Abstract
Deciphering regulatory events that drive malignant transformation represents a major challenge for systems biology. Here, we analyzed genome-wide transcription profiling of an in vitro cancerous transformation process. We focused on a cluster of genes whose expression levels increased as a function of p53 and p16INK4A tumor suppressors inactivation. This cluster predominantly consists of cell cycle genes and constitutes a signature of a diversity of cancers. By linking expression profiles of the genes in the cluster with the dynamic behavior of p53 and p16INK4A, we identified a promoter architecture that integrates signals from the two tumor suppressive channels and that maps their activity onto distinct levels of expression of the cell cycle genes, which, in turn, correspond to different cellular proliferation rates. Taking components of the mitotic spindle as an example, we experimentally verified our predictions that p53-mediated transcriptional repression of several of these novel targets is dependent on the activities of p21, NFY, and E2F. Our study demonstrates how a well-controlled transformation process allows linking between gene expression, promoter architecture, and activity of upstream signaling molecules.
Keywords: cancer, cell cycle, networks, p53, promoters
Introduction
Cellular processes are controlled by highly intricate regulatory networks (Tavazoie et al, 1999; Pilpel et al, 2001; Werner, 2001; Ihmels et al, 2002; Lee et al, 2002; Shen-Orr et al, 2002; Bar-Joseph et al, 2003; Segal et al, 2003; Sharan et al, 2003, 2004). Most successes to date in understanding such networks were obtained in lower organisms; extension to mammalian genomes is complicated in part due to the complexity of the promoter and enhancer regions and also due to the tremendous intricacy of some of the regulatory circuits. Nevertheless, initial studies, for example, in fly and in mammalian organisms, succeeded in delineating promoter elements controlling particular networks of genes (Wasserman et al, 2000; Berman et al, 2002, 2004; Halfon et al, 2002; Elkon et al, 2003; Werner et al, 2003; Thompson et al, 2004; Smith et al, 2005; Sumazin et al, 2005; Zhu et al, 2005). Recent studies (Segal et al, 2003) explored an additional level in the signaling network in yeast, namely links between gene expression profiles and activity of signaling molecules. Here too, extension to higher organisms is complicated by the considerable increase in the intricacy of network architecture.
In addition to deciphering normal physiological processes, elucidation of regulatory and signaling networks is expected to allow better understanding of pathological conditions, such as cancer (Segal et al, 2004). Monitoring gene expression changes on a genome-wide scale is a powerful method to study transcriptional programs involved in carcinogenesis (Liotta et al, 2000; Cho et al, 2001; Whitfield et al, 2002). Indeed, specific expression signatures that correlate with specific diagnosis, survival, and response to therapy were proposed (Liotta et al, 2000; Scherf et al, 2000; Rosenwald et al, 2003). Yet, associations of those signatures with specific biological processes or with distinct genetic alterations acquired by cancer cells along in vivo transformations are not obvious. The difficulties largely stem from different genetic backgrounds of patients, variable and uncharacterized mutations in tumors, and the uncontrolled contaminations by inflammatory, endothelial, and stroma cells.
Thus, in order to obtain novel and more reliable insights into genetic networks associated with oncogenesis, we have recently developed an in vitro model for cellular transformation (Milyavsky et al, 2003). The 600-day-long transformation process (Figure 1A) started with normal human fibroblasts that entered replicative senescence after 40 population doublings. In order to overcome replicative senescence, the cells were infected with human telomerase (hTERT), resulting in immortalization, which was then followed by increased proliferation rate. At that stage, cells lost expression of the p16INK4A (p16 for short) and p14ARF tumor suppressors (Milyavsky et al, 2003). To explore the transcriptional and phenotypic impact of p53 at different stages of the transformation process, the p53 protein was inactivated by expression of a dominant-negative p53 peptide (GSE56) (Ossovskaya et al, 1996). These manipulations, in conjunction with H-ras overexpression, gave rise to cells that are capable of forming tumors in nude mice (Milyavsky et al, 2005). Recent studies have described similar inactivation of p16 in additional human cell lines that overcame telomere-independent crises during immortalization (Tsutsui et al, 2002; Taylor et al, 2004). Furthermore, using various experimental models, it was shown that full transformation could be achieved by the combination of viral oncogenes together with cellular genes (Hahn et al, 1999; Voorhoeve et al, 2003). Collectively, these experimental setups generated a model of defined genetic aberrations that initiate and promote the neoplastic process.
Figure 1.
(A) Outline of the malignant transformation process. Schematic representation of the spontaneous (young, senescent, immortal, tumorigenic, INK4A methylation) and induced (hTERT, H-Ras, p53 inactivation) modifications of the WI-38 cells along the process of malignant transformation. The stages chosen for microarray profiling are indicated by boxes with numerals corresponding to columns in the expression matrix shown in (B). The time scale of the process is depicted by a horizontal axis, and the corresponding population doublings are represented by PDLs. (B) The normalized expression levels of the 168 genes in the proliferation cluster at 12 stages spanning the transformation process. Normalized expression level is color-coded according to the color bar on the right. The table below the matrix contains the following information on each sample: days in culture, geometric mean and standard deviation of expression level of the cluster's genes, doubling rate (cell cycle doublings/day) of cells at selected stages, activity of hTERT (designated as ‘+' for all samples following hTERT overexpression), activity of p53, as inferred from the application of its dominant-negative peptide, GSE56 (‘−' indicates expression of GSE56). Here and throughout the paper, the following cell line designations are introduced: cells are either young or senescent; grow slow or fast; a sample name followed by ‘G' denotes the application of GSE56; T before sample names indicates the presence of the immortalizing telomerase; R following the sample name indicates the insertion of Ras.
We have previously suggested that our in vitro cellular system reproduces some of the distinct stages that characterize tumor initiation and progression (Milyavsky et al, 2005). In the present study, we aimed at deciphering the transcriptional networks associated with malignant transformation. We utilized genome-wide mRNA expression profiling of the transformation process using the Human Genome Focus Array (Affymetrix, Santa Clara, CA) with 8500 verified human genes (Milyavsky et al, 2005). Subsequent cluster analysis of the expression profiles identified 10 stable clusters. One of them, the ‘proliferation cluster' (Milyavsky et al, 2005), showed a pronounced sensitivity to the status of p53 and p16 tumor suppressors. A large number of the genes found in this proliferation cluster also clustered in studies that analyzed human primary tumor samples (Alizadeh et al, 2000; Perou et al, 2000; Barrett et al, 2003; Rosty et al, 2005). In addition, the proliferation cluster genes were found to be significantly more highly expressed in tumors obtained from patients with bad outcome compared to patients with good outcome in breast cancer (Dai et al, 2005; Milyavsky et al, 2005). These findings strongly support the notion that the proliferation cluster genes are highly relevant to naturally occurring cancers.
Here, we revealed how the promoters of the cluster's genes generate a transcriptional program that integrates the activity of tumor suppressors. By linking expression profiles of the genes in the cluster with the dynamics of p53 and p16, we identified two promoter architectures that integrate different signals from the two tumor suppressive channels and that map their activity onto distinct levels of expression of the cell cycle genes, which, in turn, correspond to different cellular proliferation rates.
Results
The ‘proliferation cluster'
Our expression cluster analysis during the transformation process identified 10 stable clusters (Milyavsky et al, 2005). According to the superparamagnetic clustering method (Blatt et al, 1996), a stable cluster is one that is robust against perturbing the data; on the one hand, the points that belong to it are (relatively) remote from other points and, on the other hand, they constitute a well-defined entity, that is, a (relatively) contiguous region of high density. The algorithm is capable of identifying such clusters irrespective of their shape. It also provides a quantitative measure of the stability of clusters.
Here we focus on one of these clusters, termed the ‘proliferation cluster' (due to its genes' annotation; see below). The cluster has a somewhat elongated shape, yet it is stable and cannot be naturally divided into subclusters (Supplementary Figure S1). We decided to focus on this cluster since the genes that constitute it showed a complex behavior—pronounced sensitivity to the status of p53 and p16 tumor suppressors.
Figure 1B shows expression profiles of the 168 genes of the proliferation cluster along with the cells' doubling rates and telomerase and p53 activity. Four major states are distinguished. (1) The ‘young-cell period' (first and second columns in the matrix) spans the first cell cycles. During this period, the cells are young and the expression profiles of the genes in the cluster are relatively high. At the second point during the young-cell period, a separate cell culture was derived from the above culture, which was treated by introduction of the p53 dominant-negative peptide, GSE56. The cluster genes responded by upregulation (mean expression was significantly elevated from 406.5 to 479.5, P-value=3.1 × 10−23 by paired t-test). (2) The senescence period (third and fourth columns) is characterized by arrest of cell divisions. The expression level of the genes in the cluster was dramatically decreased during this period. Yet, even within this period, expression profiles of the genes in the cluster were significantly elevated at the fourth time point compared to the third point, that is, in response to p53 inhibition (P-value=7.1 × 10−28 by paired t-test). (3) The ‘slow immortalization' period (5–7th columns) is characterized by expression levels similar to those in young cells. (4) The ‘fast immortalization' stage (8–12th columns), associated with silencing of p16 (Milyavsky et al, 2005), is characterized by a significant shortening of the cell cycle period (see table below expression matrix), accompanied by further increase in the expression levels of the cluster genes. At the last two points of this period, GSE56 was reintroduced, and the genes in the cluster responded by significant (P-value=1.4 × 10−32) upregulation. Thus, the expression profiles of the genes in the proliferation cluster correlate with p53 and p16 as well as with the rate of cell proliferation.
