Abstract
Natural biological systems are selected by evolution to continue to exist and evolve. Evolution likely gives rise to complicated systems that are difficult to understand and manipulate. Here, we redesign the genome of a natural biological system, bacteriophage T7, in order to specify an engineered surrogate that, if viable, would be easier to study and extend. Our initial design goals were to physically separate and enable unique manipulation of primary genetic elements. Implicit in our design are the hypotheses that overlapping genetic elements are, in aggregate, nonessential for T7 viability and that our models for the functions encoded by elements are sufficient. To test our initial design, we replaced the left 11 515 base pairs (bp) of the 39 937 bp wild-type genome with 12 179 bp of engineered DNA. The resulting chimeric genome encodes a viable bacteriophage that appears to maintain key features of the original while being simpler to model and easier to manipulate. The viability of our initial design suggests that the genomes encoding natural biological systems can be systematically redesigned and built anew in service of scientific understanding or human intention.
Keywords: bacteriophage T7, synthetic biology, refactor
Introduction
In nature, the success of an individual organism depends directly on its ability to continue to exist and replicate. Not surprisingly, natural biological systems appear to have evolved, and continue to evolve, to meet these requirements (e.g., Block et al, 1982; Aho et al, 1988). However, should we also expect that the ‘design' of an evolved organism would be further optimized for the purposes of human understanding and interaction? Evidence drawn from fields outside biology suggests that the answer is no.
For example, consider two different approaches to programming computers and electronics: ‘genetic programming' and ‘structured design.' In genetic programming, evolutionary algorithms are used to evolve computer software or electrical hardware for a particular task (Koza et al, 2003). The absolute performance of evolved systems often meets, and sometimes exceeds, that produced by human-directed designs (Spector et al, 1999). However, so-evolved systems lack human readable descriptions and are difficult to understand, fix, and modify for new applications. By contrast, a structured design process produces systems that, in addition to functioning, are designed to be easy to understand and extend (Abelson et al, 1996). Not surprisingly, an artifact produced via structured design may not be optimal when evaluated only in terms of absolute algorithmic or physical performance. However, a structured design process can bypass two limitations, direct-descent and replication-with-error, which constrain the designs of evolved systems. Thus, we might paradoxically expect that a structured design process will, when practical, produce artifacts whose designs can ‘evolve' more quickly.
Here, we converted the genome of a natural biological system, bacteriophage T7, to a more structured design. Our work was initially motivated by past failures in modeling T7 development (below) and by a desire to better understand how the parts that comprise bacteriophage T7 work together to encode a functioning whole (Kirschner, 2005). The approach we used was inspired by the practice of ‘refactoring,' a process that is typically used to improve the design of legacy computer software (Fowler et al, 1999). In general terms, the goal of refactoring is to improve the internal structure of an existing system for future use, while simultaneously maintaining external system function.
T7 is an obligate lytic phage that infects Escherichia coli (Dunn and Studier, 1983; Studier and Dunn, 1983). T7 was twice isolated from Ward MacNeal's ‘standard anti-coli-phage' mixture (Demerec and Fano, 1945; Delbrück, 1946). MacNeal's ‘mixture' may have been cultured in series—T7 was the only identifiable isolate (Studier, 1979). One of the two original T7 isolates was reportedly chosen for future use and master cultures of ‘wild-type' T7 have been maintained since (Supplementary information). Genetics, and then biochemistry, enabled the discovery and characterization of some of the individual elements that participate in T7 development (Molineux, 2005). Sequencing of the T7 genome revealed additional elements (Dunn and Studier, 1983), not all of which have obvious functions (Molineux, 2005). As a result, T7 now provides a good model system for studying what fraction of the functional information encoded on the genome of a natural biological system has been described, and how much of what might still be understood is likely to matter (Davis, 1946).
For example, the T7 protein coding domains were first characterized by the isolation and analysis of randomly generated amber mutants. A total of 19 genes were identified by mapping mutants that disrupt T7 DNA synthesis, particle maturation, and lysis (Hausmann and Gomez, 1967; Studier, 1969; Hausmann and LaRue, 1969). Two additional genes, T7 DNA ligase and protein kinase, were isolated via loss of function and deletion, respectively (Masamune et al, 1971); the genetic analysis of ligase and kinase mutants was carried out using mutant host strains that do not support the growth of ligase- or kinase-defective phage (Studier, 1969). Up to 30 T7 proteins were observed by pulsing phage-infected cells with radioactive amino acids (Studier and Maizel, 1969; Studier, 1973a, 1973b). Further experiments, such as electrophoretic mobility shifts of amber mutants, provided evidence for up to 38 T7 proteins (Studier, 1981). Sequencing of the genome confirmed the previously constructed genetic maps (Dunn and Studier, 1983). But, analysis of the complete genome sequence also revealed that the set of protein coding domains found via mutagenesis, screening, and mapping was not exhaustive, and that additional unidentified open reading frames occupied most of the remainder of the genome. Some of these unidentified open reading frames can be labeled as putative protein coding domains based on the inferred strengths of adjacent upstream ribosome binding sites (RBSs). In all, up to 57 genes encoding 60 potential proteins have been found or postulated (Molineux, 2005). However, only 35 of these 60 proteins have at least one known function. And, of the 25 nonessential proteins, only 12 are conserved across the family of T7-like phage (Molineux, 2005). Can we safely ignore these uncharacterized protein coding domains in our models of phage infection? Should we edit the genome to remove them?
As a second example, the E. coli RNA polymerase promoters on the T7 genome (A0, A1–3, B, C, and E) were first mapped by in vitro transcription studies (Davis and Hyman, 1970; Minkley and Pribnow, 1973; Golomb and Chamberlin, 1974a, 1974b; Niles and Condit, 1975; McAllister and McCarron, 1977; Stahl and Chamberlin, 1977; Kassavetis and Chamberlin, 1979; Panayotatos and Wells, 1979) and subsequently confirmed by sequencing (Oakley and Coleman, 1977; Boothroyd and Hayward, 1979; Rosa, 1979; Carter and McAllister, 1981; Osterman and Coleman, 1981; Dunn and Studier, 1983). Results of in vitro transcription reactions using T7 genomic DNA as template agreed with the available in vivo transcription data (Studier, 1973a, 1973b; Summers et al, 1973; McAllister and Wu, 1978; McAllister et al, 1981). However, the cloning of random sections of the T7 genome into a plasmid that selected for transcription activity from the cloned fragment identified other possible promoters (Studier and Rosenberg, 1981). Sequence analysis of the cloned sections identified ∼10 regions with homology to known promoters; footprinting assays identified two additional promoters (Dunn and Studier, 1983). But, any contribution of these putative promoters to wild-type T7 infection is not now defined. As with some of the T7 genes, should we ignore these promoters in our models? Should we delete these elements from the T7 genome? Is there other information encoded on the wild-type T7 genome that we should include in our models, ignore, or actively remove?
