Abstract
The immune system provides organisms with robustness against pathogen threats, yet it also often adversely affects the organism as in autoimmune diseases. Recently, the molecular interactions involved in the immune system have been uncovered. At the same time, the role of the bacterial flora and its interactions with the host immune system have been identified. In this article, we try to reconcile these findings to draw a consistent picture of the host defense system. Specifically, we first argue that the network of molecular interactions involved in immune functions has a bow-tie architecture that entails inherent trade-offs among robustness, fragility, resource limitation, and performance. Second, we discuss the possibility that commensal bacteria and the host immune system constitute an integrated defense system. This symbiotic association has evolved to optimize its robustness against pathogen attacks and nutrient perturbations by harboring a broad range of microorganisms. Owing to the inherent propensity of a host immune system toward hyperactivity, maintenance of bacterial flora homeostasis might be particularly important in the development of preventive strategies against immune disorders such as autoimmune diseases.
Keywords: robustness, immunology, micro-organisms, bacteria flora, autoimmune disease
Introduction
Defense against pathogens is one of the most important issues for the survival of the organism. Such a function makes the organism more robust against external threats. Robustness of biological systems is gaining increasing attention these days (Kitano, 2004; Stelling et al, 2004), and a conceptual framework has been proposed that places robustness as one of the fundamental properties of living organisms (Kitano, 2004). As the immune system clearly provides robustness to the organism, interpretation of its molecular interactions and functions within the framework of biological robustness will help us to better understand the immune system within an integral framework of robust evolvable systems. It has been argued that there are specific architectural characteristics in robust evolvable systems, which are often denoted as a bow-tie structure (Csete and Doyle, 2004), and several mechanistic features such as systems control, fail–safe, modularity, and decoupling (Kitano, 2004). It is also argued that systems that are robust against specific perturbations are fragile against unexpected perturbations (Carlson and Doyle, 2000, 2002; Csete and Doyle, 2004). Aside from robustness fragility trade-offs, enhancement in robustness results in increased demands for resources and sacrifice of certain aspects of performance of the system. If these are general properties of evolvable robust systems, the immune system, which is an evolvable subsystem of the organism, is not an exception. Understanding such features in the host protection system including the immune system provides us with in-depth insights into the nature of this system and how to cope with anomalies in the system.
Two aspects of the host protection system will be discussed in this article: (1) the global architectural feature of the host defense system and its inherent trade-offs between robustness, fragility, resource limitation, and performance, and (2) the symbiotic nature of the host–microbial relationship that enhances the robustness of the host by adding a layer of adaptive defense subsystem over the host immune system. In fact, consideration of these two frameworks leads us to argue that proper control and maintenance of the bacterial flora are essential to metazoan species by preventing and mitigating autoimmune disorders.
In this article, we will first describe the global molecular interaction architecture of the immune system to illustrate its characteristic bow-tie structure and inherent trade-offs. Second, the relationship between bacterial flora and the host physiology will be discussed to demonstrate that the commensal bacterial flora is an integral part of the host system. Then, we will present a series of medical implications deduced from these observations, and examine the importance of controlling commensal bacterial flora for the prevention of autoimmune disorders. Finally, we will speculate on the characteristics of a possible bacterial flora network.
The global molecular interaction architecture of the immune system and its inherent trade-offs
In order to provide defense against a variety of pathogens, the immune system has to detect a broad range of molecular signatures for pathogens and invoke effective countermeasures. It is clear that this has to be performed under resource-limited conditions, because the number of cells for the immune system and the number of molecules that can be involved in immune reactions within each cell are not infinite.
It is conceivable that such requirements imposed selective pressure that has shaped the global architectural structure of the immune system. A typical global architecture of a bow-tie architecture comprises conserved and efficient core processes with diverse and redundant input and output processes (Csete and Doyle, 2004; Kitano, 2004). We suggest here that the molecular interactions of the immune system actually consist of a nested tandem bow-tie architecture (Figure 1). This architecture can be recognized both in intracellular signal transduction pathways and in intercellular signaling processes. The following examples will be considered: (i) Toll-like receptor (TLR) signaling in innate immunity; (ii) processing and recognition of MHC-peptides between antigen-presenting cells (APCs) and T-cells; and (iii) convergence of signaling from various cells into CD4+ T-cells to foster polarized proliferation involving a complex cytokine network. Although the bow-tie network is generally robust against attacks (i.e. removal of network components) on its core (Li et al, 2004a), a central part of the network connecting input and output networks, the bow-tie network in the immune system at both intracellular and intercellular levels is fragile against attacks on non-redundant elements within its core. Removal of such an element results in immunodeficiency, and confusions in molecular pattern recognition in MHC-peptide–TCR interaction lead to autoimmunity, both of which are manifestations of the fragility of the bow-tie architecture in the immune system.