We next examined functional annotations of the genes in the cluster, using ‘Gene Ontology' (Ashburner et al, 2000). Notably, only cell cycle-related functions are significantly over-represented (Dennis et al, 2003) in the cluster (see Supplementary Table S1). We thus termed the cluster ‘the proliferation cluster'. The genes in the cluster relate to different cell cycle phases, such as DNA replication (MCM2, MCM3, MCM5, MCM6, RRM1–2, RFC3–5, GMNN, POLA, POLD1, POLE, POLQ, PRIM1) and DNA repair (BLM, BRCA1, MSH6). G2/M phase genes represented the largest functional category. Cyclin-dependent kinase CDC2, whose function is critical for mitotic entry, and its regulators such as cyclin B2, CDC25A, and CDC25C are included, in addition to genes involved in mitotic spindle organization (CENPA, CENPF, TTK, BIRC5, kinesins), spindle checkpoint (BUB1, BUB1B, MAD2L1, CDC20), chromosome segregation (PTTG1, CENPF, ESPL1, UBE2C, PLK1, STK12), DNA packaging (HAT1, CHC1, SUV39H1, TOP2A), and chromosome organization (H1FX). Reassuringly, we also found that >50% of the genes in the cluster display high cell cycle periodicity, especially peaking at the entry into the S and M phases (Supplementary Figure S2) during HeLa cells divisions (Whitfield et al, 2002).
In addition, we note that expression of these genes correlates with poor outcome and prognosis in patients samples (Milyavsky et al, 2005), attesting to their relevance to naturally occurring cancers.
Transcriptional regulation of the proliferation cluster genes
We next turned to identify promoter regulatory motifs that drive the proliferation cluster. Rather than attempting to discover de novo promoter motifs, we assumed that transcription factors with known binding sites may be involved in regulating the genes in the cluster. Therefore, we searched within the promoters of the proliferation cluster genes for the presence of each of the 326 known vertebrate position-specific scoring matrices (PSSMs) from MatInspector (Quandt et al, 1995), using a published gene-to-binding site assignment algorithm (Elkon et al, 2003). For each PSSM, we calculated a hypergeometric P-value score (Hughes et al, 2000) to assess the extent to which it is over-represented among the cluster's genes. Noticeably, apart from VMYB, all significant motifs (Table I and Supplementary Table S5) that passed a Bonferroni test (including NFY, E2F, CHR (Cell cycle genes Homology Region), ELK1 and CDE (Cell cycle-Dependent Element)) are known to be involved in the regulation of cell cycle (Mantovani, 1998; Badie et al, 2000; Manni et al, 2001; Nevins, 2001; Matuoka et al, 2002; Bracken et al, 2004; Buchwalter et al, 2004). We therefore focused on these cell cycle motifs in all subsequent analyses. We have also examined the presence of the motifs in the 5′ UTRs of the cluster's genes and found only barely significant over-representations in the cluster (see Supplementary Table S2), and have thus decided to concentrate only on the upstream regions in all further analyses.
Table 1.
Over-represented regulatory motifs in the proliferation cluster
| Motif | Number of genes containing motif among the proliferation cluster/entire array | Over-representation P-value |
|---|---|---|
| NFY | 77/1574 | 9.92E−12 |
| E2F | 85/2617 | 3.57E−07 |
| CDE | 101/3073 | 2.20E−09 |
| ELK1 | 38/960 | 4.32E−05 |
| CHR | 10/63 | 5.43E−07 |
| CHR-NFY-CDE | 9/12 | 1.18E−13 |
| The number of genes that contain the corresponding motif in their promoter among the proliferation cluster (out of 165 genes) and among the entire array (out of 8110 genes). A hypergeometric P-value score was calculated in order to assess the extent to which a motif is over-represented in significant location among the cluster's genes compared to the rest of the genes on the array. | ||
Evolutionary conservation of the motifs
We examined promoters of mouse genes orthologous to the proliferation cluster genes and found that the same motifs are also significantly over-represented in these promoters compared to the promoters in the rest of the mouse genome (Supplementary Table S3). We have further assessed conservation at an organization level beyond the mere presence/absence of motif, namely conservation of the motif architecture between the two species. We found considerable conservation at this level too, using two criteria: first, the combinations of motifs that regulate orthologous promoters were significantly more similar to each other compared to combinations of non-orthologs (Supplementary Figure S3A). Second, we found a significant tendency to preserve the locations of the motifs relative to the transcription start site (TSS) in orthologous promoters (Supplementary Figure S3B–F). The high level of conservation observed attests to the functional role of the motifs in these promoters.
Revealing a hierarchy of regulatory motif combinations
For each motif, we identified a sequence window relative to TSS in which it is over-represented (Figure 2; see Materials and methods). Here and in subsequent analysis, a motif was considered present in a promoter if it appeared in its preferred location. We next turned to identify combinations of the motifs using ‘synergy' (Pilpel et al, 2001; Banerjee et al, 2003; Garten et al, 2005) and rate of cooccurrence (Sudarsanam et al, 2002; Elkon et al, 2003). A pair of motifs was considered ‘synergistic' if the extent of expression coherence (Pilpel et al, 2001; Lapidot et al, 2003) of genes containing both motifs in their promoters is significantly greater than that of genes containing either of the motifs alone. A pair of motifs was considered highly cooccurring if there is a significant overlap (using hypergeometric) between the set of genes containing the two motifs, given the number of genes containing each motif alone. We found that NFY, E2F, CDE, and CHR show multiple interactions with each other, many of which were supported by both synergy and cooccurrence. On the other hand, the ELK1 motif cooccurs significantly with E2F and CDE, and it has synergistic effect only E2F (Figure 3A). The above interactions were derived by considering all the genes represented on the array. Yet many of the interactions are observed also when only genes in the proliferation cluster are considered (Figure 3).
Figure 2.
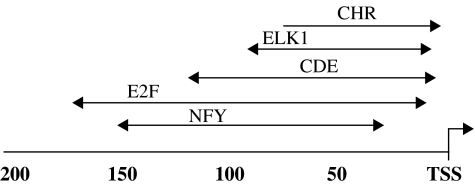
Motif positional bias in promoters of the proliferation cluster genes. The preferred window position of the five regulatory motifs, NFY.01, CDE, CHR, ELK1, and E2F.02, in the promoters of the proliferation cluster genes is shown. The CHR motif has also a clear strand bias (depicted by a directional arrow); for more details, see Supplementary Figure S6.
Figure 3.
(A) Graph depicting interaction between the five regulatory motifs measured by synergy and cooccurrence. A pair of motifs connected by a thick red line has a synergistic effect on expression and pairs connected by a thin blue line are significantly more highly cooccurring in individual promoters. These interactions were computed using all genes on the array. In addition, we calculated cooccurrence interaction given only the genes in the cluster (i.e. considering the total number of genes in the cluster and the number of cluster's genes containing each motif as a background), and found even there several significant interactions (blue dotted lines). Since for some of the motifs, the motif database contains multiple variants, we unified all variants of the same motif into one node, and an edge in the graph connects two motifs if at least one of the variants of the two motifs is either synergistic or highly cooccurring. (B, C) Combinogram analyses (Pilpel et al, 2001) using the five regulatory motifs during the transformation process (B) and HeLa cell cycle (C). All the varying genes in the respective arrays were used in the analysis. The upper and middle parts of the Combinogram are depicted as before (Pilpel et al, 2001), while the lower part is modified. Briefly, the middle section of the Combinogram depicts the motif composition of each gene set defined by motif combination (GMC; see Materials and methods). Each vertical column represents a single GMC. A black square indicates that the particular motif is present in the promoters of all the genes in that GMC. A white square indicates that none of the genes in the GMC contain the particular motif. The top section of the graph shows the dendrogram analysis that assesses the similarity in expression profiles of each GMC using normalized Euclidean distances between the average expression profiles of the genes in the GMC as a measure of distance. The lower part of this modified Combinogram displays the mean expression profiles of the genes in each GMC; color-coded as in Figure 1. The numbers of genes in each GMC appear below the dendrogram. The three main branch points in the dendrogram in (B) are represented as 1–3 (in circles). In the present analysis, only genes that contain the motifs in their preferred distance relative to TSS (as depicted in Figure 2) were considered, as genes that contain the motifs, yet away from the preferred location, display no clear expression patterns (not shown). Since CHR motif has a strong strand bias in addition, and since genes that contain this motif yet on the nonpreferred strand have a non-coherent expression profile (Supplementary Figure S6), for this motif we considered only genes that contain it in the preferred strand and distance from the TSS.