One practical test of the understanding encoded in a model of a system is to use the model to help predict what will happen when either the system or its environment is changed. In the case of T7, the experimentalists who originally discovered much of how the phage works developed the best descriptive, system-level models for T7 infection. Their models were made by integrating knowledge of the individual parts and mechanisms that act during infection, from genome entry to phage particle formation (Studier and Dunn, 1983). Two features specific to T7 biology made the construction of system-level models easier. First, compared to other phage, T7 is relatively independent of complex host physiology. For example, the optical density of T7-infected cultures stops increasing at the time of infection, T7 encodes phage-specific RNA and DNA polymerases, and E. coli mRNA and protein synthesis is inhibited within the first ∼6 min of T7 infection. Second, RNA polymerase pulls most of the T7 genome into the newly infected cell (Zavriev and Shemyakin, 1982; Garcia and Molineux, 1995). Polymerase-mediated genome entry is a relatively slow process that results in the direct physical coupling of gene expression dynamics to gene position. For example, a gene cannot be expressed until its coding domain enters the newly infected cell.
Building on this work, others and we developed computational, quantitative models of T7 infection in order to explore questions related to the organization of genetic elements on the T7 genome, and the timing and control of gene expression across uncertain physical environments (Endy et al, 1997; Endy et al, 2000; You and Yin, 2002; You et al, 2002). Initially, our models were used to test the hypothesis that the results of 60 years of research on bacteriophage T7, conducted by many researchers across many labs, could be integrated to produce a T7-like computer simulation. The resulting model and simulation recapitulated the apparent molecular details and dynamics of T7 development quite well. Unfortunately, the model itself was of little interest, as it turned out to be overfitted to limited experimental data—changes to the model led to predictions that were unbelievable (D Endy, unpublished). A subsequent revision, in which care was taken to only include known facts and mechanisms, produced a model of T7 that matched the available system-wide data less well, but that was more useful as a tool for exploring how changes to the phage genome and the host cell environment impact phage development (Endy et al, 1997). However, in using these computational models, some predictions did not agree with experiments (Endy and Brent, 2001). For example, a mutant phage expected to grow faster than the wild type grew slower (Endy et al, 2000).
Upon inspection, disagreements between model-based prediction and experiment could have arisen for at least three reasons. First, our models could not meaningfully include unknown functions. For example, disruption of an uncharacterized nonessential gene, 1.7, appeared to impact DNA replication (Endy et al, 2000). While differences between expectation and observation can suggest follow-on science, a lack of complete component-level understanding debased our system-level analyses. Second, the boundaries of genetic elements on the T7 genome are more complex than our models of the genome. For example, genes 2.8 and 3 are most easily modeled as separable genetic elements even though the actual genes 2.8 and 3 overlap (Figure 1A and Supplementary Figure S3). Element overlap may also encode uncharacterized function(s) having to do with the regulation of protein synthesis or the coupling of selective pressures during evolution. For example, bioinformatic analysis of microbial genomes suggests that gene overlaps are conserved across evolutionary distance (e.g., Johnson and Chisholm, 2004). Element overlap also prohibits independent element manipulation. For example, on the wild-type genome, we cannot change the gene 3 RBS without at least changing the codon usage of gene 2.8. Third, a computer model built with separable parts that encode independent functions can be overmanipulated relative to the actual physical system. For example, while we could simulate the expected behavior of large sets of permuted genomes, we could not easily move a single open reading frame to another arbitrary position on the actual T7 genome (Endy et al, 2000).
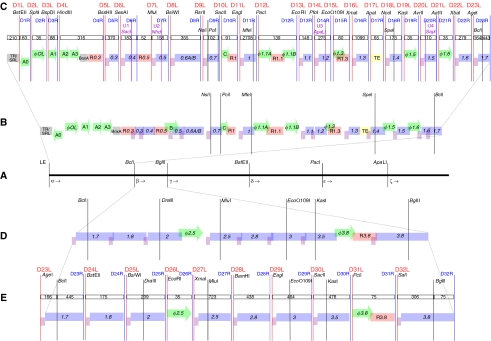
Figure 1.
Element decompression and part design. (A) The coding regions of genes 2.8 and 3 overlap in the wild-type T7 genome. The RBS of gene 3 (underlined) is encoded within gene 2.8. (B) Distinct genetic parts make up the T7.1 genome. The natural RBS and start codon (green) for gene 3 are disrupted by point mutations (capitals); mutations do not change the amino-acid sequence of the 2.8 protein. Parts 28 and 29 are separated by bracketing restriction sites, BamHI (blue) and EagI (orange). Supplementary Figure S3 lists all changes in the DNA sequence of T7.1 relative to wild-type T7.
Wild-type T7 is a superb organism for discovering the primary components of a natural biological system (Studier, 1972). However, is the original T7 isolate also best suited for understanding how all parts of the phage are organized to encode a functioning whole? Given our experiences, we decided to attempt to engineer a surrogate genome, which we designated T7.1.
Results
Design goals
Six goals drove our design of the T7.1 genome. First, we wanted to define a set of components that function during T7 development and, for each element, choose an exact DNA sequence that we could use to encode element function. Second, we wanted the DNA sequence encoding the function of any one element to not overlap with the DNA sequence encoding any other element. Third, we wanted the DNA sequence of each element to encode only the function assigned to that element and not any other functions. Fourth, we wanted to enable the precise and independent manipulation of each element. Fifth, we needed to be able to construct the T7.1 genome. Sixth, we needed the T7.1 genome to encode viable bacteriophage; at the start of this work, we were uncertain how many simultaneous changes the wild-type genome could tolerate.
Design process
A general algorithm describing our genome refactoring process is given in Supplementary Figure S1. Briefly, we began design of the T7.1 genome by reannotating the genome of wild-type T7. The wild-type T7 genome is a 39 937 base pair (bp) linear double-stranded DNA molecule (Dunn and Studier, 1983). We annotated the genome by specifying the boundaries of the following functional genetic elements: 57 open reading frames with 57 putative RBSs encoding 60 proteins, and 51 regulatory elements controlling phage gene expression, DNA replication, and genome packaging.
To specify the architecture of T7.1, we organized the functional genetic elements into 73 ‘parts.' Each part contains one or more elements. While the DNA sequence of elements within parts may overlap, there is no overlap across part boundaries (Figure 1B). Next, we organized contiguous parts into ‘sections' with section boundaries defined by restriction endonuclease sites found only once in the sequence of the wild-type genome. Six sections, alpha through zêta, make up the T7.1 genome (Figure 2A and Supplementary Figure S2). Sections were used to compartmentalize changes across the genome. In addition, sections can be built, tested, and manipulated independently.
Figure 2.