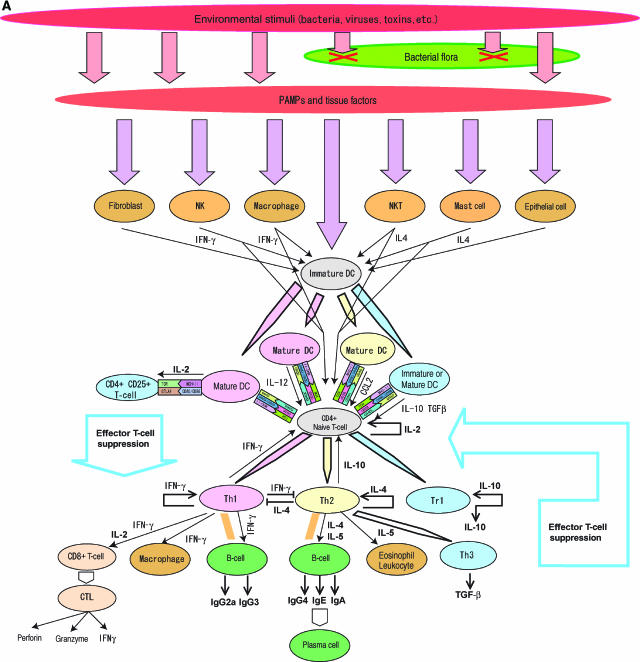
Figure 1a.
Nested bow-tie architecture in immune system. Salient features of the immune system network are nested bow-tie structures and extensive feedback loops. (A) The bow-tie structure of intercellular interactions. The CD4+ T-cells are the hub of this interaction network. Various stimuli from pathogens are transmitted to dendritic cells (DC) that polarize CD4+ T-cells. Stimuli trigger the differentiation of naïve CD4+ T-cells and effector cytokine releases to follow. The whole behavior of this subsystem is controlled by complex signal transductions, and the cytokine network has adapted to the pathogenic environment to which it was exposed during evolution. Upon recognition of appropriate peptide–MHC complex and/or cytokine stimuli, naïve CD4+ T-cells polarize into either Th1, Th2, or Tr1 cells depending upon the cytokine stimuli (Moser and Murphy, 2000; Luther and Cyster, 2001; Murphy and Reiner, 2002) that are provided by polarized DC and a variety of innate immune cells (Kapsenberg, 2003). Th1 cells are induced by IFN-γ, IL-12, and IL-18, and secrete IFN-γ and IL-2, whereas Th2 cells are induced by IL-4 and secrete IL-4, IL-5, IL-13, and IL-10 (Abbas et al, 1996). Chromatin remodeling by GATA3 and T-bet is pivotal in Th1/Th2 polarization (Murphy and Reiner, 2002). Effector cytokines secreted from Th1 and Th2 cells affect various cells. For example, IFN-γ activates B-cells to secrete IgG2a and IgG3, IL-2 activates cytotoxic T lymphocytes (CTL), and IL-4 and IFN-γ mutually inhibit the growth of Th1 and Th2 T-cells, respectively (Liew, 2002). Among these cytokines, IL-2 plays an important role in shaping the dynamics of T-cell response because it promotes growth and activation of CD4+ CD25+ regulatory T-cells (Horak et al, 1995), which suppress autoreactive T-cells whether Th1 or Th2 (Shevach, 2002; Malek and Bayer, 2004). The source of IL-2 involved in CD4+ CD25+ T-cell activation has yet to be fully determined, but DC (Granucci et al, 2001) and autoreactive T-cells (Malek and Bayer, 2004) are considered to be involved. CD4+ CD25+ regulatory T-cells are considered to interact with mature DC and suppress helper and effector T-cell activities by an as yet unidentified mechanism (Mills, 2004). Th3 and Tr1 are induced by IL-4 in the presence of TGF-β and IL-10, respectively (Weiner, 2001). Tr1 may also be induced by immature DC or IL-10-modulated DC under TGF-β stimulation (Mahnke et al, 2002; Kapsenberg, 2003). Tr1 secretes IL-10 and TGF-β in a CTLA-4-dependent manner (Roncarolo et al, 2001) and Th3 secretes TGF-β. Repeated stimulation of naïve T-cells in the presence of IL-10 induces Th1 T-cells, and a high dose of IL-10 suppresses the growth of both Th1 and Th2 cells (Read and Powrie, 2001). The mechanism of suppression of CD4+ CD25+ T-cells is actively being investigated, but is considered to involve TGF-β release and binding of CTLA-4 to CD80 and CD86 on effector T-cells (von Boehmer, 2005). The negative feedback loop is mediated by Tr1 and Th3, and CD4+ CD25+ regulatory T-cells constitute feedforward control via mature DC and negative feedback control via autoreactive T-cells that are critical in the proper control of adaptive immune response to prevent autoimmune diseases.