In order to gain more insight into such motif interactions, we used the Combinogram analysis (Pilpel et al, 2001). We searched for the above five regulatory motifs within the promoters of all varying genes represented on the array. We partitioned the array genes into up to 25=32 gene sets, each defined by a unique binary signature that reflects the presence or absence of each of the five motifs in their promoters, and grouped together genes with identical binary signatures. The Combinogram depicts the motif content of each gene set (binary black and white matrix), the similarity between the average expression profiles of all pairs of gene sets (upper part of the dendrogram), and the averaged expression profiles of the genes in each set (expression matrix at the bottom part). The Combinogram in Figure 3B, which was obtained with 18 gene sets that were populated with genes (14 other potential sets were not populated with genes), reveals several clear trends. First, although the analysis was applied to all the genes represented on the array, that is, without a preceding clustering stage, a division (corresponding to the main branching point of the dendrogram, marked ‘1') into genes that contain some of the motifs, and genes that do not, appears. Gene sets that are to the left of branch point ‘1' largely represent the proliferation cluster signature, whereas gene sets that are on the right branch display no particular trend. The motifs that appear to determine this split are mainly ELK1, or NFY and CHR. Genes that contain none of the five motifs (column #14 from left) display the flat profile, as do genes that contain one or two of the motifs, but not ELK1 (columns 11–18 from left). The left branch, which largely shows the proliferation signature, may be further divided (branch point ‘2') into gene sets that contain at least ELK1 (columns 1–8) versus gene sets that contain NFY and CHR, but not ELK1. These differences in motif composition reflect themselves at the level of expression pattern—without ELK1, a clear decrease in expression in the senescence (40) and senescence GSE (4) time points (third and fourth rows of the expression matrix) is seen, whereas an increase in expression in the last two time points is evident too. The presence of ELK1 appears to be both necessary and sufficient for its typical dictated expression pattern, as genes that contain only ELK1 (column 5) are members of the ELK1 cluster and genes that do not contain ELK1 (columns 9–18) are not in the cluster. Branch point ‘3' further sharpens the ELK1-dictated signal. Genes that contain ELK1 and NFY (columns 6–8) are located to the right of branch point ‘3', as they display an intermediate between the pure ELK1 pattern and the NFY and CHR pattern, while genes to the left of this branch point display a distinct pattern. In general, this analysis shows a striking correspondence between motif content and gross and fine differences in expression, akin to a previous, yet simpler, observation made in yeast (Pilpel et al, 2001). It allows the dissection of the role of individual motifs and their combinations (e.g. the effect of CHR on the background of NFY and CDE is clear from the difference in expression patterns between columns 10 and 11 in which CHR is present and absent, respectively). This analysis strongly suggests that indeed the five regulatory motifs examined here govern gene expression profiles during the transformation process and that the proliferation signature represents a response genuinely regulated by these motifs.
Next, we examined whether it is possible to trace the regulatory effect of these motifs down to the relatively microscopic time scale of single cell cycle divisions. We tested whether the motif architecture we discovered here also governs the expression of these genes during cell cycle divisions. To this end, we constructed a Combinogram based on the five motifs together with cell cycle expression data derived from the HeLa cell cycle experiment described above (Whitfield et al, 2002; Figure 3C). Notably, we observed similar relationships between motif combinations and their effects on expression in both the transformation and the cell cycle experiments. In both experiments, NFY appears to interact synergistically with CHR, an interaction that gives rise to a clear G2–M phase expression pattern in cell cycle. Here, the presence of the CDE motif further amplifies this pattern. The resemblance between the transformation and the cell cycle experiments indicates that the transcriptional regulation of the cluster during the complex and largely uncharacterized 600-day-long transformation process can be reduced to the more ‘atomistic' level of the regulation of cell cycle. Interestingly though, in the cell cycle experiment, we did not detect any clear pattern dictated by E2F, alone or through combinations with other motifs, suggesting either that despite intensive research on this transcription factor, an accurate description of its binding site is still missing, or that its regulatory role is too complex and diverse (Bracken et al, 2004). This too is consistent with a recent observation (Elkon et al, 2003) that although E2F is over-represented in the promoters of cell cycle genes, it is not restricted to genes that peak at a specific cell cycle phase. Likewise, the ELK1 motif does not seem to affect gene expression throughout the cell cycle. The observation that the E2F and ELK1 motifs affect transcription mainly in the transformation experiment and less so in cell cycle may indicate that their role in the transformation process is not mediated through a direct effect on the cell cycle, but rather on a potentially higher level. Another potential explanation for the fact that presence of either ELK1 motif or the combination NFY and E2F was significant in the transformation experiment but not in HeLa cell cycle experiment may be related to the fact that in HeLa cells, both p53 and pRb are inactivated by the viral oncoproteins E6 and E7. Thus, if the ELK1 motif or the combination of NFY and E2F potentially mediates growth restrictive effects of these major tumor suppressive pathways, we would not expect these effects to be manifested in HeLa cells.
The proliferation cluster genes integrate information from two tumor suppressive channels
Our knowledge of the detailed molecular history of the transformation process in the experiment allowed us to extend our analysis beyond the formation of links between regulatory motifs and expression profiles. Since the activity of upstream tumor suppressors was manipulated and monitored during the transformation, we could link gene expression patterns, mediated by various regulatory motifs, to activity of tumor suppressors. In particular, we followed the activity levels of two prime tumor suppressor genes, p53 and p16, that varied throughout the transformation process. Since p53 was inactivated at the protein level, we used as a surrogate for its activity mRNA levels of its regulated target, p21, which is indeed downregulated in response to p53 inactivation (Figure 4A).
Figure 4.
(A) Normalized expression profiles of the tumor suppressors p21 (brown line) and p16 (black line), along with a profile that represents the average of their profiles (green area) and a profile that represents the mean expression profiles of the genes in the proliferation cluster (blue area). (B) Average expression profiles of all genes in the genome that contain Elk1 in their promoters (left) and the NFY and CHR motifs (right). Each bar represents an average over the samples that corresponds to low expression (<−0.3) denoted by ‘L' and high expression (>0.2) denoted by ‘H' of the normalized mRNA expression levels of p21 and p16.
First, we observed that while the averaged mRNA profiles of the genes in the cluster do not correlate with the mRNA levels of either p21 or p16 alone, they show high negative correlation (r=−0.85) with a profile obtained by summing the mRNA expression profiles of these two genes (Figure 4A). This is a significant correlation, as the probability of pairs of random genes that are summed up to obtain such correlation or lower with the proliferation cluster's average is <1% as estimated by 10 000 random samples. Simple logical functions such as AND or OR gates, applied to the combination of p21 and p16, were unable to describe the activity levels of the genes in the cluster. The promoters of these genes thus appear to sum up the expression levels of the two tumor suppressors and produce a corresponding output in the form of expression profiles. Recently, similar ‘sum-gated' designs were shown in Escherichia coli (Setty et al, 2003; Kalir et al, 2004).
More importantly, we identified the promoter motifs that likely mediate such integrative function. We analyzed separately all the genes represented on the array that contain in their promoters the ELK1 motif, and all genes that contain a combination of NFY and CHR. Figure 4B shows that genes that are regulated by CHR and NFY clearly depend on both tumor suppressors, and their expression levels map the presence/absence of the two suppressors onto four distinct expression levels (multiple t-tests on all six pairwise comparisons yielded a significant P-value of 0.14 × 10−6 or lower). In contrast, the ELK1 motif mainly mediates a response to the presence/absence of p16. The expression of ELK1-regulated genes is significantly upregulated following p16 inactivation. Although the ELK1 transcription factor was previously implicated in the regulation of expression of proliferation genes (Ullrich et al, 1990; Gille et al, 1995), its potential regulatory interaction with either p16 or p53 was not addressed before.
Three-way linkage of expression, promoter architecture, and tumor suppressor activity
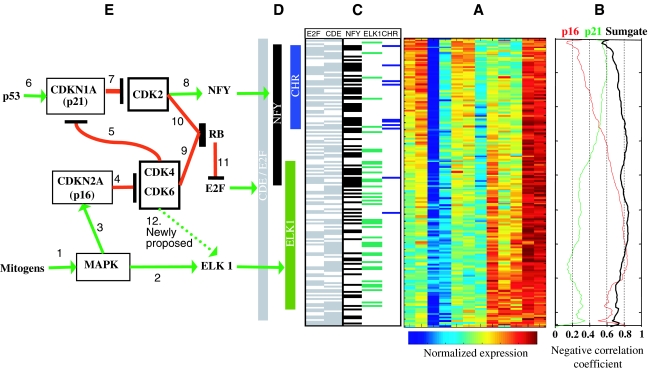
In order to gain further insights into the relationship between mRNA expression profiles and promoter architecture, we sorted the proliferation cluster genes using SPIN (Tsafrir et al, 2005), a sorting algorithm that positions genes with similar expression profiles in adjacent rows of an expression matrix (Figure 5A). We also examined the presence of the regulatory motifs in promoters of genes in the cluster along the sorted expression matrix (Figure 5C and D). Interestingly, the CDE and E2F motifs are relatively evenly scattered along the cluster. This, together with the observation that they are significantly highly specific to the proliferation cluster suggests that these motifs are major characteristics of the cluster. On the other hand, the CHR motif and, to a smaller extent, the NFY motif are mainly concentrated in promoters of genes in the ‘upper' part of the sorted cluster, while ELK1 mainly shows a preference toward the genes in the ‘lower' part. We thus conclude that while the presence of the CDE and E2F motifs defines the cluster and are present in the majority of its genes, CHR, NFY, and ELK1 serve to modulate the general expression patterns dictated by CDE and E2F.
Figure 5.
Three-way linkage between expression profiles, promoter architecture, and tumor suppressor pathways. (A) The entire expression matrix of the proliferation cluster genes, sorted with SPIN (Tsafrir et al, 2005), revealing an ‘elongated' shape for this cluster. (B) Correlation coefficient between p21 and p16 expression profiles to each of the genes in the cluster; color-coded according to color bar shown. (C) Cumulative distribution of CHR, ELK1, and NFY along the sorted list of genes in (A). (D) Bars depicting main areas of density of regulatory motifs along the sorted expression matrix in (A) are based on cumulative appearance in (C). (E) Arrows in the networks represent positive (green) or negative (red) interactions. Reviewed: 1—Ullrich et al (1990); 2—Gille et al (1995); 3—Serrano et al (1997), Lin et al (1998), Zhu et al (1998); 4, 5, 7, 9–11—Weinberg (1995), Sherr (1996), Sherr et al (1999), Hahn et al (2002); 6—el-Deiry et al (1993); 8—Yun et al (2003); 12—newly proposed interaction (this paper).