Genome design. (A) We split the wild-type T7 genome into six sections, alpha through zêta, using five restriction sites unique across the natural sequence. (B) Wild-type section alpha genetic elements: protein coding regions (blue), RBSs (purple), promoters (green), RNaseIII recognition sites (pink), a transcription terminator (yellow), and others (gray). Elements are labeled by convention (Dunn and Studier, 1983). Images are not to scale, but overlapping boundaries indicate elements with shared sequence. The five useful natural restriction sites across section alpha are shown (black lines). (C) T7.1 section alpha parts. Parts are given integer numbers, 1–73, starting at the left end of the genome. Unique restriction site pairs bracket each part (red/blue lines, labeled D[part #]L/R). Added unique restriction sites (purple lines, U[part #]) and part length (# base pairs, open boxes) are shown. We do not know if sequence changes in and around parts 6 and 7 destroy the minor E. coli promoter, B. (D) Wild-type section beta genetic elements. (E) T7.1 section beta parts. Supplementary Figure S2 depicts the six sections, alpha through zêta, which make up the T7.1 genome.
To specify the DNA sequence of T7.1, we eliminated sequence overlap across part boundaries. Overlaps were eliminated by exact duplication of the wild-type DNA sequence; subsequent sequence editing produced a single instance of any duplicated element (Figure 1B and Supplementary Figure S3). All sequence edits within open reading frames were silent and maintained the wild-type tRNA specification or, when necessary, specified a higher abundance tRNA (Ikemura, 1981). We also added bracketing restriction endonuclease sites to insulate and enable the independent manipulation of each part (Figure 2C and E and Supplementary Figure S2). Bracketing sites are not used elsewhere in the sequence of any one section but are reused across sections. The DNA sequence of T7.1 changes or adds 1424 bp to the wild-type genome (Supplementary Figure S3).
Construction and testing
The sections that comprise the T7.1 genome can be built and tested independently. We constructed the first two sections, alpha and beta (Materials and methods). Alpha and beta contain the first 32 of 73 parts of the T7.1 genome, replacing the left 11 515 bp of the wild-type genome with 12 179 bp of redesigned DNA, and encoding the entire T7 early region, the primary origins of DNA replication, most of the T7 middle genes, and the control architecture that regulates T7 gene expression. Alpha and beta also contain the highest density of elements across the genome. We combined alpha and beta with the remainder of the wild-type (WT) genome to produce three chimeric phage: alpha-WT, WT-beta-WT, and alpha-beta-WT.
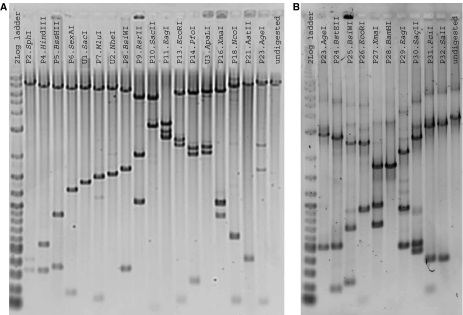
We tested and recovered viable chimeric phage by transfection and plating. All three chimeric phages are viable. We isolated DNA and performed restriction digests across alpha and beta to confirm that individual parts could be independently manipulated. A total of 30 of 32 parts in sections alpha and beta can be cut out as designed (Figure 3). We also sequenced alpha and beta. Sequencing revealed differences between the design of T7.1 and the actual ‘as-built' sections. Relevant sequence differences in section alpha include a single-base deletion in gene 0.4 and in the E. coli terminator TE. Differences in section beta include a single amino-acid substitution in both genes 1.8 and 2, a single-base deletion in gene 2.5, and an 82-base truncation in gene 2.8. All differences were due to errors or limitations in construction (Supplementary Tables S1 and S2).
Figure 3.
Cutting parts from T7.1. (A) Restriction enzymes specific to the sites that bracket parts (P#.Enzyme) and added unique restriction sites (U#.Enzyme) were used to cut section alpha (Supplementary information). A subset of the digests is shown. As built, part 1 cannot be removed. (B) Restriction digests cutting out all parts in section beta. As built, part 28 cannot be removed.
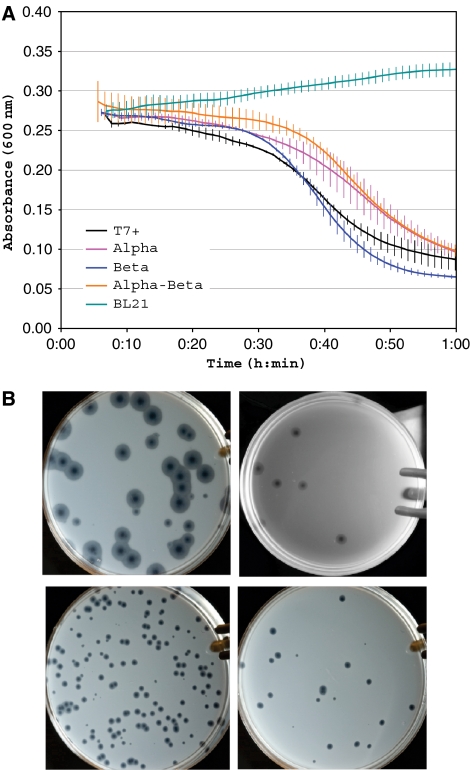
Finally, we characterized some growth properties of the chimeric phage by liquid culture lysis and plating. Phage-induced lysis of log-phase 30°C liquid cultures indicated a 20, −1.4, and 22% increase in the half-lysis times of the alpha, beta, and alpha-beta chimeras, respectively, relative to the wild type (Figure 4A). Plaques were indistinguishable early during plaque growth and at 30°C (not shown). At 37°C, the chimeric phage plaques appeared to stop growing as the bacterial lawns developed; after 24 h at 37°C, plaque sizes relative to the wild type were smaller for each of the chimeric phage, with the alpha-beta chimera being smallest (Figure 4B).
Figure 4.
Characterization of T7.1 (A) Lysis of log-phase liquid cultures of E. coli BL21 (30°C) by wild-type T7 (black), alpha-WT chimera (red), WT-beta-WT chimera (blue), alpha-beta-WT chimera (orange); absorbance of 0.275 is ∼2E8 cells/ml. Vertical bars show standard deviation at each time point (based on four replicates) (Supplementary information). (B) T7 plaques on E. coli BL21 (24 h, 37°C, 10 cm Petri dish). Clockwise from top left: wild-type (WT) T7, alpha-WT chimera, WT-beta-WT chimera, alpha-beta-WT chimera (Supplementary information).
Discussion
A system that is partially understood can continue to be studied in hope of exact characterization. Or, if enough is known about the system, a surrogate can be specified to help study or extend the original. Here, we decided to redesign the genome of a natural biological system, bacteriophage T7, in order to specify an engineered biological system that is easier to study and manipulate. The new genome, T7.1, is based on our incomplete understanding of the information encoded in the wild-type genome and our desire to insulate and independently manipulate known primary genetic elements. We constructed the first two sections of T7.1, making over 600 simultaneous changes or additions to the wild-type DNA, and observed that the resulting chimeric phage are viable.
Phage viability demonstrates the following for sections alpha and beta. First, our parts as chosen can be separated by exogenous DNA sequence. Second, any functions encoded by genetic element overlap are, in aggregate, nonessential under standard laboratory conditions. Third, our current understanding of T7 is not insufficient to specify a viable bacteriophage. Viability does not demonstrate sufficiency because (i) if the chimeric phage had not been viable, then our current understanding would have been demonstrably insufficient, and (ii) while T7.1 is based on our current understanding, we do not have an exact understanding of all functions encoded in the T7.1 genome (e.g., genes of unknown function). Finally, viability, combined with the observed similarities in lysis times, suggests that T7.1 preserves polymerase-mediated genome entry and remains relatively independent of host cell physiology.
