Blagoev et al, 2004) and subjected to immunoprecipitation with a general antibody against phosphotyrosine residues.
Peptide synthesis and pull-downs
Desthiobiotinylated peptides were synthesized on a solid-phase peptide synthesizer using amide resin (Intavis, Germany). Peptides were designed as 15-mers bearing an N-terminal biotin, the tetrapeptide linker SGSG (Ward et al, 1996). The identity and purity of the synthesized peptides was confirmed by mass spectrometric analysis. Peptides were synthesized as pairs in an 'active' and 'control' form. For affinity pull-downs, 50 nmol of immobilized peptide was added to an average of 2 mg of cell lysate.
Dynabeads MyOne StreptavidinTM were saturated with biotinylated peptide prior to incubation with cell lysates. Cells were lysed in 1% (v/v) Nonident P-40, 150 mM sodium chloride, 50 mM Tris-HCl pH 7.5, protease inhibitors (Complete Tablets, Roche), and 1 mM sodium orthovanadate as phosphatase inhibitor. An equal amount of protein was incubated with the respective immobilized peptides at 4°C for 4 h. After six rounds of washing with lysis buffer, beads of pull-down pairs with 'control' and 'phosphorylated' were combined (Schulze and Mann, 2004) and bound proteins were eluted using 20 mM biotin. Eluted proteins were precipitated and subsequently digested with trypsin for LC-MS/MS analysis as described.
Immunoprecipitation
HeLa cells were lysed in 1% (v/v) Nonident P-40, 150 mM sodium chloride, 50 mM Tris-HCl pH 7.5, 1 mM sodium orthovanadate and protease inhibitors (Complete Tablets, Roche), and incubated at 4°C with respective primary antibodies. Depending on the host animal of the antibody, protein-A-sepharose or protein-G-sepharose beads were added after 2 h and incubated for an additional 4 h. After three rounds of washes, coprecipitated proteins were eluted in sample buffer for SDS gel electrophoresis. For peptide competition analyses, 250 μg (125 nmol) of phosphorylated or nonphosphorylated peptide was added to the immunoprecipitation reaction. STAT5 was immunprecipitated with an antibody directed against STAT5a and b isoforms (Santa Cruz), EGFR was immunoprecipitated with a polyclonal antibody (Santa Cruz). For immunoprecipitation of tyrosine-phosphorylated proteins, we used the 4G10-anti-phosphotyrosine antibody (Upstate Cell Signaling Technologies).
LC/MS/MS, database searching and quantitation
Eluted proteins were in-solution digested with 1 μg trypsin after reduction in 1 μg DTT, alkylation with 5 μg iodoacetamide and dilution of the sample with 4 volumes of 50 mM NH4HCO3. Tryptic peptide mixtures were then desalted on STAGE tips (Rappsilber et al, 2003) and loaded onto reversed phase analytical columns for liquid chromatography (Ishihama et al, 2002). Peptides were eluted from the analytical column by a multistep linear gradient running from 5 to 30% acetonitrile in 90 min and sprayed directly into the orifice of a QSTAR-Pulsar quadrupole Time-Of-Flight hybrid mass spectrometer (PE-Sciex, USA) or a LTQ-FT (Thermo Electron, Germany). Proteins were identified by tandem mass spectrometry (MS/MS) by information-dependent acquisition of fragmentation spectra of multiple-charged peptides that were then searched against the human International Protein Index Database (IPI; http://srs6.ebi.ac.uk) using the Mascot algorithm (Perkins et al, 1999) with search parameters as described in (Foster et al, 2003) for QSTAR data and as in Olsen et al (2004) for LTQ-FT data. For quantitation and spectra validation, the software MSQuant (http://msquant.sourceforge.net) was used.
Determination of significant binding partners
Ratios of labeled to unlabeled forms of each validated tryptic peptide, and the associated average ratio for the whole protein were obtained by MSQuant. A plot of the distribution of all protein ratios from one experiment revealed the highest number of proteins with a ratio of around one. Those ratios being significantly (2σ) different from the average ratio of the majority (covering mean±σ) of identified proteins were considered as significant. Furthermore, we used crossover experiments (Schulze and Mann, 2004) in which specific interaction partners were required to have inverse ratios compared to the first experiment. In the pull-down experiments, 89-153 proteins were quantified. Cutoff thresholds were calculated for each experiment individually. The range of average ratios for unspecific binders ranged from 0.96±0.52 to 1.86±0.87 in different pull-down experiments.