Although the averaged expression profile of the cluster genes is strongly negatively correlated with the summed expression profiles of p16 and p21, and not with the individual tumor suppressors (Figure 4A), it is still possible that part of the genes correlate only with p21 while others correlate only with p16. We have thus measured (Figure 5B) for each gene in the (sorted) cluster the correlation of its mRNA expression profile during the transformation process with the expression profiles of p16 and p21 and with the summed profiles of p21 and p16 (Figure 5B). Strikingly, for the majority of the genes in the cluster, the negative correlation with the summed profile is stronger than with the individual tumor suppressors. This is predominantly true for the genes in the ‘upper' part of the sorted cluster. Although for these genes, there exists also a negative correlation with p21 alone, they are more strongly (negatively) correlated with the sum-gate, suggesting that the motifs regulating these genes are indeed integrating, by summing up, the information from the two suppressor channels. The correlation with p21 gradually diminishes with genes that are located toward the ‘lower' part of the cluster, and on the other hand the correlations with p16 level show the opposite trend, namely a high correlation with genes in the lower part of the cluster. We stress that the data suggest no obvious point where the cluster can be subdivided into two clusters based on correlations with the two tumor suppressors.
It was found recently that DNA-binding activity of NFY transcription factor is positively regulated by CDK2 phosphorylation. This may explain the higher sensitivity of NFY-containing genes to p21 level, as it specifically inhibits CDK2 (Weinberg, 1995; Sherr, 1996; Sherr et al, 1999; Hahn et al, 2002). On the other hand, p16 specifically inhibits CDK4 and CDK6 (Weinberg, 1995; Sherr, 1996; Sherr et al, 1999; Hahn et al, 2002). Thus, the increased sensitivity of ELK1-containing promoters to p16 levels enables us to propose novel role for CDK4/6 in ELK1 regulation.
The integration of these findings together with published experimental data allowed us to propose a network linking three layers of data—mRNA expression, promoter regulatory motifs/transcription factors, and the upstream tumor suppressors and signaling molecules (Figure 5E). It is entirely possible though that additional tumor suppressors and transcription factors participate in the network, and future analysis may reveal their identity and role.
Experimental validation of computational predictions
Our data suggested that the proliferation cluster genes are subject to p53- and p16-mediated transcriptional repression. Notably, many cluster genes including TOP2A, CCNB2, CCNA2, BIRC5, CDC2, CDC25C, PRC1, POLD1, PLK, and others were previously shown to be downregulated by p53 (Yamamoto et al, 1994; Wang et al, 1997; Yun et al, 1999; Krause et al, 2000; Hoffman et al, 2001; Tang et al, 2001; Manni et al, 2001; Burns et al, 2003; Li et al, 2004; St Clair et al, 2004), validating our analysis and enabling us to propose numerous novel p53 transrepression targets. Interestingly, multiple components of the kinetochore complex and most of the known spindle checkpoint genes are found in our proliferation cluster. Since p53 was not previously implicated in the regulation of this group of genes, we decided to test for p53-mediated transcriptional repression of several genes from this category. Importantly, the regulatory network we proposed, based on the microarray experiment conducted under basal unstressed conditions, is expected to hold for cases where the upstream tumor suppressors are induced either by forced overexpression or by stress. We therefore tested whether a stress-induced p53 will repress the expression of several kinetochore/spindle genes. To this end, we treated normal and GSE56-expressing WI-38 cells with doxorubicin, a DNA-damaging agent and a potent p53 activator, and measured the levels of several proliferation cluster-derived genes by quantitative real-time PCR (qPCR). Confirming our hypothesis, we found that following DNA damage, Cdc20, Bub1, CCNF, and Mad2L1, all of which are cluster members, were downregulated in normal WI-38 cells, but not in their isogenic counterparts, in which p53 was inactivated (Figure 6). Thus, these kinetochore- and spindle checkpoint-related genes represent novel targets of p53-mediated transcriptional repression.
Figure 6.
p53 represses proliferation cluster genes expression following DNA damage. Normal and GSE56-expressing WI-38 cells were treated with 0.2 μg/ml doxorubicin (dox) for 48 h. mRNA levels for the indicated genes (y-axis) were measured by qPCR and normalized to the GAPDH housekeeping control.
Since our computational analysis revealed that the proliferation cluster genes display a negative correlation with p21 mRNA profile, we tested whether p53 exerts repression of proliferation cluster genes via p21 induction. To this end, we treated the HCT-116 colon carcinoma cells and their p53-null and p21-null derivatives with doxorubicin. We measured the expression levels of several proliferation cluster genes by qPCR and calculated the fold repression for each gene as the ratio of expression level in nontreated cells to that in doxorubicin-treated cells (Table II). Notably, only cells that contained both functional p53 and p21 (HCT-116 p53+/+) displayed downregulation of these genes following DNA damage. This supports the notion that the proliferation cluster genes are transcriptionally repressed by p53, and suggests that this repression is mediated through p21.
Table 2.
p53- and p21-dependent repression of the proliferation cluster genes expression
| Fold repression | |||
|---|---|---|---|
| Gene symbol | HCT116 p53+/+ | HCT116 p53−/− | HCT116 p21−/− |
| Cdc20 | 2.6 | 1.2 | 0.7 |
| BIRC5 | 1.6 | 0.6 | 0.7 |
| NEK2 | 1.8 | 0.5 | 0.9 |
| Mad2L1 | 2.4 | 0.7 | 0.8 |
| Bub1B | 1.7 | 0.8 | 1.1 |
| PRC1 | 1.7 | 0.9 | 0.9 |
| CENPF | 1.7 | 0.7 | 1.2 |
| HCT-116 cells containing wild-type p53 and their p53-null and p21-null derivatives were treated with 0.2 μg/ml doxorubicin for 48 h. The expression level for each gene was measured by qPCR and normalized to GAPDH expression level. The numbers in the table represent fold repression, calculated for each gene as the ratio of expression level in nontreated cells to doxorubicin-treated cells. | |||
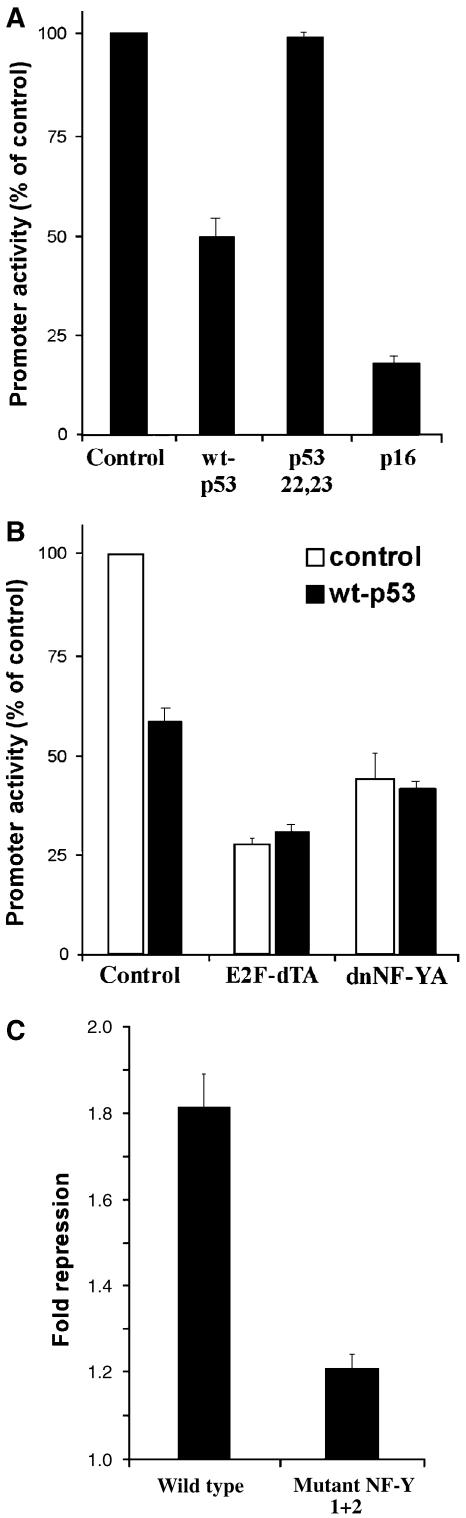
In order to gain further insights into the mechanism of p53-dependent repression of the proliferation cluster genes, we decided to focus our efforts on the cdc20 gene as a representative member of the cluster. We cloned the cdc20 promoter into a luciferase reporter vector and transfected it into HCT-116 p53−/− cells with or without a p53 expression plasmid. As indicated in Figure 7A, in the presence of wild-type p53, the activity of cdc20 promoter was significantly repressed. In contrast, a p53 mutant lacking a functional transactivation domain (L22Q/W23S) did not repress cdc20. The requirement for a functional transactivation domain supports our conclusion that cdc20 repression by p53 is indirect and is mediated by induction of a mediator gene. Cotransfection of cdc20 promoter reporter with the p16 expression vector also resulted in repression of promoter activity (Figure 7A), supporting our prediction that the proliferation cluster genes integrate signals from both p53 and p16.
Figure 7.
(A) The cdc20 promoter is repressed by wild-type p53 and by p16, but not by a transactivation-deficient p53 mutant. Normalized luciferase activity of the p-cdc20-luc reporter in HCT-116 p53−/− cells is shown. cdc20 promoter activity in the absence (control) or presence of either wild-type p53 (wt-p53), the L22Q/W23S p53 mutant (p53 22,23), or p16. (B) cdc20 promoter is regulated by E2F and NF-Y. Normalized luciferase activity of p-cdc20-luc reporter in HCT-116 p53−/− cells. cdc20 promoter activity in the absence or presence of wild-type p53 and in the presence of either control vector (control), dominant-negative E2F1 (E2F-dTA), or dominant-negative NF-YA (dnNF-YA). Fold repression was calculated as the ratio of promoter activity in the absence of wt-p53 to the level in its presence, and was significantly lower in the presence of dominant-negative E2F1 and dominant-negative NF-YA compared to control (paired t-test P-values=0.03 and 0.01, respectively). (C) NF-Y motifs are important for p53-mediated repression of the cdc20 promoter. The wild-type promoter construct p-cdc20-luc and a construct with both NF-Y motifs mutated (mutant NF-Y 1+2) were cotransfected into HCT116 p53+/+ cells in the presence or absence of a dominant-negative p53 (p53-DD). Fold repression was calculated as the ratio of promoter activity in the presence of p53-DD to control and was significantly lower for the mutant NF-Y 1+2 construct (P-value=0.005). Data represent the average of three independent experiments, each performed in triplicate.