The TLR-mediated innate immune system has a bow-tie architecture in which a wide variety of pathogens and their molecules are represented by much smaller numbers of ligands, called pathogen-associated molecular patterns (PAMPs), recognized by 11 TLRs (Akira et al, 2001; Medzhitov, 2001; K Oda and H Kitano, in preparation) in mammalians. Only a handful of adaptor proteins, including MyD88, MAL/TIRAP, TRIF, and TRAM, and two primary kinases (TBK1 and IRAK) mediate the many responses from TLR activation. MyD88 is a non-redundant core element of the main bow-tie network. Stimulation of TLR signaling culminates with massive transcriptional activation of a broad range of secreted cytokines and chemokines (Beutler, 2004). Details of this organization have been documented in a comprehensive molecular interaction map of TLR signaling (K Oda and H Kitano, in preparation). In addition to this bow-tie structure, several positive and negative feedback loops exert important regulatory actions. Thus, positive feedback loops involving NF-κB play an important role in the dynamics of the immune system in boosting the innate immune response (Li and Verma, 2002) and possibly contributing to its propensity to become hyperactivated. TLR signaling network is fragile against failure of the non-redundant core of this bow-tie structure, which is MyD88. While there are collateral pathways such as MyD88-independent pathway, MyD88 dominates the activation of TLR-mediated responses. Collateral pathways such as MyD88-independent pathways, small GTPase-mediated network, and PIPs seem to modulate different responses to various stimuli (K Oda and H Kitano, in preparation). Observation of MyD88 knockout mice indicates that impeding one of these hub proteins is fatal to the organism because it seriously undermines the function of the innate immune system that fails to detect pathogen-associated molecular signatures (Adachi et al, 1998; Kawai et al, 1999; Csete and Doyle, 2004).
The adaptive immune subsystem displays a clear bow-tie structure at both the cellular interaction and signal transduction levels. As shown in Figure 1A, signals triggered by detection of potential threats are transmitted to naïve CD4+ T-cells by APCs to stimulate their differentiation and cytokines release. Upon recognition of an appropriate peptide–MHC complex and/or cytokine stimuli, naïve CD4+ T-cells polarize into either Th1, Th2, or Tr1 cells depending upon the cytokine stimuli (Moser and Murphy, 2000; Luther and Cyster, 2001; Murphy and Reiner, 2002) that are provided by polarized dendritic cell and a variety of innate immune cells (Kapsenberg, 2003). Owing to the structure of the intercellular interactions that form the bow-tie structure, with naïve CD4+ T-cells as the core of the network, the system is vulnerable to attack on CD4+ helper T-cells. AIDS is the result of an attack on this fragility, as HIV selectively infects CD4+ helper T-cells (McCune, 2001), eventually causing the entire immune system to collapse and make the patient prone to opportunistic infection (McCune, 2001).