Supplementary Material
Supplementary Table 1
Legend to Supplementary Figure 1
Supplementary Figure 1
Acknowledgments
We thank members of CEBI for critical comments to the manuscript. Special thanks go to Jesper Olsen for setting up acquisition methods for the LTQ-FT. This work was supported by 'Interaction Proteome' a grant from the European Commission in the 6th framework program. Work at CEBI is also supported by grants from the Danish National Research Foundation.
References
- Andersen JS, Wilkinson CJ, Myoru T, Mortensen P, Nigg EA, Mann M (2003) Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426: 570–574 [DOI] [PubMed] [Google Scholar]
- Birge RB, Fajardo JE, Mayer BJ, Hanafusa H (1992) Tyrosine-phosphorylated epidermal growth factor receptor and cellular p130 provide high affinity binding substrates to analyze Crk-phosphotyrosine-dependent interactions in vitro. J Biol Chem 267: 10588–10595 [PubMed] [Google Scholar]
- Blagoev B, Kratchmarova I, Ong S-E, Nielsen M, Foster L, Mann M (2003) A proteomics strategy to elucidate functional protein-protein interactions applied to EGF signaling. Nat Biotechnol 21: 315–318 [DOI] [PubMed] [Google Scholar]
- Blagoev B, Ong S-E, Kratchmarova I, Mann M (2004) Temporal ordering of signaling networks by quantitive proteomics. Nat Biotechnol 22: 1139–1145 [DOI] [PubMed] [Google Scholar]
- Blume-Jensen P, Hunter T (2001) Oncogenic kinase signalling. Nature 411: 355–365 [DOI] [PubMed] [Google Scholar]
- Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, Cruciat C, Eberhard D, Gagneur J, Ghidelli S, Hopf C, Huhse B, Mangano R, Michon AM, Schirle M, Schlegl J, Schwab M, Stein MA, Bauer A, Casari G, Drewes G, Gavin AC, Jackson DB, Joberty G, Neubauer G, Rick J, Kuster B, Superti-Furga G (2004) A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol 6: 97–105 [DOI] [PubMed] [Google Scholar]
- Cesareni G, Panni S, Nardelli G, Castagnoli L (2002) Can we infer peptide recognition specificity mediated by SH3 domains? FEBS Lett 513: 38–44 [DOI] [PubMed] [Google Scholar]
- Cole PA, Shen K, Qiao Y, Wang D (2003) Protein tyrosine kinases Src and Csk: a tail's tale. Curr Opin Chem Biol 7: 580–585 [DOI] [PubMed] [Google Scholar]
- Fedi P, Pierce JH, Di Fiore PP, Kraus MH (1994) Efficient coupling with phosphatidylinositol 3-kinase, but not phospholipase C gamma or GTPase-activating protein, distinguishes ErbB-3 signaling from that of other ErbB/EGFR family members. Mol Cell Biol 14: 492–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiddes RJ, Campbell DH, Janes PW, Siversten SP, Sasaki H, Wallasch C, Daly RJ (1998) Analysis of Grb7 recruitment by heregulin-activated erbB receptors reveals a novel target selectivity for erbB3. J Biol Chem 273: 7717–7724 [DOI] [PubMed] [Google Scholar]
- Foster LJ, de Hoog C, Mann M (2003) Unbiased quantitive proteomics of lipid rats reveals high specificity for signaling factors. Proc Natl Acad Sci USA 100: 5813–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin A-C, Bösche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon A-M, Cruciat CM, Remor M, Höfert C, Schelder M, Brajenovic M, Ruffner H, Merino A, Klein K, Hudak M, Dickson D, Rudi T, Gnau V, Bauch A, Rybin V, Drewes G, Raida M, Bouwmeester T, Bork P, Seraphin B, Kuster B, Neubauer G, Superti-Furga G (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147 [DOI] [PubMed] [Google Scholar]
- Goehler H, Lalowski M, Stelzl U, Waelter S, Stroedicke M, Worm U, Droege A, Lindenberg KS, Knoblich M, Haenig C, Herbst M, Suopanki J, Scherzinger E, Abraham C, Bauer B, Hasenbank R, Fritzsche A, Ludewig AH, Buessow K, Coleman SH, Gutekunst CA, Landwehrmeyer BG, Lehrach H, Wanker EE (2004) A protein interaction network links GIT1, an enhancer of huntingtin aggregation, to Huntington's disease. Mol Cell 15: 853–865 [DOI] [PubMed] [Google Scholar]