Since promoters of the proliferation cluster genes are highly enriched with E2F motifs, we tested whether cdc20 promoter activity is affected by the presence of an E2F1 dominant-negative protein (E2F-dTA) that is capable of DNA binding but defective in its transactivation and RB-binding domain. Overexpression of this construct displaces the endogenous E2F proteins from the DNA, abolishing both activation and repression by E2F family members (Hofmann et al, 1996). As demonstrated in Figure 7B, cdc20 promoter activity decreased in the presence of a dominant-negative E2F1, and p53 did not repress it further under those conditions (see figure legend for details). The most significantly enriched motif in the proliferation cluster promoters is NF-Y, suggesting the involvement of its cognate transcription factor in the regulation of the cluster's genes. To validate this hypothesis, we tested whether cdc20 reporter activity is affected by the presence of an NF-Y dominant-negative protein (Mantovani et al, 1994). Indeed, overexpression of a dominant-negative NF-YA (dnNF-YA) resulted in reduction of cdc20 promoter activity and in strong attenuation of the p53-dependent repression of this promoter. These results indicate that both the E2F family of transcription factors and the NF-Y transcription factor participate in cdc20 regulation, and that p53-dependent repression of cdc20 is mediated through these proteins.
Finally, we addressed the significance of the NF-Y motifs found in the cdc20 promoter for p53-mediated repression. Two NF-Y motifs reside in cdc20 promoter within the first 100 bp relative to the TSS. We generated cdc20 promoter reporter constructs that harbor mutations in each of the motifs and an additional construct with both NF-Y motifs mutated. These constructs, together with the wild-type promoter, were tested for their responsiveness to p53 status by cotransfecting them into HCT-116 p53+/+ cells in the presence or absence of a dominant-negative p53. While mutation of each NF-Y site alone did not affect p53-mediated repression (data not shown), mutations in both NF-Y motifs resulted in significant attenuation of the repression (Figure 7C). These results support our computational prediction that NF-Y motifs, enriched in the promoters of the cluster genes, are involved in the regulation of these genes by p53.
Discussion
This study describes the analysis of genome-wide expression profiles of an in vitro transformation process. Focusing on a well-defined expression cluster that consists predominantly of core cell cycle genes, we identified promoter motifs and their combinations that regulate the transformation process. We suggest that at least part of such regulation can be explained by a direct effect on cell cycle progression. Working with a controlled transformation process allowed us, for the first time, to not only establish a connection between gene expression and promoter architecture, but also to identify links to the activity of upstream tumor suppressors. Such a three-way connection was most revealing, as it identified promoter motifs that likely ‘count' the number of active tumor suppressive channels and map them onto distinct expression states. Finally, detailed experimental analyses of selected genes experimentally established many of the suggested components of the network.
The two tumor suppressors studied here, namely p53 and p16, mainly respond on cell intrinsic and environmental signals, respectively. Thus, the promoter architecture discovered in this study integrates internal and external signals that affect core cell cycle genes. Such integration is performed by summing up activity from the two suppressive channels and mapping the result onto distinct expression levels. The intermediate expression level states, which correspond to precisely one active suppressive channel, may represent an ‘undecided' state. Such a state might be followed by either high or low expression states of the cell cycle genes that may ensue after inactivation or activation of the second channel, respectively. Residing in such intermediate state can facilitate more rapid transition to one of the two extreme stages in response to addition or removal of intrinsic or environmental suppressive signal. In this respect, it is crucial to note that the expression levels of the cluster's genes are correlated with proliferation rate (Figure 1B); thus, promoter tuning of transcription of at least some of the genes may affect proliferation.
It is well known that activation of p53 leads to induction of p21 and inhibition of CDK2 activity (Weinberg, 1995; Sherr, 1996; Sherr et al, 1999; Hahn et al, 2002). As depicted in Figure 5E, CDK2 controls E2F indirectly (through inactivation of RB by phosphorylation) and NFY directly (through CDK2-mediated phosphorylation). The CHR (cell cycle genes homology region) and the CDE (cell cycle-dependent element) represent ‘orphan' binding sites, as the factors that bind these motifs are still uncloned (Zwicker et al, 1995). These two elements are often found in close proximity to each other and these CDE/CHR ‘tandems' were shown to be important for the cell cycle-dependent expression of many genes. However, not always these two sites appear together. For example, a single CHR was shown to control cell cycle-dependent transcription of the cdc25C phosphatase gene and to cooperate with E2F or Sp1/3 sites (Haugwitz et al, 2002). Our genome-wide analysis strongly suggests the existence of a novel strong functional cooperation between CHR and NFY elements. According to our Combinogram analysis, the presence of these two sites in the proximal 200 bp upstream of the TSS is sufficient to dictate a G2/M expression pattern of multiple genes (Figure 3C). In addition, we discover here that the same promoter architecture, namely the combination of NFY and CHR, is responsible for the integration of inputs from p21 and p16 during the in vitro transformation process.
ELK1 transcription factor is a known downstream target of the MAP kinase pathway. It was demonstrated that proliferative inputs from deregulated MAP kinase pathway are counteracted by a negative feedback loop involving p16 activation with subsequent inhibition of CDK4/6 activities (Serrano et al, 1997; Lin et al, 1998; Zhu et al, 1998). Interestingly, our results indicate a strong negative correlation between the activities of ELK1-containing promoters and the expression level of p16, suggesting a possibility that p16 inhibits the activity of ELK1. To the best of our knowledge, this relationship was not reported previously. Since p16 specifically inhibits CDK4 and CDK6, it is possible that phosphorylation by these kinases plays a role in ELK1 regulation.
Many genes in the proliferation cluster represent previously identified targets of p53-mediated transcriptional repression. Our results significantly broaden the list of potential p53 transrepression targets. Here, for example, we identified an entire set of kinetochore/spindle genes, the expression of which is negatively regulated by p53. The functional significance of this finding is still unclear but it is tempting to speculate that loss of this transcriptional control contributes to aneuploidy formation, which is frequently found in tumors with mutated p53.
An additional important conclusion of our study relates to the mechanism of p53-mediated transcriptional repression. Unlike transactivation by p53, which clearly requires p53 binding to the regulatory sequences of targets, the mechanisms of repression by p53 are less well understood. The promoters of repressed genes usually do not contain p53-binding sites. Various mechanisms of p53 transrepression were proposed (for review, see Ho et al, 2003). In addition, it was recently demonstrated for several genes that p53-mediated transcriptional repression requires the induction of p21 (Lohr et al, 2003). Our study addresses systematically this point using the three-way linkage of expression, promoter architecture, and tumor suppressor activity. We found that transcriptional repression by p53 is in most cases indirect, mediated by p21 induction. The signal is then transduced to E2F/CDE, NF-Y, and CHR motifs in the promoters of target genes.
Finally, it is crucial to note that the proliferation signature has clear relationship with naturally occurring tumors. Rosty et al (2005) have identified a cluster of genes whose expression levels were predictive of outcome in samples derived from human patients with cervical cancer; low levels of expression characterized a subset of the patients with good outcome. In our previous work (Milyavsky et al, 2005), we have shown that there is a significant overlap between our proliferation cluster and that reported by Rosty et al, and we mentioned there other indications of additional proliferation cluster genes that constitute good predictors of relapse. We are aware, however, of the fact that such common features should be carefully evaluated using additional natural malignancies. In addition, in the future, similar transformation processes, performed with additional cell lines, may be important for further establishing the generality of the signatures derived here. In this respect, we note that in our previous work (Milyavsky et al, 2005) we addressed this issue by monitoring similar molecular events, such as INK4A locus inactivation in two additional cultures, supporting the generality of our findings.
Materials and methods
Promoter sequence
DNA sequences upstream of human ORFs were downloaded from the GoldenPath server at UCSC http://genome.ucsc.edu/goldenPath/hg16/bigZips/. Putative regulatory regions (1000 bp upstream of the TSS) for the different genes were obtained. We used for the original experiment (Milyavsky et al, 2005) the GeneChip Human Genome Focus Array (Affymetrix, Santa Clara, CA) that represents over 8500 verified human sequences from the NCBI RefSeq database. We identified promoters for 8110 genes out of the 8500. Of the 8110 genes, we have selected 5582 genes that had a ‘present call' (according to Affymetrix calling procedure) in the two duplicates of at least one sample. Of the 168 genes in the proliferation cluster, 141 probe sets had a promoter in GoldenPath. When more than one probe set on the array corresponded to same genomic locus (e.g. owing to alternative splicing), we considered the corresponding regulatory region only once.
While the present analysis covers only the 8500 genes represented on the GeneChip Focus Array that was used in our original experiment (Milyavsky et al, 2005), we have also examined the promoter motif content of all ∼33 000 genes that were represented on the U133 Array (Affymetrix, Santa Clara, CA). We found additional 2316 genes that were not represented on the Focus Array that contain at least two of the discovered transcriptional modules; 36 of them contain four of the motifs analyzed here (see Supplementary Table S4). These genes may represent additional candidates for the network discovered here.