At the signal transduction level, the most critical part of the architecture of adaptive immunity is an MHC–TCR-centered bow-tie network structure (Figure 1B). Various exogenous materials are captured by APCs through phagocytosis, macropinocytosis, and fluid-phase endocytosis. APCs also express a broad range of receptors that induce receptor-mediated endocytosis. Captured exogenous materials undergo peptide processing before being loaded onto MHC II and trimmed. The size of loaded peptides on MHC II ranges between 13 and 17 (Rudensky et al, 1991), and only a core of peptides with a length of about 9–10 amino acid epitope binds to a receptor on CD4+ helper T-cells (Brown et al, 1993). A proper binding of TCR to MHC II activates signal transduction pathways, where adaptors such as LAT (a linker for activation of T-cells), Src family PTK, and ZAP-70 serve as critical mediators (Jordan et al, 2003), triggering cytokine secretion and polarization. MHC class I is yet another bow-tie structure where a wide variety of peptides of endogenous origin and with a length of 8–10 amino acids are processed for loading and trimming on MHC I and are recognized by CD8+ T-cells (Yewdell and Bennink, 2001) followed by various responses.
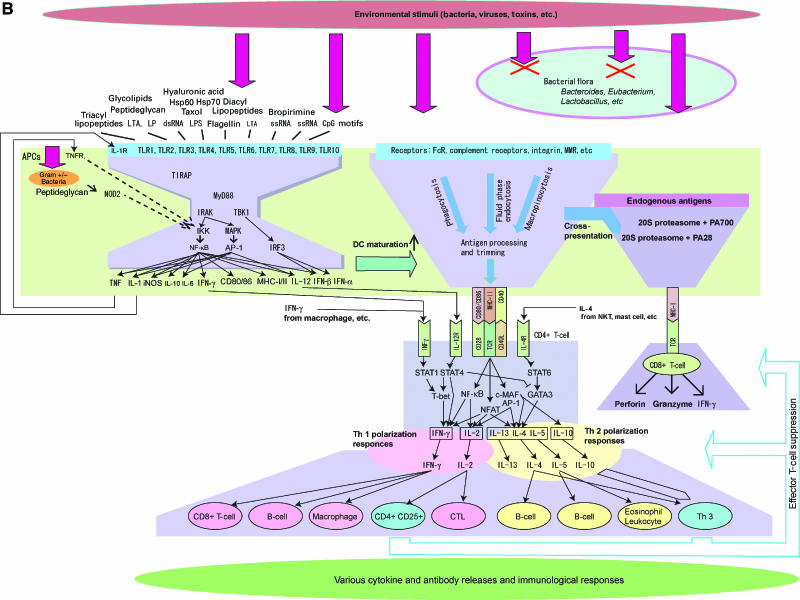
Figure 1b.
(B) The bow-tie architecture also exists at the signal transduction level. There are bow-tie structures in the TLR signaling network and adaptive immunity where antigen presenting and recognition is the hub of the network. APCs express a diverse range of receptors to recognize a wide variety of pathogens via PAMPs, etc. Signaling pathways and capturing processes converge into a smaller variety of core processes. For TLRs, only a handful of adaptor proteins and kinases mediate the signaling process that results in diverse responses including cytokine release, transcription, and other events. The innate immune system comprises TLRs and their downstream cascades that form the characteristic shape of the network called a bow-tie, or hour-glass, structure (Csete and Doyle, 2004; Kitano, 2004). A wide variety of pathogens and their molecules are represented by much smaller numbers of ligands, called PAMPs, recognized by IL-1R and 10 TLRs (Akira et al, 2001; Medzhitov, 2001). Only a handful of adaptor proteins, such as MyD88, MAL/TIRAP, TRIF, and TRAM, and two primary kinases (TBK1 and IRAK) mediate various responses and trigger massive changes in a broad range of transcription of target genes including secretion of cytokines and chemokines (Beutler, 2004). MyD88 has special importance as it constitutes a non-redundant core element of the main bow-tie network. Although there are collateral pathways such as MyD88-independent pathway, MyD88 is largely responsible for activation of TLR-mediated responses. Collateral pathways play roles in modulating different responses. This is a versatile architecture that enables organisms to evolutionarily add a new kind of receptor for new molecular signatures that eventually activate TBK1 and IRAK to bring about some responses. This is a bow-tie architecture at the signaling pathway, and exists in a broad range of somatic cells, but most saliently in macrophage and DC. In this bow-tie structure, there are nested bow-tie structures as well as positive feedback loops. For example, NF-κB and TNF form bow-tie structures having extensive input signals and changes in transcription and cytokine production (Li and Verma, 2002). Positive feedback loops involving NF-κB play an important role in the dynamics of the immune system in boosting innate immune response. An antigen-processing and presentation process captures a variety of peptides and presents them on MHC-I or MHC-II molecules that are recognized by TCRs. This antigen presentation and recognition part is a critical core process in adaptive immunity. A breach in proper presentation and recognition leads to improper immune reactions such as autoimmune diseases. Details of TLR signaling network are reported elsewhere (K Oda and H Kitano, in preparation).