The T7.1 genome is easier to model and study. For example, by removing genetic element overlap, the T7.1 genome better matches the understanding of T7 biology encoded in our models, relative to the wild-type phage. However, more work is needed to demonstrate that the dynamic behavior of the system encoded by the T7.1 genome is easier to predict. Such work will benefit from the fact that the parts of T7.1 can be independently manipulated.
Our design of T7.1 was constrained by fears of producing a nonviable DNA fragment that would have been difficult to analyze and rescue. Given our initial success with T7.1, we have decided to revisit and extend our original design goals. For example, the design of our next phage, T7.2, will include (i) reduced gene sets that eliminate nonessential and nonconserved protein coding domains, (ii) codon shuffling of protein coding domains in order to disrupt secondary and cryptic regulatory elements, and putative mRNA secondary structure, and (iii) standard regulatory elements and regulatory element spacing. By actively removing all of the uncharacterized elements that we know about, as well as taking steps to disrupt any uncharacterized elements as yet unknown, we will be able to better study how the parts of T7 work to encode a functioning whole.
We constructed sections alpha and beta manually. Concurrent advances in de novo DNA synthesis technology have recently enabled the rapid automatic synthesis of DNA fragments the size of the T7.1 genome sections (Stemmer et al, 1995; Yount et al, 2000; Kodumal et al, 2003; Smith et al, 2003; Tian et al, 2004). As genome synthesis and engineering technologies continue to improve (Carlson, 2003), the use of bracketing restriction sites for manipulating each part should become less important, but the physical separation of parts by the elimination of sequence overlap will remain useful. In general, we expect that the ability to ‘write' long fragments of synthetic DNA will directly accelerate the engineering of biology, and impact the science of biology at least as much as large-scale automated DNA sequencing technology.
Our work with T7 suggests that the genomes encoding other natural, evolved biological systems could be redesigned and built anew in support of scientific discovery or human intention. For systems beyond model laboratory organisms, pursuing such work will require the widespread societal acceptance of responsibility for the direct manipulation of genetic information.
Materials and methods
Reannotation of the wild-type bacteriophage T7 genome
Summary
We used past experiments and observations to define specific boundaries of functional genetic elements on the bacteriophage T7 genome. We followed the standard naming conventions developed by Studier and Dunn (Dunn and Studier, 1983; Studier and Dunn, 1983).
Protein coding domains
The definition of a protein coding domain that we used here is a contiguous stretch of DNA that, when transcribed, produces an mRNA that specifies the amino-acid sequence of a protein. We described the discovery and mapping of the T7 protein coding domains above (Introduction). In the few cases where multiple possible start codons exist, we used the most upstream start codon to define the beginning of the protein coding domain.
Ribosome binding sites
The definition of an RBS that we used here is a contiguous stretch of DNA that, when transcribed, produces a region of RNA that interacts with the ribosome and allows for the initiation of protein synthesis. The T7 RBSs were first postulated by analysis of the sequence data upstream of protein coding domain start codons; DNA sequence complementary to the E. coli 16S rRNA suggested a functioning RBS. Direct observation of proteins during T7 infection provides additional support for the function of a subset of RBSs (Studier and Maizel, 1969).
RNA polymerase promoters
The definition of a promoter that we used here is a contiguous stretch of DNA that interacts with an RNA polymerase molecule and allows for the initiation of mRNA synthesis. At least 22 RNA polymerase promoters help to coordinate transcription dynamics during T7 infection. The discovery and mapping of the RNA polymerase promoters is given above (Introduction). The T7 RNA polymerase promoter was determined by sequencing the 23 bp region common to the late T7 promoters (Boothroyd and Hayward, 1979). Here, we used a 35 bp region to define T7 promoters; our broader definition of T7 promoter elements hoped to include conserved regions beyond the initial 23 bp (Dunn and Studier, 1983). The E. coli RNA polymerase promoter is less well defined. Here, we used regions of at least 60 bp, ranging from the −50 to +10 positions, to define the major and minor E. coli promoters (A0, A1, A2, A3, B, C, and E). Also, a boxA recognition site located between A3 and gene 0.3 is thought to be involved with antitermination of polymerases that initiate from the three strong early promoters, A1, A2, and A3 (Olson et al, 1982). Finally, the cloning of random sections of the T7 genome into a plasmid that selected for transcription activity from the cloned fragment identified other possible promoters (Studier and Rosenberg, 1981). Sequence analysis in regions containing these sections identified regions of homology to other known promoters (Dunn and Studier, 1983). Any contribution of these additional promoters to wild-type T7 infection is not now defined. While we annotated these additional promoters, we did not incorporate them as functional genetic elements of T7.1.
RNA polymerase terminators
The definition of a terminator that we used here is a contiguous stretch of DNA that, during transcription, produces a region of mRNA that stops the process of transcription (at some efficiency). The first T7 transcription termination site was identified by mapping the end points of mRNA starting from E. coli promoters (Studier, 1972). Later, it was shown that termination occurred at the same place in vivo and in vitro (Dunn and Studier, 1973). The termination site was later mapped precisely, sequenced, and subsequently named ‘TE' (Studier et al, 1979; Dunn and Studier, 1980). A second terminator specific to T7 RNA polymerase was suggested by in vitro transcription studies on digested T7 DNA (Golomb and Chamberlin, 1974a, 1974b; Niles and Condit, 1975). The terminator, named ‘Tø,' was shown to function in situ (Dunn and Studier, 1980) and on plasmids (McAllister et al, 1981). Both TE and Tø have stem loop structures that are thought to set termination efficiency (Dunn and Studier, 1973). The stem loop and flanking sequence, which includes a poly-uridine tract, were taken together to define the element we used here. While other terminators have been postulated, their precise location and function, if any, during wild-type infection are tenuous (Dunn and Studier, 1983), and thus we did not include them in our annotation.
RNaseIII recognition sites
The definition of an RNaseIII recognition site that we used here is a contiguous stretch of DNA that, when transcribed, produces a region of mRNA that is recognized and cleaved (at some efficiency) by RNaseIII. Sites for specific cleavage of T7 RNA by RNaseIII were first shown in vitro and then correlated to in vivo data (Dunn and Studier, 1973). In time, 10 RNaseIII sites were mapped and their sites of cleavage identified (Dunn and Studier, 1983). The sites are thought to stabilize the 3′ end of T7 transcripts by providing a stem loop that prevents the binding of scanning single-stranded RNases. A downstream gene often immediately follows an RNaseIII site. Thus, we kept the RNaseIII recognition site elements as short as possible—with a minimum boundary set by the probable stem loop structures (Dunn and Studier, 1983).