A collection of position-specific scoring matrices
We used the MatInspector library of 326 PSSMs maintained by Genomatix (Release 4.1) (Quandt et al, 1995) and a customary promoter to PSSM assignment score (Elkon et al, 2003). We then identified a threshold on this score, above which a PSSM is considered assigned to a promoter. For this, we used the genes in a cluster and for a range of potential values of the threshold score we calculated, using the hypergeometric statistic, the groups specificity score (Hughes et al, 2000) of the motif relative to the genes in the cluster. We identified and adopted the threshold score that minimizes the hypergeometric probability function. See Supplementary Figure S5 for examples for threshold score determination for a selection of motifs. Only motifs that passed the Bonferroni correction for multiple hypotheses testing (that considered the multiple attempted thresholds) were retained.
Assessing motif positional bias
Positional bias was previously defined as the extent to which a motif that is assigned to a set of promoters is enriched in a sequence window (defined in terms of distance relative to the TSS) of a fixed length (e.g. 50 bp) with the maximal number of promoter (Hughes et al, 2000). Although efficient and simple, this algorithm has a limitation of having to define a fixed length window, without a priori knowledge about the relevant window width. We thus devised the following alternative procedure that learns the window's width from the data. We search for the window that is most enriched with occurrences of the motif using the following procedure:
Assume we have N occurrences of the motif in the promoters of the cluster's genes. Denote their (ordered) distances from the TSS (of each gene) by Ci (i=1,…,N), such that 0⩽C1⩽⋯⩽CN<1000.
A window is defined as a subinterval [a,b]⊂[0.1000]. We search for the window that is most enriched with motifs, compared to a random background model. For each window [a,b], we denote by M(a,b) the number of motifs Ci with distance of at least a and no more than b from the TSS. The background distribution of the number of motifs in the window [a,b] is M(a,b)∼Binomial(N, (b−a+1)/1000). Since windows overlap, an enrichment of a specific window leads to enrichment of windows overlapping it. Thus, we defined the most enriched window to be the one with smallest background probability, that is, the interval [a,b] minimizing Pr(M(a,b)) under this background model. Obviously, the densest window is [Ci, Cj] for some i⩽j; therefore, we can restrict our search only for intervals of the form [Ci, Cj]. Thus, it is defined as Wmin=argmini,j Pr(M(Ci, Cj)⩾j−i+1), with Pmin=mini,j Pr(M(Ci, Cj)⩾j−i+1)).
To test statistical significance of the densest window, the distribution of Pmin in the background model is required. This was calculated by simulations. For N from 2 to 300, we have performed 100 000 simulations, each time selecting N points randomly in [0.1000] and then computing Pmin. This gave an empirical distribution denoted FN,min. The P-value for the observed most enriched window is simply FN,min(Pmin).
Combinogram analyses
The analysis was initiated with a set of N motifs. Each of the 5582 genes was assigned with a binary signature of length N with a 1 at the ith position if the gene contains motif i in its promoter, and a 0 otherwise. Thus, 2N gene sets (that constitute the ‘power set' of the set N), termed genes defined by motif combinations (GMCs), were generated where all the genes in a given GMC share the same subset of the N motifs. The averaged expression profile of all the genes in each GMC was determined. The normalized Euclidian distance between averaged expression profiles for all pairs of GMCs was calculated and served as the input for the dendrogram analyses that were generated with the Cluster Analysis module in Matlab 7 (Mathworks) using the average-linkage option.
Cell lines
WI-38 cells were maintained in MEM supplemented with 10% FCS, 1 mM sodium pyruvate, 2 mM L-glutamine, and antibiotics. Cells were passaged and counted once in 5–7 days. The HCT-116 cells and their p53-null and p21-null derivatives were a gift from B Vogelstein (The John Hopkins University, Baltimore, MD) and were described by Bunz et al (1998). HCT-116 cells were maintained in McCoy's medium supplemented with 10% FCS, 1 mM sodium pyruvate, 2 mM L-glutamine, and antibiotics. All cell lines were grown at 37°C in a humidified atmosphere of 5% CO2 in air.
Plasmids
The construct p-cdc20-luc was generated by cloning cdc20 promoter and 5′-UTR into a luciferase reporter. Briefly, a genomic fragment of the cdc20 promoter, spanning from −1002 to +229 relative to the TSS, was amplified by PCR with the primers 5′-tccacctctgagcacattcat-3′ and 5′-tccttgcagttggtgcct-3′, using Expand Long Template PCR system (Roche). The amplified region was cloned into pGEM-T easy vector (Promega) and then transferred into pGL3 super basic vector (gift from M Oren, Weizmann Institute of Science) using the restriction enzymes NdeI and NcoI. Mutations in NF-Y motifs were generated on the template of p-cdc20-luc using the QuikChange Site-Directed Mutagenesis kit (Stratagene) with the following primers (mutations are in uppercase): for mutation of NF-Y motif at position (−83), cccttcgccggagaggTAGTAgggctagggcaacg gttgc, and for mutation of NF-Y motif at position (−38), gacggttggattttgaaggagAAGTAaggcgctcg gagcggagagt. Expression plasmids for wild-type human p53 and mutants L22Q/W23S were gifts of C Hurris (NIH, Bethesda, USA) and were described by Zhou et al (1999). Expression plasmid for the p53 dominant-negative peptide (p53-DD) was a gift of M Oren and was described by Shaulian et al (1992). Expression plasmid for p16 was kindly provided by R Agami (Netherlands Cancer Institute). E2F-dTA expression plasmid pRcCMVE2F1-(1–363) was as described by Hofmann et al (1996). dnNF-YA expression plasmid NF-YA13m29 was described by Mantovani et al (1994).
Transfections and reporter assays
HCT-116 cells were plated at 3 × 104 cells/well in a 24-well plate 48 h before transfection. Cells were transfected using JetPEI (Polyplus Transfection), with 150 ng/well of luciferase reporter, 50 ng/well of pCMV-β-galactosidase expression vector, and appropriate expression plasmids for a total DNA amount of 1 μg/well. The p53 expression plasmids were transfected at 10 ng/well. The p16, dnNF-YA, and E2F-dTA expression plasmids were transfected at 300 ng/well. Cell extracts were prepared 48 h after transfection, and luciferase and β-galactosidase activities were determined using commercial reagents and procedures (Promega). Statistical significance was determined by paired t-test.
RNA preparation, cDNA synthesis, and qPCR
Total RNA was extracted with the Versagene RNA cell kit and was treated with the Versagene DNase treatment kit (Gentra Systems Inc.). A 2 μg aliquot of the RNA was reverse transcribed using MMLV-RT (Promega) and random hexamer primers (Roche Applied Science). qPCR was performed using SYBR Green PCR Master Mix (Applied Biosystems). The expression level for each sample was normalized to that of the GAPDH housekeeping gene in the same sample. Primer sequences were as follows:
GAPDH, 5′-agcctcaagatcatcagcaatg-3′ and
5′-cacgataccaaagttgtcatggat-3′;
cdc20, 5′-gagggtggctgggttcctct-3′ and
5′-cagatgcgaatgtgtcgatca-3′;
CCNF, 5′-catctgcacccggtttatca-3′ and
5′-cttccaaggcggagacga-3′;
BIRC5, 5′-tcatccactgccccactga-3′ and
5′-agaagaaacactgggccaagtc-3′;
MAD2L1, 5′-gttggaagtttcttgttcatttgatct-3′ and
5′-ggtcccgactcttcccattt-3′;
CENPF, 5′-agaaagcagtcatgagtggtattca-3′ and
5′-gcaggatatatgggctagtctttcc-3′;
PRC1, 5′-acaaaccgaggaggaaatcttct-3′ and
5′-caattcgtgccttcaactcttct-3′;
Bub1b, 5′-tacactggaaatgaccctctggat-3′ and
5′-tataatatcgtttttctccttgtagtgctt-3′.
Supplementary Material
Supplementary Figures
Legend to supplementary data
Supplemental Table 1
Supplemental Table 2
Supplemental Table 3
Supplemental Table 4
Supplemental Table 5
Supplemental Table 6
Acknowledgments
We thank all members of the Domany, Rotter and Pilpel labs for stimulating discussions. This research was supported by grants from the Israel Academy of Sciences, the Minerva Foundation and the Ben May Foundation (YP), the Leo and Julia Forchheimer Center for Molecular Genetics (YP), the FAMRI foundation (VR), the Ridgefield Foundation, and by the NIH (grant #5 POI CA 65930-06). VR holds the Norman and Helen Asher Professorial Chair in Cancer Research at the Weizmann Institute. ED is the incumbent of the Henry J Leir Professorial Chair. YP is an incumbent of the Aser Rothstein Career Development Chair in Genetic Diseases, and is a Fellow of the Hurwitz Foundation for Complexity Sciences.