On the one hand, this architecture enhances the robustness of the system by enabling it to cope efficiently with a broader range of pathogens with limited resources. On the other hand, it entails inherent trade-offs when the core of the bow-tie architecture is breached. Any confusion at the stage of MHC presentation and TCR recognition results in systemic disorders of the immune system. Thus, deficient removal of crossreactive T-cells by peripheral tolerance mechanisms may result in autoimmunity. The use of relatively short antigen sequences (less than 10 core amino acids) by the adaptive immune system inevitably results in cases of molecular mimicry when a presented antigen closely matches the host's own signature. Pathogens may trigger autoimmune responses if their molecular signature is sufficiently similar to one of the host tissue signatures. In fact, some autoimmune diseases are now identified as being triggered by bacterial infection, such as Crohn's disease (CD) (Kobayashi et al, 2005; Maeda et al, 2005), which is an intestine autoimmune disease that causes chronic inflammation. Dilated cardiomyopathy is associated with cardiotropic virus infection triggering dendritic cell-induced autoimmune heart failure (Eriksson et al, 2003). It has been argued that the broad range of autoimmune disorders discussed above, as well as rheumatoid arthritis (RA), systemic lupus erythematosus, and others, are due to multiple exposure to pathogenic bacteria and viruses (von Herrath et al, 2003). This class of autoimmune diseases can be attributed to breaches of the immune response against pathogens, by which invaded pathogens trigger a sustained immune response by molecular mimicry (Baum et al, 1996), possibly with TCR-dependent bystander activation (von Herrath et al, 2003).
Theoretically, cross-activation via molecular mimicry should have a significantly lower rate if longer peptides are presented to TCR. However, a significantly large number of T-cells need to be prepared to properly react to a greater variety of peptides presented. Otherwise, serious immunodeficiency will take place, as only a fraction of peptides can be recognized by the limited number of T-cells. Thus, MHC-peptide–TCR recognition is constrained by trade-offs between robustness against a broader range of pathogens, fragility against misrecognition, and resource limitations of T-cells that can be supplied.
Commensal bacterial flora as an integral part of the host defense system
Bacteria have been traditionally considered as ‘non-self', and thus subject to rejection by the immune system. However, the fact is that certain species of bacteria successfully colonize our body, and even provide us with essential functions without which we could not survive. Commensal bacterial flora are ubiquitously observed in various metazoan species, including termites (Schmitt-Wagner et al, 2003), cockroaches (Bracke et al, 1979), prawns (Oxley et al, 2002), and mammalians, and have established inseparable relationships with the host organisms, and are even considered to have co-evolved (Backhed et al, 2005). In human beings, the intestinal microbiota contains 500–1000 species of diverse microorganisms and about 1014 bacteria, totaling about 1.5 kg of biomass (Xu and Gordon, 2003). It is breathtaking to realize that the human as a symbiotic system consists of approximately 90% prokaryotes and 10% eukaryotes (Savage, 1977), and that a random shotgun sequencing of the whole human symbiotic system would result in predominantly bacterial genome readouts of about 2 million genes with sporadic mammalian genes (Hooper et al, 2002). Such commensal intestinal bacteria play a critical role in various aspects of host physiology.