DNA replication origins
The definition of a DNA replication origin that we used here is a stretch of DNA that is used to initiate the copying of phage DNA during T7 infection. The primary replication origin was mapped to the dual promoter region downstream of ø1.1A and ø1.1B by analysis of replication bubbles in electron micrographs (Dressler et al, 1972; Wolfson et al, 1972) and subsequently sequenced (Saito et al, 1980). The secondary origin at øOL was identified using mutants that lacked the primary origin (Tamanoi et al, 1980; Studier and Rosenberg, 1981). Finally, plasmids containing cloned fragments of T7 DNA were used to screen for regions that act as replication origins during T7 infection; these experiments revealed that øOR and ø13 have origin activity (Dunn and Studier, 1983). While the precise boundaries of the replication origins are unknown, each appears to be linked to a functioning RNA polymerase promoter (Zhang and Studier, 2004). Here, we only annotate and define an element for the primary origin. While we do not include other replication origins as elements, we do preserve the RNA polymerase promoters that are associated with these secondary origins as elements, and thus possibly the secondary origins as well.
Terminal repeats and short repeats
The definition of a terminal repeat that we used here is a contiguous stretch of DNA present at both ends of the T7 genome, and a short repeat is a series of direct repeats of DNA near the end of the genome. Both the left and right ends of the T7 genome contain exact 160 bp direct repeats (Ritchie et al, 1967). Also, adjacent to the direct repeats on both ends of the genome are regions of DNA that contain 12 regularly arranged and highly conserved 7 bp sequences termed the short repeats left, SRL, and right, SRR (Dunn and Studier, 1981). The terminal repeats and SRL/R are thought to be involved in concatemer formation, DNA packaging, and particle maturation (Kelly and Thomas, 1969). However, the mechanisms by which the direct repeats and the SRL/R act are unclear. Thus, we treated each end's direct repeat and SRL/R as a monolithic element (the design of T7.1 does not make any changes to the DNA sequence of these elements).
Design of T7.1 genome
Overview
The design of T7.1 genome uses six sections, alpha through zêta. Each section contains parts that are amalgamations of one or more functional genetic elements (Supplementary Figure S2). In our design, the modification of parts on the full T7.1 genome is a two-stage process. First, we can manipulate parts to construct a section. Second, we can combine sections to assemble a full genome. We improved upon the design of sections beta through zêta based on our experience constructing section alpha.
Definitions
#-Cutter—a restriction enzyme that cuts a particular DNA sequence # times;
Functional genetic element—a promoter, protein coding domain, RBS, etc., defined during our reannotation of the T7 genome;
Part—a piece of DNA that encodes one or more functional genetic elements and is bracketed by a pair of identical restriction sites;
Construct—any amalgamation of functional genetic elements or parts;
Section—a segment of the T7.1 genome the boundaries of which are 1-cutters on the wild-type T7 genome.
T7.1 genome sections
We used sections to limit the number of simultaneous changes to the wild-type T7 sequence and to make the construction process more manageable. Two practical considerations drove our choice of section boundaries. First and foremost, the boundaries of the sections had to be compatible with the sparse distribution of 1-cutter sites across the wild-type genome. (The use of 1-cutter sites for section boundaries allows refactored sections to be easily combined with other sections or with wild-type DNA.) Second, the number of parts per section was limited by the number of ‘useful' 0-cutters across the DNA sequence of each wild-type section. Useful 0-cutters are specific, free or smaller recognition sites, dam/dcm insensitive, and leave sticky-end overhangs.
From functional genetic elements to T7.1 parts
Parts are made up of one or more functional genetic elements. Parts were sometimes defined to have more than one element in order to maintain the natural proximity of elements known, or likely, to be physically or functionally coupled. For example, we grouped most RBSs and downstream protein coding domains into two-element parts. Also, some functional genetic elements overlap so severely as to prevent efficient separation (e.g., the genes 4A, 4B, 4.1, and 4.2). Finally, some functional genetic elements were very short (<150 bp) such that variants containing deletions or separations of the individual elements could be easily constructed (e.g., the E. coli promoter C and RNaseIII site R1). In total, we combined the elements that make up T7.1 into 73 parts. We numbered parts, 1–73, starting from the genetic left end.
The arrangements of parts on the wild-type T7 DNA sequence sometimes resulted in the overlapping of the DNA sequence specifying parts. To remove part–part overlap, we duplicated the DNA sequence of the overlap, providing both parts with an independent copy of the previously overlapping sequence. If, as a result of sequence duplication, either of the parts encoded a function specific to an element in the other part, we mutated the sequence to eliminate the duplicate function. All mutations to protein coding domains were silent and result in either no change in the tRNA or, when necessary, specify a higher abundance tRNA (Ikemura, 1981). Parts separation is detailed in Supplementary Figure S2.
We surrounded each part with a restriction site pair that is not contained elsewhere in that part's section. Typically, we added bracketing restriction sites to the DNA sequence of each part, but, when appropriate, we integrated the sites into the natural DNA sequence. Also, to help reduce the length of T7.1, where possible, we chose adjacent restriction sites to have overlapping sequence with one another.
One of the most significant differences between the design of section alpha and the other sections was in our choice of bracketing restriction sites. In section alpha, we picked restriction enzymes that did not cut within section alpha only. However, as the construction of alpha proceeded, and cloning directly into the phage became useful, we adjusted our design strategy to use restriction enzymes that did not cut within the entire genome wherever possible.
Design features
Deletion and insertion: The design of the T7.1 genome allows for the simple deletions of parts. Generally, we isolate the section containing the part by digesting with the bracketing restriction enzyme. We ligate the fragments to reform the section minus the deleted part, and then join the section to the rest of the genome. Insertion of a new part can be more involved. Most simply, if there is a pre-existing restriction site due to a deletion operation, then we can insert a new part in its place. If no such site exists, another method involves using two restriction enzymes, NgoMIV and BspEI, that are 0-cutters across both the wild-type T7 and all refactored sections. NgoMIV and BspEI have different recognition sequences but produce the same overhang upon digestion. This allows for ligation of a product into these sites, while simultaneously preventing the restriction sites from being reformed. Thus, we can replace a part adjacent to the desired insertion site with the same part that has an NgoMIV site appended to it. Then, we amplify the part to be inserted with bracketing BspEI sites and insert the part into the NgoMIV site. Since neither restriction site is reformed upon insertion, this method is idempotent.
Unstuffing hooks: Since we did not know how a phage made of separated parts would function (e.g., would it form plaques?), we thought that it would be prudent to be able to easily revert to the wild-type T7 sequence for purposes of comparison and debugging. Thus, we used silent mutations to add additional 1-cutter restriction sites to section alpha. These new restriction sites, labeled U1–4, are useful if we desired to replace refactored regions with wild-type sequence. In sections beta through zêta, such extra sites were superfluous because we used 0-cutters to bracket parts; 0-cutters can also be used to revert refactored regions to wild-type sequence.
Scaffolds: We used scaffolds to build sections alpha and beta. A scaffold is essentially the sequence that remains when all parts are removed from the section. As such, the scaffold contains all the restriction sites required to assemble the parts to form the section. In addition, if a fully refactored phage was not viable, we could use the scaffold to incrementally revert the sequence back to wild type in an attempt to restore function.