References
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, Powell JI, Yang L, Marti GE, Moore T, Hudson J Jr, Lu L, Lewis DB, Tibshirani R, Sherlock G, Chan WC, Greiner TC, Weisenburger DD, Armitage JO, Warnke R, Levy R, Wilson W, Grever MR, Byrd JC, Botstein D, Brown PO, Staudt LM (2000) Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403: 503–511 [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badie C, Bourhis J, Sobczak-Thepot J, Haddada H, Chiron M, Janicot M, Janot F, Tursz T, Vassal G (2000) p53-dependent G2 arrest associated with a decrease in cyclins A2 and B1 levels in a human carcinoma cell line. Br J Cancer 82: 642–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee N, Zhang MQ (2003) Identifying cooperativity among transcription factors controlling the cell cycle in yeast. Nucleic Acids Res 31: 7024–7031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Joseph Z, Gerber GK, Lee TI, Rinaldi NJ, Yoo JY, Robert F, Gordon DB, Fraenkel E, Jaakkola TS, Young RA, Gifford DK (2003) Computational discovery of gene modules and regulatory networks. Nat Biotechnol 21: 1337–1342 [DOI] [PubMed] [Google Scholar]
- Barrett MT, Pritchard D, Palanca-Wessels C, Anderson J, Reid BJ, Rabinovitch PS (2003) Molecular phenotype of spontaneously arising 4N (G2-tetraploid) intermediates of neoplastic progression in Barrett's esophagus. Cancer Res 63: 4211–4217 [PubMed] [Google Scholar]
- Berman BP, Nibu Y, Pfeiffer BD, Tomancak P, Celniker SE, Levine M, Rubin GM, Eisen MB (2002) Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc Natl Acad Sci USA 99: 757–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman BP, Pfeiffer BD, Laverty TR, Salzberg SL, Rubin GM, Eisen MB, Celniker SE (2004) Computational identification of developmental enhancers: conservation and function of transcription factor binding-site clusters in Drosophila melanogaster and Drosophila pseudoobscura. Genome Biol 5: R61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatt M, Wiseman S, Domany E (1996) Superparamagnetic clustering of data. Phys Rev Lett 76: 3251–3254 [DOI] [PubMed] [Google Scholar]
- Bracken AP, Ciro M, Cocito A, Helin K (2004) E2F target genes: unraveling the biology. Trends Biochem Sci 29: 409–417 [DOI] [PubMed] [Google Scholar]
- Buchwalter G, Gross C, Wasylyk B (2004) Ets ternary complex transcription factors. Gene 324: 1–14 [DOI] [PubMed] [Google Scholar]
- Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, Sedivy JM, Kinzler KW, Vogelstein B (1998) Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 282: 1497–1501 [DOI] [PubMed] [Google Scholar]
- Burns TF, Fei P, Scata KA, Dicker DT, El-Deiry WS (2003) Silencing of the novel p53 target gene Snk/Plk2 leads to mitotic catastrophe in paclitaxel (taxol)-exposed cells. Mol Cell Biol 23: 5556–5571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho RJ, Huang M, Campbell MJ, Dong H, Steinmetz L, Sapinoso L, Hampton G, Elledge SJ, Davis RW, Lockhart DJ (2001) Transcriptional regulation and function during the human cell cycle. Nat Genet 27: 48–54 [DOI] [PubMed] [Google Scholar]
- Dai H, van't Veer L, Lamb J, He YD, Mao M, Fine BM, Bernards R, van de Vijver M, Deutsch P, Sachs A, Stoughton R, Friend S (2005) A cell proliferation signature is a marker of extremely poor outcome in a subpopulation of breast cancer patients. Cancer Res 65: 4059–4066 [DOI] [PubMed] [Google Scholar]
- Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA (2003) DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 4: P3. [PubMed] [Google Scholar]
- el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B (1993) WAF1, a potential mediator of p53 tumor suppression. Cell 75: 817–825 [DOI] [PubMed] [Google Scholar]
- Elkon R, Linhart C, Sharan R, Shamir R, Shiloh Y (2003) Genome-wide in silico identification of transcriptional regulators controlling the cell cycle in human cells. Genome Res 13: 773–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garten Y, Kaplan S, Pilpel Y (2005) Extraction of transcription regulatory signals from genome-wide DNA–protein interaction data. Nucleic Acids Res 33: 605–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille H, Kortenjann M, Thomae O, Moomaw C, Slaughter C, Cobb MH, Shaw PE (1995) ERK phosphorylation potentiates Elk-1-mediated ternary complex formation and transactivation. EMBO J 14: 951–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn WC, Stewart SA, Brooks MW, York SG, Eaton E, Kurachi A, Beijersbergen RL, Knoll JH, Meyerson M, Weinberg RA (1999) Inhibition of telomerase limits the growth of human cancer cells. Nat Med 5: 1164–1170 [DOI] [PubMed] [Google Scholar]
- Hahn WC, Weinberg RA (2002) Modelling the molecular circuitry of cancer. Nat Rev Cancer 2: 331–341 [DOI] [PubMed] [Google Scholar]
- Halfon MS, Grad Y, Church GM, Michelson AM (2002) Computation-based discovery of related transcriptional regulatory modules and motifs using an experimentally validated combinatorial model. Genome Res 12: 1019–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugwitz U, Wasner M, Wiedmann M, Spiesbach K, Rother K, Mossner J, Engeland K (2002) A single cell cycle genes homology region (CHR) controls cell cycle-dependent transcription of the cdc25C phosphatase gene and is able to cooperate with E2F or Sp1/3 sites. Nucleic Acids Res 30: 1967–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho J, Benchimol S (2003) Transcriptional repression mediated by the p53 tumour suppressor. Cell Death Differ 10: 404–408 [DOI] [PubMed] [Google Scholar]
- Hofmann F, Martelli F, Livingston DM, Wang Z (1996) The retinoblastoma gene product protects E2F-1 from degradation by the ubiquitin–proteasome pathway. Genes Dev 10: 2949–2959 [DOI] [PubMed] [Google Scholar]
- Hoffman WH, Biade S, Zilfou JT, Chen J, Murphy M (2001) Transcriptional repression of the anti-apoptotic survivin gene by wild type p53. J Biol Chem 19: 19. [DOI] [PubMed] [Google Scholar]
- Hughes JD, Estep PW, Tavazoie S, Church GM (2000) Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J Mol Biol 296: 1205–1214 [DOI] [PubMed] [Google Scholar]
- Ihmels J, Friedlander G, Bergmann S, Sarig O, Ziv Y, Barkai N (2002) Revealing modular organization in the yeast transcriptional network. Nat Genet 31: 370–377 [DOI] [PubMed] [Google Scholar]
- Kalir S, Alon U (2004) Using a quantitative blueprint to reprogram the dynamics of the flagella gene network. Cell 117: 713–720 [DOI] [PubMed] [Google Scholar]
- Krause K, Wasner M, Reinhard W, Haugwitz U, Dohna CL, Mossner J, Engeland K (2000) The tumour suppressor protein p53 can repress transcription of cyclin B. Nucleic Acids Res 28: 4410–4418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapidot M, Pilpel Y (2003) Comprehensive quantitative analyses of the effects of promoter sequence elements on mRNA transcription. Nucleic Acids Res 31: 3824–3828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, Zeitlinger J, Jennings EG, Murray HL, Gordon DB, Ren B, Wyrick JJ, Tagne JB, Volkert TL, Fraenkel E, Gifford DK, Young RA (2002) Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298: 799–804 [DOI] [PubMed] [Google Scholar]
- Li C, Lin M, Liu J (2004) Identification of PRC1 as the p53 target gene uncovers a novel function of p53 in the regulation of cytokinesis. Oncogene 23: 9336–9347 [DOI] [PubMed] [Google Scholar]
- Lin AW, Barradas M, Stone JC, van Aelst L, Serrano M, Lowe SW (1998) Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Genes Dev 12: 3008–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liotta L, Petricoin E (2000) Molecular profiling of human cancer. Nat Rev Genet 1: 48–56 [DOI] [PubMed] [Google Scholar]
- Lohr K, Moritz C, Contente A, Dobbelstein M (2003) p21/CDKN1A mediates negative regulation of transcription by p53. J Biol Chem 278: 32507–32516 [DOI] [PubMed] [Google Scholar]
- Manni I, Mazzaro G, Gurtner A, Mantovani R, Haugwitz U, Krause K, Engeland K, Sacchi A, Soddu S, Piaggio G (2001) NF-Y mediates the transcriptional inhibition of the cyclin B1, cyclin B2, and cdc25C promoters upon induced G2 arrest. J Biol Chem 276: 5570–5576 [DOI] [PubMed] [Google Scholar]
- Mantovani R (1998) A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acids Res 26: 1135–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani R, Li XY, Pessara U, Hooft van Huisjduijnen R, Benoist C, Mathis D (1994) Dominant negative analogs of NF-YA. J Biol Chem 269: 20340–20346 [PubMed] [Google Scholar]
- Matuoka K, Chen KY (2002) Transcriptional regulation of cellular ageing by the CCAAT box-binding factor CBF/NF-Y. Ageing Res Rev 1: 639–651 [DOI] [PubMed] [Google Scholar]
- Milyavsky M, Shats I, Erez N, Tang X, Senderovich S, Meerson A, Tabach Y, Goldfinger N, Ginsberg D, Harris CC, Rotter V (2003) Prolonged culture of telomerase-immortalized human fibroblasts leads to a premalignant phenotype. Cancer Res 63: 7147–7157 [PubMed] [Google Scholar]
- Milyavsky M, Tabach Y, Shats I, Erez N, Cohen Y, Tang X, Kalis M, Kogan I, Buganim Y, Goldfinger N, Ginsberg D, Harris CC, Domany E, Rotter V (2005) Transcriptional programs following genetic alterations in p53, INK4A, and H-Ras genes along defined stages of malignant transformation. Cancer Res 65: 4530–4543 [DOI] [PubMed] [Google Scholar]
- Nevins JR (2001) The Rb/E2F pathway and cancer. Hum Mol Genet 10: 699–703 [DOI] [PubMed] [Google Scholar]