First, it is essential for gut development. Germ-free mice that have no commensal bacterial flora have an undeveloped mucosal immune system that has hypoplastic Peyer's patches, as well as significantly reduced numbers of IgA-producing plasma cells and lamina propria CD4+ T-cells (Macpherson et al, 2001, 2002). A recent study on one commensal bacterial species, Bacteroides thetaiotaomicron, revealed that it stimulates angiogenesis during postnatal intestine development to enhance the nutrient-absorbing capability (Stappenbeck et al, 2002).
Second, commensal bacteria even function antagonistically against pathogenic bacteria, a phenomenon known as colonization resistance. Continuous-flow experiments using a smaller subset of commensal bacteria revealed that this effect is due to competition for nutrients and spaces that are sustained by the dynamics of an intermicrobial metabolic network (Ushijima and Ozaki, 1986, 1988). Perturbation of commensal bacteria due to the use of antibiotics causes antibiotic-associated diarrhea (Bergogne-Berezin, 2000) by allowing pathogenic bacteria such as Clostridium difficile to proliferate. As yet another example, inflammatory bowel diseases are considered to be caused by excessive activation of the cell-mediated immune system to commensal enteric bacteria (Sartor, 2003). This is perhaps due to reduced biodiversity of the flora, as indicated by a dramatic increase in certain types of bacteria (Swidsinski et al, 2002).
Third, intestinal bacterial flora contributes to the host's nutrient supply by, for example, contributing to starch digestion, amino-acid homeostasis, and vitamin synthesis (Hooper et al, 2002). Certain metabolic interactions are so tightly coupled that metabolites are produced in which neither host nor bacteria alone can produce (Nicholson and Wilson, 2003; Nicholson et al, 2005). As the composition of bacterial flora changes dynamically as a function of changes in the dietary and nutrient contents (Harmsen et al, 2000; Mai and Morris, 2004), it is essential for the host to be able to harbor a broad range of bacteria to enable its flora to cope with environmental perturbations (Figure 2).
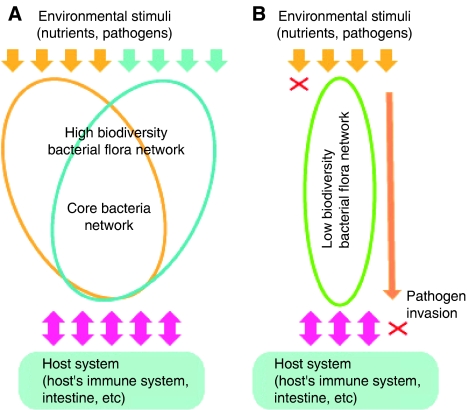
Figure 2.
Bacterial flora biodiversity improves robustness against environmental perturbations. (A) Upon changes in environmental conditions, such as changes in types of foods and pathogens, the host that can accommodate highly diverse bacterial flora is able to cope with such changes by composition changes of bacterial flora. (B) The host that only allows a narrow set of bacteria, hence low biodiversity bacterial flora, may not be able to cope with changes in food compositions and pathogens. This results in failure to digest certain types of foods and invasion of pathogenic bacteria to the host tissues.
A genomic study revealed that the flora can even manipulate host gene expression to establish mutually advantageous partnerships (Hooper et al, 2001; Xu et al, 2003). Not only does the bacterial flora affect the host, but the host's genotype (Zoetendal et al, 2001) and immunological responses (Hornef et al, 2002) also reciprocally affect the activity and composition of the flora. Therefore, the host immune system does not ignore the bacterial flora, but, rather, it maintains complex and dynamic interactions with the commensal bacteria, which results in a mutually beneficial symbiotic state (Hornef et al, 2002). Owing to the intricate relationship between bacterial flora and the host, some believe that flora should even be considered as an ‘organ', rather than an unwanted guest (Hooper et al, 2002). The symbiotic association actually provides enhanced robustness against environmental perturbations by stimulating immune system development, nutrient processing and biosynthesis, and resistance to pathogenic microbes. These findings suggest that bacterial flora are an essential part of the system, and that the immune system might have co-evolved to allow diverse symbionts to reside on the host without compromising the defense of the host against pathogens. The fact that the defense system is a symbiotic system implies that host defense functions are distributed between the host system and the symbiont system, so that perturbations of either one of these systems impedes the overall function of the immune system. It is critically important to obtain a better understanding of this complex ecosystem in order to improve our understanding of its relevance in disease outbreak, prevention, and therapeutic control.