Construction of section alpha
The design of the scaffold for section alpha included all functional genetic elements from the left end of T7 through R0.3, R0.5, parts 17, 19, 21, plus the restriction sites required to add all remaining parts. The section alpha scaffold does not contain any known protein coding domains. We sent the scaffold sequence (1334 bp) to Blue Heron Biotechnology for synthesis (http://www.blueheronbio.com/). Blue Heron could not assemble the scaffold using the standard cloning plasmids then in use (we have since worked with Blue Heron to fix this problem—below). Blue Heron agreed to ship the section alpha scaffold as four fragments with point mutations in each fragment. The point mutations were
- Fragment 1:
single-base changes at 89(G-T), 168(A-T), 169(C-A), 245(G-A), and 249(C-A) as well as single-base deletions at 138 and 159;
a single-base deletion in the −35 box of the A1 promoter;
a four-base deletion between the −35 and −10 boxes of the A3 promoter;
a single-base deletion in the loop of TE.
- Fragment 2:
a single-base deletion in the −35 box of the A1 promoter;
a four-base deletion between the −35 and −10 boxes of the A3 promoter;
a single-base deletion in the loop of TE.
- Fragment 3:
a four-base deletion between the −35 and −10 boxes of the A3 promoter;
a single-base deletion in the loop of TE.
- Fragment 4:
a single-base deletion in the loop of TE.
We decided to discard Fragment 1 but to correct and make use of Fragments 2, 3, and 4. We built a new vector, pREB, to facilitate the assembly of section alpha. pREB (for rebuild) started as a chimera of the inducible copy control system of pSCANS-5 and the insulated multicloning site (MCS) of pSB2K3-1 (below). We completed pREB by adding a smaller MCS containing PstI, BstBI, and BclI restriction endonuclease sites and by removing 19 other restriction sites from the plasmid backbone.
To build section alpha, we first cloned parts 5, 6, 7, 8, 12, 13, 14, 15, 16, 18, 20, 22, and 24 into pSB104. We cloned part 11 into pSB2K3. We cloned each part with its part-specific bracketing restriction sites surrounded by additional BioBrick restriction sites (Knight, 2002). We used site-directed mutagenesis on parts 6, 7, 14, and 20 to introduce the sites U1, U2, U3, and U4, respectively. Our site-directed mutagenesis of part 20 failed.
We used site-directed mutagenesis to remove a single Eco0109I restriction site from the vector pUB119BHB carrying the scaffold Fragment 4. We cloned part 15 into this modified vector. We then cloned scaffold Fragment 4 into pREB and used serial cloning to add the following parts: 7, 8, 12, 13, 14, 16, 18, 20, 22, and 23. We digested the now-populated scaffold Fragment 4 with NheI and BclI and purified the resulting DNA.
Next, we cloned parts 5 and 6 into pUB119BHB carrying scaffold Fragment 3. We used the resulting DNA for in vitro assembly of a construct spanning from the left end of T7 to part 7. To do this, we cut wild-type T7 genomic DNA with AseI, isolated the 388 bp left-end fragment, and ligated this DNA to scaffold Fragment 2. We selected the correct ligation product by PCR. We fixed the mutation in part 3 (A1) via a two-step process. First, PCR primers with the corrected sequence for part 3 were used to amplify the two halves of the construct to the left and right ends of part 3. Second, a PCR ligation joined the two constructs. We added scaffold Fragment 3 to the above left-end construct once again by PCR ligation as described above. We repaired the mutation in part 4 (A2, A3, and R0.3) following the same procedure as with part 3. We used a right-end primer containing an MluI site to amplify the entire construct, and used the MluI site to add part 7. We used PCR to select the ligation product, digested the product with NheI, and purified the resulting DNA.
We isolated the right arm of a BclI digestion of wild-type T7 genomic DNA and used ligation to add the populated left-end construct and the populated Scaffold Fragment 4. We transfected the three-way ligation product into IJ1127. We purified DNA from liquid culture lysates inoculated from single plaques. We used restriction enzymes to digest the DNA and isolate the correct clones.
Next, we added part 11 via three-way ligation and transfection. Because the restriction sites that bracket part 9 (RsrII) also cut wild-type T7 DNA, we needed to use in vitro assembly to add this part to a subsection of section alpha. To do this, we used PCR to amplify the region spanning parts 5–12 from the refactored genome. We cut the PCR product with RsrII and ligated part 9. We used PCR to select the correct ligation product; this PCR reaction also added a SacII site to the fragment. We digested the PCR product with SacI and SacII and cloned onto the otherwise wild-type phage. Lastly, we used the SacII site to clone part 10 onto the phage.
Construction of section beta
We constructed section beta using a process similar to that used with alpha. A scaffold with all restriction sites as well as part 26 was made by Klenow extension of overlapping primers. We digested this DNA with BstBI and cloned it onto pREB. We then added the following parts: 23, 24, 27, 28, 30, 31, and 32. We had to clone part 32 (containing gene 3.8) as a truncation since we were unable to clone the full-length part, probably due to the apparent toxicity of gene 3.8 product. The truncated version of part 32 still included the BglII site to allow for assembly of section beta onto phage. We added parts 25 and 29, also previously reported to be toxic, in vitro. To insert part 25, we amplified a region spanning parts 23–27 by PCR. We cut this fragment with BsiWI. Part 25 was then ligated to each of these fragments separately and selected for by PCR. We cut both PCR products with DraIII, a restriction site internal to part 25, ligated and then selected for full-length part 25 by PCR. We cut part 25 with BclI and MluI, purified, and ligated it to wild-type fragments. We used a similar approach to insert part 29 (using the EcoO109I site internal to this part). Lastly, we cut both phage genomes with MluI; we ligated the left fragment of the genome containing the refactored region spanning parts 23–27 to the right fragment of a genome containing the refactored region spanning parts 27–32.
Synthesis and construction errors
Differences between the designed and constructed sections alpha and beta are detailed in Supplementary Tables S1 and S2.
Protocol, strains, and media
Detailed laboratory protocols, strain information, and media recipes are provided as Supplementary information.
Accession numbers
The DNA sequence encoding the as-built sections alpha and beta are available via GenBank (DQ100054, DQ100055). The entire T7.1 design and our reannotation of the wild-type T7 genome are also available online:
http://web.mit.edu/endy/www/ncbi/
Competing interests
The authors have declared that no competing interests exist.
Author contributions
DE conceived the project. LYC, SK, and DE designed the experiments. LYC and SK designed the T7.1 genome, developed all software, and performed the experiments. LYC, SK, and DE wrote the paper.