- Ossovskaya VS, Mazo IA, Chernov MV, Chernova OB, Strezoska Z, Kondratov R, Stark GR, Chumakov PM, Gudkov AV (1996) Use of genetic suppressor elements to dissect distinct biological effects of separate p53 domains. Proc Natl Acad Sci USA 93: 10309–10314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D (2000) Molecular portraits of human breast tumours. Nature 406: 747–752 [DOI] [PubMed] [Google Scholar]
- Pilpel Y, Sudarsanam P, Church GM (2001) Identifying regulatory networks by combinatorial analysis of promoter elements. Nat Genet 29: 153–159 [DOI] [PubMed] [Google Scholar]
- Quandt K, Frech K, Karas H, Wingender E, Werner T (1995) MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res 23: 4878–4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenwald A, Wright G, Wiestner A, Chan WC, Connors JM, Campo E, Gascoyne RD, Grogan TM, Muller-Hermelink HK, Smeland EB, Chiorazzi M, Giltnane JM, Hurt EM, Zhao H, Averett L, Henrickson S, Yang L, Powell J, Wilson WH, Jaffe ES, Simon R, Klausner RD, Montserrat E, Bosch F, Greiner TC, Weisenburger DD, Sanger WG, Dave BJ, Lynch JC, Vose J, Armitage JO, Fisher RI, Miller TP, LeBlanc M, Ott G, Kvaloy S, Holte H, Delabie J, Staudt LM (2003) The proliferation gene expression signature is a quantitative integrator of oncogenic events that predicts survival in mantle cell lymphoma. Cancer Cell 3: 185–197 [DOI] [PubMed] [Google Scholar]
- Rosty C, Sheffer M, Tsafrir D, Stransky N, Tsafrir I, Peter M, de Cremoux P, de La Rochefordiere A, Salmon R, Dorval T, Thiery JP, Couturier J, Radvanyi F, Domany E, Sastre-Garau X (2005) Identification of a proliferation gene cluster associated with HPV E6/E7 expression level and viral DNA load in invasive cervical carcinoma. Oncogene, advance online publication 27 June 2005; doi:10.1038/sj.onc.1208854 [DOI] [PubMed] [Google Scholar]
- Scherf U, Ross DT, Waltham M, Smith LH, Lee JK, Tanabe L, Kohn KW, Reinhold WC, Myers TG, Andrews DT, Scudiero DA, Eisen MB, Sausville EA, Pommier Y, Botstein D, Brown PO, Weinstein JN (2000) A gene expression database for the molecular pharmacology of cancer. Nat Genet 24: 236–244 [DOI] [PubMed] [Google Scholar]
- Segal E, Friedman N, Koller D, Regev A (2004) A module map showing conditional activity of expression modules in cancer. Nat Genet 36: 1090–1098 [DOI] [PubMed] [Google Scholar]
- Segal E, Shapira M, Regev A, Pe'er D, Botstein D, Koller D, Friedman N (2003) Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat Genet 34: 166–176 [DOI] [PubMed] [Google Scholar]
- Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW (1997) Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88: 593–602 [DOI] [PubMed] [Google Scholar]
- Setty Y, Mayo AE, Surette MG, Alon U (2003) Detailed map of a cis-regulatory input function. Proc Natl Acad Sci USA 100: 7702–7707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharan R, Ben-Hur A, Loots GG, Ovcharenko I (2004) CREME: Cis-Regulatory Module Explorer for the human genome. Nucleic Acids Res 32: W253–W256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharan R, Ovcharenko I, Ben-Hur A, Karp RM (2003) CREME: a framework for identifying cis-regulatory modules in human–mouse conserved segments. Bioinformatics 19 (Suppl 1): i283–i291 [DOI] [PubMed] [Google Scholar]
- Shaulian E, Zauberman A, Ginsberg D, Oren M (1992) Identification of a minimal transforming domain of p53: negative dominance through abrogation of sequence-specific DNA binding. Mol Cell Biol 12: 5581–5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen-Orr SS, Milo R, Mangan S, Alon U (2002) Network motifs in the transcriptional regulation network of Escherichia coli. Nat Genet 31: 64–68 [DOI] [PubMed] [Google Scholar]
- Sherr CJ (1996) Cancer cell cycles. Science 274: 1672–1677 [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM (1999) CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev 13: 1501–1512 [DOI] [PubMed] [Google Scholar]
- Smith AD, Sumazin P, Zhang MQ (2005) Identifying tissue-selective transcription factor binding sites in vertebrate promoters. Proc Natl Acad Sci USA 102: 1560–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Clair S, Giono L, Varmeh-Ziaie S, Resnick-Silverman L, Liu WJ, Padi A, Dastidar J, DaCosta A, Mattia M, Manfredi JJ (2004) DNA damage-induced downregulation of Cdc25C is mediated by p53 via two independent mechanisms: one involves direct binding to the cdc25C promoter. Mol Cell 16: 725–736 [DOI] [PubMed] [Google Scholar]
- Sudarsanam P, Pilpel Y, Church GM (2002) Genome-wide co-occurrence of promoter elements reveals a cis-regulatory cassette of rRNA transcription motifs in Saccharomyces cerevisiae. Genome Res 12: 1723–1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumazin P, Chen G, Hata N, Smith AD, Zhang T, Zhang MQ (2005) DWE: discriminating word enumerator. Bioinformatics 21: 31–38 [DOI] [PubMed] [Google Scholar]
- Tang Z, Bharadwaj R, Li B, Yu H (2001) Mad2-independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev Cell 1: 227–237 [DOI] [PubMed] [Google Scholar]
- Tavazoie S, Hughes JD, Campbell MJ, Cho RJ, Church GM (1999) Systematic determination of genetic network architecture. Nat Genet 22: 281–285 [DOI] [PubMed] [Google Scholar]
- Taylor LM, James A, Schuller CE, Brce J, Lock RB, Mackenzie KL (2004) Inactivation of p16INK4a, with retention of pRB and p53/p21cip1 function, in human MRC5 fibroblasts that overcome a telomere-independent crisis during immortalization. J Biol Chem 279: 43634–43645 [DOI] [PubMed] [Google Scholar]
- Thompson W, Palumbo MJ, Wasserman WW, Liu JS, Lawrence CE (2004) Decoding human regulatory circuits. Genome Res 14: 1967–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsafrir D, Tsafrir I, Ein-Dor L, Zuk O, Notterman DA, Domany E (2005) Sorting points into neighborhoods (SPIN): data analysis and visualization by ordering distance matrices. Bioinformatics 21: 2301–2308 [DOI] [PubMed] [Google Scholar]
- Tsutsui T, Kumakura S, Yamamoto A, Kanai H, Tamura Y, Kato T, Anpo M, Tahara H, Barrett JC (2002) Association of p16(INK4a) and pRb inactivation with immortalization of human cells. Carcinogenesis 23: 2111–2117 [DOI] [PubMed] [Google Scholar]
- Ullrich A, Schlessinger J (1990) Signal transduction by receptors with tyrosine kinase activity. Cell 61: 203–212 [DOI] [PubMed] [Google Scholar]
- Voorhoeve PM, Agami R (2003) The tumor-suppressive functions of the human INK4A locus. Cancer Cell 4: 311–319 [DOI] [PubMed] [Google Scholar]
- Wang Q, Zambetti GP, Suttle DP (1997) Inhibition of DNA topoisomerase II alpha gene expression by the p53 tumor suppressor. Mol Cell Biol 17: 389–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman WW, Palumbo M, Thompson W, Fickett JW, Lawrence CE (2000) Human–mouse genome comparisons to locate regulatory sites. Nat Genet 26: 225–228 [DOI] [PubMed] [Google Scholar]
- Weinberg R (1995) The retinoblastoma protein and cell cycle control. Cell 81: 323–330 [DOI] [PubMed] [Google Scholar]
- Werner T (2001) The promoter connection. Nat Genet 29: 105–106 [DOI] [PubMed] [Google Scholar]
- Werner T, Fessele S, Maier H, Nelson PJ (2003) Computer modeling of promoter organization as a tool to study transcriptional coregulation. FASEB J 17: 1228–1237 [DOI] [PubMed] [Google Scholar]
- Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, Matese JC, Perou CM, Hurt MM, Brown PO, Botstein D (2002) Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell 13: 1977–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Yoshida M, Ono K, Fujita T, Ohtani-Fujita N, Sakai T, Nikaido T (1994) Effect of tumor suppressors on cell cycle-regulatory genes: RB suppresses p34cdc2 expression and normal p53 suppresses cyclin A expression. Exp Cell Res 210: 94–101 [DOI] [PubMed] [Google Scholar]
- Yun J, Chae HD, Choi TS, Kim EH, Bang YJ, Chung J, Choi KS, Mantovani R, Shin DY (2003) Cdk2-dependent phosphorylation of the NF-Y transcription factor and its involvement in the p53–p21 signaling pathway. J Biol Chem 278: 36966–36972 [DOI] [PubMed] [Google Scholar]
- Yun J, Chae HD, Choy HE, Chung J, Yoo HS, Han MH, Shin DY (1999) p53 negatively regulates cdc2 transcription via the CCAAT-binding NF-Y transcription factor. J Biol Chem 274: 29677–29682 [DOI] [PubMed] [Google Scholar]
- Zhou X, Wang XW, Xu L, Hagiwara K, Nagashima M, Wolkowicz R, Zurer I, Rotter V, Harris CC (1999) COOH-terminal domain of p53 modulates p53-mediated transcriptional transactivation, cell growth, and apoptosis. Cancer Res 59: 843–848 [PubMed] [Google Scholar]
- Zhu J, Woods D, McMahon M, Bishop JM (1998) Senescence of human fibroblasts induced by oncogenic Raf. Genes Dev 12: 2997–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Shendure J, Church GM (2005) Discovering functional transcription-factor combinations in the human cell cycle. Genome Res 15: 848–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwicker J, Lucibello FC, Wolfraim LA, Gross C, Truss M, Engeland K, Muller R (1995) Cell cycle regulation of the cyclin A, cdc25C and cdc2 genes is based on a common mechanism of transcriptional repression. EMBO J 14: 4514–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Legend to supplementary data
Supplemental Table 1
Supplemental Table 2
Supplemental Table 3
Supplemental Table 4
Supplemental Table 5
Supplemental Table 6