Medical implications
There are two theoretical frameworks that we must recognize when discussing the medical implications of the arguments developed so far. First, there is an inherent and inescapable trade-off among robustness, resource limitations, performance, and fragility of the system (Carlson and Doyle, 2000, 2002; Csete and Doyle, 2004; Kitano, 2004) that is exemplified in two typical problems of the immune system: (1) the trade-off between the robustness required to cope with a broad range of pathogens and the limited resources available to the system results in the risk of misrecognition due to short peptide length for MHC presentation in adaptive immunity, and (2) breaches of hubs within the bow-tie architecture render the mammalian immune systems susceptible to attacks of CD4+ T-cells by HIV or to mutations of MyD88. Second, if the commensal bacterial flora constitutes an integral part of the host protection system together with the host immune system, it is possible that the perturbation of bacterial flora homeostasis may undermine the overall host defense immune function. It is important to link these issues to illustrate a global perspective of the immune system because problems of host immune systems, such as autoimmunity and fragility against specific component failures, are deeply embedded within the architectural feature of the immune system. In fact, these observations indicate that maintenance of homeostasis of commensal bacterial flora and host immunity should be a high-priority target for the prevention and mitigation of immunological disorders.
In fact, there are increasing reports indicating a close relationship between infections and pathogenesis and mitigation of immunological disorders (Bach, 2002, 2005), as well as specific observations on bacterial flora. For example, a recent study revealed that the diversity of bacterial flora in CD patients and ulcerative colitis was reduced by 50 and 30% compared to the healthy control group, respectively, and that such reduction of diversity was attributed to the loss of normal anaerobic bacteria including Bacteroides species, Eubacterium species, and Lactobacillus species (Ott et al, 2004). These bacteria that are significantly lost in the population are consistent with specific species that are observed to have high intradivision biodiversity (Backhed et al, 2005), implying that this could disrupt the essential part of the bacterial flora network. An extensive concentration of bacteria such as Mycobacterium paratuberculosis and Listeria monocytogenes has been observed by biopsy of CD patients using 16S rDNA PCR and DNA hybridization analysis (Tiveljung et al, 1999). Combined with the genetic susceptibility of the subpopulation of CD patients with NOD2 mutation (Kobayashi et al, 2005; Maeda et al, 2005), perturbations of bacterial flora might have impeded the ability of bacterial flora to suppress such pathogenic bacteria and allowed them to grow and invade. However, due to the highly interactive nature of bacterial flora and the mucosal immune system, it is unclear whether such reduction in biodiversity of flora is part of the cause or the result of disease.
Nevertheless, enhancing the robustness of bacterial flora for disease prevention and cure may be a viable approach, as it is tightly coupled with mucosal immune responses. One way to accomplish this is to use probiotics to restore the balance of commensals (Sartor, 2005; Sullivan and Nord, 2005). Promising results have been reported for refractory pouchitis, an inflammation in the reservoirs, where clinical studies have documented that chronic administration of VSL, a combination of four Lactorbacillus species, three Bifidobacterium species, and Streptococcus salivarium subspecies, has been effective in preventing relapse of refractory pouchitis (Gionchetti et al, 2000, 2003). For CD-related experiments, it has been reported that production of IL-10 has been found to be elevated with the administration of VSL in vitro, and ex vivo treatment of mucosal tissues from active CD patients shows reduced TNF production with some Lactobacillus species (Sartor, 2003). However, the clinical outcome of probiotics treatment of CD patients has had mixed results (Sartor, 2005). On the contrary, control of bacterial flora could be a more effective means of prevention by mitigating the risk of invasion of pathogenic bacteria by enhancing intestinal epithelial barrier functions (Madsen et al, 2001) and suppressing the proliferation and attachment of pathogenic bacteria to the intestine (Sartor, 2005).