- Graus-Porta D, Beerli RR, Daly JM, Hynes NE (1997) ErbB2, the preferred heterodimerization partner of a ErbB receptors, is a mediator of lateral signaling. EMBO J 16: 1647–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Kozlosky CJ, Ericsson LH, Daniel TO, Cerretti DP, Johnson RS (2003) Studies of ligand-induced site-specific phosphorylation of epidermal growth factor receptor. J Am Soc Mass Spectrometry 14: 1022–1031 [DOI] [PubMed] [Google Scholar]
- Gustafson TA, He W, Craparo A, Schaub CD, O'Neill TJ (1995) Phosphotyrosine dependent interaction of SHC and insulin receptor substrate 1 with the NPEY motif of the insulin receptor via a novel non-SH2 domain. Mol Cell Biol 15: 2500–2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackel PO, Zwick E, Prenzel N, Ulirich A (1999) Epidermal growth factor receptors: critical mediators of multiple receptor pathways. Curr Opin Cell Biol 11: 184–189 [DOI] [PubMed] [Google Scholar]
- Hellyer NJ, Kim MS, Koland JG (2001) Heregulin-dependent activation of phosphoinositide 3-kinase and Akt via the ErbB2/ErbB3 co-receptor. J Biol Chem 276: 42153–42161 [DOI] [PubMed] [Google Scholar]
- Hinsby AM, Olsen JV, Mann M (2004) Tyrosine phosphoproteomics of fibroblast growth factor signaling: a role for insulin receptor substrate-4. J Biol Chem 279: 46438–46447 [DOI] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams S-L, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C, Dewar D, Lin Z, Michalickova K, Willems AR, Sassi H, Nielsen PA, Rasmussen KJ, Andersen JR, Johansen LE, Hansen LH, Jespersen H, Podtelejinkov A, Nielsen E, Craword J, Poulsen V, Sorensen BD, Matthiesen J, Hendrickson RC, Gleeson F, Pawson T, Moran MF, Durocher D, Mann M, Houge CWV, Figeys D, Tyers M (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415: 180–183 [DOI] [PubMed] [Google Scholar]
- Hynes NE, Horsch K, Olayioye MA, Badache A (2001) The ErbB receptor tyrosine family as signal integrators. Endocr-Related Cancer 8: 151–159 [DOI] [PubMed] [Google Scholar]
- Ishihama Y, Rappsilber J, Andersen JS, Mann M (2002) Microcolumns with self-assembled particle fits for proteomics. J Chromatogr A 979: 233–239 [DOI] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y (2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA 98: 4569–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Tashiro K, Muta S, Ozawa R, Chiba T, Nishizawa M, Yamamoto K, Kuhara S, Sakaki Y (2000) Toward a protein-protein interaction map of the budding yeast: a comprehensive system to examine two-hybrid interactions in all possible combinations between yeast proteins. Proc Natl Acad Sci USA 97: 1143–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones FE, Welte T, Fu X-Y, Stem DF (1999) ErbB4 signaling in the mammary gland is required for lobuloalveolar development and Stat5 activation during lactation. J Cell Biol 147: 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keilhack H, Tenev T, Nyakatura E, Godovac-Zinimermann J, Nielsen L, Seedorf K, Bohmer FD (1998) Phosphotyrosine 1173 mediates binding of the protein-tyrosine phosphatase SHP-1 to the epidermal growth factor receptor and attenuation of receptor signaling. J Biol Chem 273: 24839–24846 [DOI] [PubMed] [Google Scholar]
- Kloth MT, Laughlin KK, Biscardi JS, Boerner JL, Parsons SJ, Silva CM (2003) STAT5b, a mediator of synergism between c-Src and the epidermal growth factor receptor. J Biol Chem 278: 1671–1679 [DOI] [PubMed] [Google Scholar]
- Landgraf C, Paimi S, Montecchi-Palazzi L, Castagnoli L, Schneider-Mergener J, Volkmer-Engert R, Cesareni G (2004) Protein interaction networks by proteome peptide scanning. PLoS Biol 2: E14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo CR, Consler TG, Kassel DB (1995) In vitro phosphorylation of the epidermal growth factor receptor autophosphorylation domain by c-src: identification of phosphorylation sites and c-src SH2 domain binding sites. Biochemistry 34: 16456–16466 [DOI] [PubMed] [Google Scholar]