Supplementary Material
Supplementary Tables S1 and S2
Supplementary Information
Supplementary Figures
Acknowledgments
We thank Ian Molineux, Priscilla Kemp, and Heather Keller for discussions and advice throughout the work. We thank John Dunn and Barbara Lade for the pSCANS-5 vector. We thank Roger Brent, Eric Eisenstadt, Tom Knight, and members of the Endy group for additional discussions and sustained encouragement. We thank Jorge Borges and Adolfo Casares for ‘On Exactitude in Science' (Davis, 1946). We thank Austin Che, Heather Keller, Alex Mallet, Kathleen McGinness, Samantha Sutton, Ty Thomson, Elizabeth Vesilind, and Rebecca Ward for comments on the manuscript. We thank Felice Frankel for plaque photography and encouragement. This work was funded by grants to DE from the US Office of Naval Research, DARPA, and NIH. SK was supported by an NIH MIT BPEC training fellowship. Additional support was provided by MIT.
References
- Abelson H, Sussman GJ, Sussman J (1996) Structure and Interpretation of Computer Programs, 2nd edn. Cambridge, MA, USA: MIT Press [Google Scholar]
- Aho A-C, Donner K, Hyden C, Larsen LO, Reuter T (1988) Low retinal noise in animals with low body temperature allows high visual sensitivity. Nature 334: 348–350 [DOI] [PubMed] [Google Scholar]
- Block SM, Segall JE, Berg HC (1982) Impulse responses in bacterial chemotaxis. Cell 31: 215–226 [DOI] [PubMed] [Google Scholar]
- Boothroyd JC, Hayward RS (1979) New genes and promoters suggested by the DNA sequence near the end of the coliphage T7 early operon. Nucleic Acids Res 7: 1931–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson R (2003) The pace and proliferation of biological technologies. Biosecur Bioterror 1: 203–214 [DOI] [PubMed] [Google Scholar]
- Carter AD, McAllister WT (1981) Sequences of three class II promoters for the bacteriophage T7 RNA polymerase. J Mol Biol 153: 825–830 [DOI] [PubMed] [Google Scholar]
- Davis BL (1946) Del rigor en la ciencia. Los Anales de Buenos Aires año 1 [Google Scholar]
- Davis RW, Hyman RW (1970) Physical locations of the in vitro RNA initiation site and termination sites of T7 DNA. Cold Spring Harb Symp Quant Biol 35: 269–282 [Google Scholar]
- Delbrück M (1946) Bacterial viruses or bacteriophages. Biol Rev Camb Philos Soc 21: 30–40 [PubMed] [Google Scholar]
- Demerec M, Fano U (1945) Bacteriophage-resistant mutants in Escherichia coli . Genetics 30: 119–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressler D, Wolfson J, Magazin M (1972) Initiation and reinitiation of DNA synthesis during replication of bacteriophage T7. Proc Natl Acad Sci USA 69: 998–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn JJ, Studier FW (1973) T7 early RNAs are generated by site-specific cleavages. Proc Natl Acad Sci USA 70: 1559–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn JJ, Studier FW (1980) The transcription termination site at the end of the early region of bacteriophage T7 DNA. Nucleic Acids Res 8: 2119–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn JJ, Studier FW (1981) Nucleotide sequence from the genetic left end of bacteriophage T7 DNA to the beginning of gene 4. J Mol Biol 148: 303–330 [DOI] [PubMed] [Google Scholar]
- Dunn JJ, Studier FW (1983) Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol 166: 477–535 [DOI] [PubMed] [Google Scholar]
- Endy D, Brent R (2001) Modelling Cellular Behavior. Nature 409: 391–395 [DOI] [PubMed] [Google Scholar]
- Endy D, Kong D, Yin J (1997) Intracellular kinetics of a growing virus: a genetically structured simulation for bacteriophage T7. Biotechnol Bioeng 55: 375–389 [DOI] [PubMed] [Google Scholar]
- Endy D, You L, Yin J, Molineux IJ (2000) Computation, prediction, and experimental tests of fitness for bacteriophage T7 mutants with permuted genomes. Proc Natl Acad Sci USA 97: 5375–5380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler M, Beck K, Brant J, Opdyke W, Roberts D (1999) Refactoring: Improving the Design of Existing Code. Boston, MA, USA: Addison-Wesley Professional [Google Scholar]
- Garcia LR, Molineux IJ (1995) Rate of translocation of bacteriophage T7 DNA across the membranes of Escherichia coli . J Bacteriol 177: 4066–4076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golomb M, Chamberlin M (1974a) A preliminary map of the major transcription units read by T7 RNA polymerase on the T7 and T3 bacteriophage chromosomes. Proc Natl Acad Sci USA 71: 760–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golomb M, Chamberlin M (1974b) Characterization of T7-specific ribonucleic acid polymerase. IV. Resolution of the major in vitro transcripts by gel electrophoresis. J Biol Chem 249: 2858–2863 [PubMed] [Google Scholar]
- Hausmann R, Gomez B (1967) Amber mutants of bacteriophages T3 and T7 defective in phage-directed deoxyribonucleic acid synthesis. J Virol 1: 779–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausmann R, LaRue K (1969) Variations in sedimentation patterns among deoxyribonucleic acids synthesized after infection of Escherichia coli by different amber mutants of bacteriophage T7. J Virol 3: 278–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura T (1981) Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J Mol Biol 151: 389–409 [DOI] [PubMed] [Google Scholar]
- Johnson ZI, Chisholm SW (2004) Properties of overlapping genes are conserved across microbial genomes. Genome Res 14: 2268–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis GA, Chamberlin MJ (1979) Mapping of class II promoter sites utilized in vitro by T7-specific RNA polymerase on bacteriophage T7 DNA. J Virol 29: 196–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly TJ Jr, Thomas CA Jr (1969) An intermediate in the replication of bacteriophage T7 DNA molecules. J Mol Biol 44: 459–475 [DOI] [PubMed] [Google Scholar]
- Kirschner MW (2005) The meaning of systems biology. Cell 20: 503–504 [DOI] [PubMed] [Google Scholar]
- Knight T (2002) Idempotent vector design for standard assembly of biobricks. MIT Synthetic Biology Working Group Technical Report 0 [http://web.mit.edu/synbio/release/d ocs/biobricks.pdf] [Google Scholar]
- Kodumal SJ, Patel KG, Reid R, Menzella HG, Welch M, Santi DV (2003) Total synthesis of long DNA sequences: synthesis of a contiguous 32-kb polyketide synthase gene cluster. Proc Natl Acad Sci USA 101: 15573–15578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koza JR, Keane MA, Streeter MJ, Mydlowec W, Yu J, Lanza G (2003) Genetic Programming IV: Routine Human-Competitive Machine Intelligence. Dordrecht, Netherlands: Kluwer Academic Publishers [Google Scholar]
- Masamune Y, Frenkel GD, Richardson CC (1971) A mutant of bacteriophage T7 deficient in polynucleotide ligase. J Biol Chem 246: 6874–6879 [PubMed] [Google Scholar]
- McAllister WT, McCarron RJ (1977) Hybridization of the in vitro products of bacteriophage T7 RNA polymerase to restriction fragments of T7 DNA. Virology 82: 288–298 [DOI] [PubMed] [Google Scholar]
- McAllister WT, Morris C, Rosenberg AH, Studier FW (1981) Utilization of bacteriophage T7 late promoters in recombinant plasmids during infection. J Mol Biol 153: 527–544 [DOI] [PubMed] [Google Scholar]
- McAllister WT, Wu HL (1978) Regulation of transcription of the late genes of bacteriophage T7. Proc Natl Acad Sci USA 75: 804–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minkley EG, Pribnow D (1973) Transcription of the early region of bacteriophage T7: selective initiation with dinucleotides. J Mol Biol 77: 255–277 [DOI] [PubMed] [Google Scholar]
- Molineux IJ (2005) The T7 Group. In The Bacteriophages, Calendar RL (ed) Chapter 20. Oxford: Oxford University Press [Google Scholar]
- Niles EG, Condit RC (1975) Translational Mapping of Bacteriophage T7 RNAs synthesized in vitro by purified T7 RNA polymerase. J Mol Biol 98: 57–67 [DOI] [PubMed] [Google Scholar]
- Oakley JL, Coleman JE (1977) Structure of a promoter for T7 RNA polymerase. Proc Natl Acad Sci USA 74: 4266–4270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson ER, Flamm EL, Friedman DI (1982) Analysis of nutR: a region of phage lambda required for antitermination of transcription. Cell 31: 61–70 [DOI] [PubMed] [Google Scholar]
- Osterman HL, Coleman JE (1981) T7 ribonucleic acid polymerase-promoter interactions. Biochemistry 20: 4884–4892 [DOI] [PubMed] [Google Scholar]
- Panayotatos N, Wells RD (1979) Recognition and initiation site for four late promoters of phage T7 is a 22-base pair DNA sequence. Nature 280: 35–39 [DOI] [PubMed] [Google Scholar]
- Ritchie DA, Thomas CA Jr, MacHattie LA, Wensink PC (1967) Terminal repetition in non-permuted T3 and T7 bacteriophage DNA molecules. J Mol Biol 23: 365–376 [DOI] [PubMed] [Google Scholar]
- Rosa MD (1979) Four T7 RNA polymerase promoters contain an identical 23 bp sequence. Cell 16: 815–825 [DOI] [PubMed] [Google Scholar]
- Saito H, Tabor S, Tamanoi F, Richardson CC (1980) Nucleotide sequence of the primary origin of bacteriophage T7 DNA replication: relationship to adjacent genes and regulatory elements. Proc Natl Acad Sci USA 77: 3917–3921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HO, Hutchison CA III, Pfannkoch C, Venter JC (2003) Generating a synthetic genome by whole genome assembly: phiX174 bacteriophage from synthetic oligonucleotides. Proc Natl Acad Sci USA 100: 15440–15445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector L, Barnum H, Bernstein HJ, Swamy N (1999) Finding a better-than-classical quantum AND/OR algorithm using genetic programming. In IEEE Proceedings of the 1999 Congress on Evolutionary Computation, pp 2239–2246. Piscataway, NJ: IEEE Press. For additional examples, see http://www.genetic-programming.com/ humancompetitive.html [Google Scholar]
- Stahl SJ, Chamberlin MJ (1977) An expanded transcriptional map of T7 bacteriophage. Reading of minor T7 promoter sites in vitro by Escherichia coli RNA polymerase. J Mol Biol 112: 577–601 [DOI] [PubMed] [Google Scholar]
- Stemmer WP, Crameri A, Ha KD, Brennan TM, Heyneker HL (1995) Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene 164: 49–53 [DOI] [PubMed] [Google Scholar]
- Studier FW (1969) The genetics and physiology of bacteriophage T7. Virology 39: 562–574 [DOI] [PubMed] [Google Scholar]
- Studier FW (1972) Bacteriophage T7. Science 176: 367–376 [DOI] [PubMed] [Google Scholar]
- Studier FW (1973a) Genetic analysis of non-essential bacteriophage T7 genes. J Mol Biol 79: 227–236 [DOI] [PubMed] [Google Scholar]
- Studier FW (1973b) Analysis of bacteriophage T7 early RNAs and proteins on slab gels. J Mol Biol 79: 237–248 [DOI] [PubMed] [Google Scholar]
- Studier FW (1979) Relationships among different strains of T7 and among T7-related bacteriophages. Virology 95: 70–84 [DOI] [PubMed] [Google Scholar]
- Studier FW (1981) Identification and mapping of five new genes in bacteriophage T7. J Mol Biol 153: 493–502 [DOI] [PubMed] [Google Scholar]
- Studier FW, Dunn JJ (1983) Organization and expression of bacteriophage T7 DNA. Cold Spring Harb Symp Quant Biol 47 (Part 2): 999–1007 [DOI] [PubMed] [Google Scholar]
- Studier FW, Maizel JV (1969) T7-directed protein synthesis. Virology 39: 575–586 [DOI] [PubMed] [Google Scholar]
- Studier FW, Rosenberg AH (1981) Genetic and physical mapping of the late region of bacteriophage T7 DNA by use of cloned fragments of T7 DNA. J Mol Biol 153: 503–525 [DOI] [PubMed] [Google Scholar]
- Studier FW, Rosenberg AH, Simon MN, Dunn JJ (1979) Genetic and physical mapping in the early region of bacteriophage T7 DNA. J Mol Biol 135: 917–937 [DOI] [PubMed] [Google Scholar]
- Summers WC, Brunovskis I, Hyman RW (1973) The process of infection with coliphage T7. VII. Characterization and mapping of the major in vivo transcription products of the early region. J Mol Biol 74: 291–300 [DOI] [PubMed] [Google Scholar]
- Tamanoi F, Saito H, Richardson CC (1980) Physical mapping of primary and secondary origins of bacteriophage T7 DNA replication. Proc Natl Acad Sci USA 77: 2656–2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J, Gong H, Sheng N, Zhou X, Gulari E, Gao X, Church G (2004) Accurate multiplex gene synthesis from programmable DNA microchips. Nature 432: 1050–1054 [DOI] [PubMed] [Google Scholar]
- Wolfson J, Dressler D, Magazin M (1972) Bacteriophage T7 DNA replication: a linear replicating intermediate (gradient centrifugation-electron microscopy-E. coli-DNA partial denaturation). Proc Natl Acad Sci USA 69: 499–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You L, Suthers PF, Yin J (2002) Effects of Escherichia coli physiology on growth of phage T7 in vivo and in silico . J Bacteriol 184: 1888–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You L, Yin J (2002) Dependence of epistasis on environment and mutation severity as revealed by in silico mutagenesis of phage T7. Genetics 160: 1273–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B, Curtis KM, Baric RS (2000) Strategy for systematic assembly of large RNA and DNA genomes: transmissible gastroenteritis virus model. J Virol 74: 10600–10611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavriev SK, Shemyakin MF (1982) RNA polymerase-dependent mechanism for the stepwise T7 phage DNA transport from the virion into E. coli . Nucleic Acids Res 10: 1635–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Studier FW (2004) Multiple roles of T7 RNA polymerase and T7 lysozyme during bacteriophage T7 infection. J Mol Biol 340: 707–730 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables S1 and S2
Supplementary Information
Supplementary Figures