- Margolis B, Li N, Koch A, Mohammadi M, Hurwitz DR, Zilberstein A, Ullrich A, Pawson T, Schlessinger J (1990) The tyrosine phosphorylated carboxyterminus of the EGF receptor is a binding site for GAP and PLC-gamma. EMBO J 13: 4375–4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moro L, Dolce L, Cabodi S, Bergatto E, Erba EB, Smeriglio M, Turco E, Retta SF, Giuffrida MG, Venturino M, Godovac-Zimmermaim J, Conti A, Schaefer E, Beguinot L, Tacchetti C, Gaggini P, Silengo L, Tarone G, Defilippi P (2002) Integrin-induced epidermal growth factor (EGF) receptor activation requires c-Src and pl3OCas and leads to phosphorylation of specific EGF receptor tyrosines. J Biol Chem 277: 9405–9414 [DOI] [PubMed] [Google Scholar]
- Olayioye M, Neve RM, Lane HA, Hynes NE (2000) The ErbB signaling network: receptor heterodimerization in development and cancer. EMBO J 19: 3159–3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olayioye MA, Beuvink I, Horsch K, Daly JM, Hynes NE (1999) ErbB receptor-induced activation of Stat transcription factors is mediated by Src tyrosine kinases. J Biol Chem 274: 17209–17218 [DOI] [PubMed] [Google Scholar]
- Olayioye MA, Graus-Porta D, Beerli RR, Rohrer J, Gay B, Hynes NE (1998) ErbB-1 and ErbB-2 acquire distinct signaling properties dependent upon their dimerization partner. Mol Cell Biol 18: 5042–5051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, Ong S-E, Mann M (2004) Trypsin cleaves exclusively C-terminal to arginine and lysine residues. Mol Cell Proteomics 3: 608–614 [DOI] [PubMed] [Google Scholar]
- Ong S-E, Kratchmarova I, Mann M (2003) Properties of 13C-substituted arginine in stable isotope labeling by amino acids in cell culture (SILAC). J Proteome Res 2: 173–181 [DOI] [PubMed] [Google Scholar]
- Pawson T (2004) Specificity in signal transduction: from phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell 116: 191–203 [DOI] [PubMed] [Google Scholar]
- Pawson T, Nash P (2003) Assembly of cell regulatory systems through protein interaction domains. Science 300: 445–452 [DOI] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJC, Creasy DM, Cottrell JS (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20: 3551–3567 [DOI] [PubMed] [Google Scholar]
- Prigent SA, Gullick WJ (1994) Identification of c-erbB-3 binding sites for phosphatidylinositol 3′-kinase and SHC using an EGF receptor/c-erbB-3 chimera. EMBO J 12: 2831–2841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranish JA, Yi EC, Leslie DM, Purvine SO, Goodlett DR, Eng J, Aebersold R (2003) The study of macromolecular complexes by quantitative proteomics. Nat Genet 33: 349–355 [DOI] [PubMed] [Google Scholar]
- Rappsilber J, Ishihama Y, Mann M (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem 75: 663–670 [DOI] [PubMed] [Google Scholar]
- Robinson MS, Bonifacino JS (2001) Adaptor-related proteins. Curr Opin Cell Biol 13: 443–453 [DOI] [PubMed] [Google Scholar]
- Rotin D, Margolis B, Mohammadi M, Daly RJ, Daum G, Li N, Fischer EH, Burgess WH, Ullrich A, Schlessinger J (1992) SH2 domains prevent tyrosine dephosphorylation of the EGF receptor: identification of Tyr992 as the high-affinity binding site for SH2 domains of phospholipase C gamma. EMBO J 11: 559–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlessinger J, Lemmon MA (2003) SH2 and PTB domains in tyrosine kinase signaling. Sci STKE 2003 Jul 15; 2003(191): RE12. Review [DOI] [PubMed] [Google Scholar]
- Schulze W, Mann M (2004) A novel proteomic screen for peptide-protein interactions. J Biol Chem 279: 10756–10764 [DOI] [PubMed] [Google Scholar]
- Schumacher C, Knudsen BS, Ohuchi T, Di Fiore PP, Glassman RH, Hanafusa H (1995) The SH3 domain of Crk binds specifically to a conserved proline-rich motif in Epsl5 and Epsl5R. J Biol Chem 270: 15341–15347 [DOI] [PubMed] [Google Scholar]
- Smith BL, Chin D, Maltzman W, Crosby K, Hortobagyi GN, Bacus SS (2004) The efficacy of Herceptin therapies is influenced by the expression of other erbB receptors, their ligands and the activation of downstream signalling proteins. Br J Cancer 91: 1190–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songyang Z, Shoelson SE, McGlade J, Olivier P, Pawson T, Bustelo XR, Barbacid M, Sabe H, Hanafusa H, Yi T (1994) Specific motifs recognized by the SH2 domains of Csk, 3BP2, fps/fes, GRB-2, HCP, SHC, Syk, and Vav. Mol Cell Biol 14: 2777–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorkin A, Mazzotti M, Sorkina T, Scotto L, Beguinot L (1996) Epidermal growth factor receptor interaction with clathrin adaptors is mediated by the Tyr974-containing internalization motif. J Biol Chem 271: 13377–13384 [DOI] [PubMed] [Google Scholar]
- Stover DR, Becker M, Liebetanz J, Lydon NB (1995) Src phosphorylation of the epidermal growth factor receptor at novel sites mediates receptor interaction with Src and p85alpha. J Biol Chem 270: 15591–15597 [DOI] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadmodar G, Yang M, Johnston M, Fields S, Rothberg JM (2000) A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403: 623–627 [DOI] [PubMed] [Google Scholar]
- van Delft S, Schumacher C, Hage W, Verkleij AJ, van Bergen en Henegouwen PM (1997) Association and colocalization of Epsl5 with adaptor protein-2 and clathrin. J Cell Biol 136: 811–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mering C, Krause R, Snel B, Cornell M, Oliver SG, Fields S, Bork P (2002) Comparative assessment of large-scale data sets of protein-protein interactions. Nature 417: 339–403 [DOI] [PubMed] [Google Scholar]
- Ward CW, Gough KH, Rashke M, Wan SS, Tribbick G, Wang JX (1996) Systematic mapping of potential binding sites for Shc and Grb2 SH2 domains on insulin receptor substrate-1 and the receptors for insulin, epidermal growth factor, platelet-derived growth factor, and fibroblast growth factor. J Biol Chem 271: 5603–5609 [DOI] [PubMed] [Google Scholar]
- Xia L, Wang L, Chung AS, Ivanov SS, Ling MY, Dragoi AM, Platt A, Gilmer TM, Fu XY, Chin YE (2002) Identification of both positive and negative domains within the epidermal growth factor receptor COOH-terminal region for signal transducer and activator of transcription (STAT) activation. J Biol Chem 277: 30716–33023 [DOI] [PubMed] [Google Scholar]
- Yaffe MB (2002) Phosphotyrosine-binding domains in signal transduction. Nat Rev Mol Cell Biol 3: 177–186 [DOI] [PubMed] [Google Scholar]
- Yan KS, Kuti M, Zuou M-M (2002) PTB or not PTB–that is the question. FEBS Lett 513: 67–70 [DOI] [PubMed] [Google Scholar]
- Yarden Y, Sliwkowski MX (2001) Untangling the ErbB signalling network. Nat Rev Mol Cell Biol 2: 127–137 [DOI] [PubMed] [Google Scholar]
- Zanzoni A, Montecci-Palazzi L, Quondam M, Ausiello G, Helmer-Citterich M, Cesareni G (2002) MINT: a Molecular INTeraction database. FEBS Lett 513: 135–140 [DOI] [PubMed] [Google Scholar]
- Zrihan-Licht S, Deng B, Yarden Y, McShan G, Keydar I, Avraham H (1998) Csk homologous kinase, a novel signaling molecule, directly associates with the activated ErbB2 receptor in breast cancer cells and inhibits their proliferation. J Biol Chem 273: 4065–4072 [DOI] [PubMed] [Google Scholar]
- Zwick E, Hackel PO, Prenzel N, Ullrich A (1999) The EGF receptor as central transducer of heterologous signalling systems. Trends Pharmacol Sci 20: 408–412 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1
Legend to Supplementary Figure 1
Supplementary Figure 1