Such approaches are worth pursuing, as current therapies focus on suppression of immunological response by suppressing TNF-α function, which was shown to be effective in mitigating chronic inflammation and autoimmune diseases including RA (Feldmann et al, 2004) and CD, but is associated with the risk of side effects (Day, 2002; Feldmann and Maini, 2002). The effectiveness and risk of TNF-α target therapy are predictable, as TNF-α is one of the major cytokine-activating immune systems secreted by cells such as macrophage for innate immune responses, as well as a major component of a positive feedback loop in cytokine networks activating TLR-driven immune responses. However, the problem is that the hyperactive immune response is caused by, at least in certain subpopulations of patients, compromised immune response (due to reduced biodiversity of bacterial flora and mutations in innate immune systems) followed by molecular mimicry. Thus, TNF-α therapy leaves this root cause of the disease untouched. With the discovery of TLRs, a range of therapies to selectively inhibit TLRs have been proposed (Zuany-Amorim et al, 2002). Although this provides us with a variety of approaches to achieve higher levels of specificity, it is not without side effects, because these therapies essentially reduce the immune responses to a certain range of pathogenic targets. Owing to the specificity of perturbations, side effects could be less serious than TNF-α inhibitors, but they still undermine normal immune functions. An even better approach for neutralizing hyperactivity is to selectively mitigate the inflammatory reaction itself without suppressing immune reactions, but such work is still in the early stage (Nathan, 2002; Gilroy et al, 2004) and would not remedy the root cause. If autoimmune responses are viewed as an inherent architectural problem of the immune system, preventive strategies might be more appropriate to successfully tackle the causes of this class of immunological disorders. Therefore, approaches to prevent and mitigate autoimmune disorders by controlling biodiversity of bacterial flora have the potential to provide significant benefits for the health of patient and potential patient populations.
The structure and dynamics of the bacterial flora network have to be identified
Although bacterial flora affects the pathogenesis and treatment of inflammatory diseases, the major problem is that the complexity and dynamics of bacterial flora and their interaction with pathogens and the mucosal immune system are largely unknown. Unexpected and counterintuitive dynamics often associated with complex networks may be a cause of the mixed results.
First, the composition of microorganisms constituting bacterial flora has yet to be identified. It was only in the last 5 years that genomic analysis using cloned 16S rRNA gene (rDNA) sequences has become available to identify the composition of bacteria for the significant number of bacterial species that cannot be cultured (Suau et al, 1999). Although enormous biodiversity involving over 500–1000 species is well accepted, a recent study revealed the existence of extremely dense biodiversity within selected divisions such as Cytophage–Flavobacterium–Bacteroides and the Firmicutes (Backhed et al, 2005). This implies a highly redundant network that ensures robustness upon reduction of biodiversity, as postulated by the insurance hypothesis (Yachi and Loreau, 1999).
Second, the kinds of interaction networks that bacterial flora form have not yet been identified. Although it is understood that diverse species of bacteria constitute bacterial flora and that they interact by exchanging metabolic products and nutrients as well as competing for space and resources, the structure of the networks that such interactions generate is largely unknown. Furthermore, interactions are taking place not only among bacteria, but also between bacteria and host tissues. Depending upon the structure of the network, its dynamics can be dramatically different (Li et al, 2004b). There are some cues that enable us to speculate on the possible network structures. First, a partial profile of the diversity of bacteria composition has recently been revealed, as discussed above. Second, there is resource competition between bacterial species over nutrition and spaces. Substances that are preferred for metabolism by a broader range of bacteria are quickly consumed, precluding limitless growth of interactions over such substances; hence preferential attachment essential for a scale-free network (Barabasi and Oltvai, 2004) may not take place. In fact, the food web that also involves resource competition and metabolic flow was shown to be neither a random network nor a small-world scale-free network (Dunne et al, 2002). Third, pressure exists for optimization of the bacterial flora network toward its stable functioning. The metabolic network of bacterial flora has to efficiently digest incoming substances and convert them into a form suitable for absorption by the gut, synthesize nutrients that cannot be produced by the host alone, and resist invasion by pathogenic microbes. Failure to accomplish such tasks may cause inflammation and diarrhea that impede growth of the bacterial flora. This implies that only bacterial flora networks that are optimized, or suboptimized, for certain functions can persist, whereas networks that fail to function properly will be impeded and reconstructed.
Although it is not possible to infer the possible structure conclusively, it is often the case that the global structure of a network that is being optimized, whether by evolution or by design, entails highly optimized tolerance (HOT) properties (Carlson and Doyle, 2000, 2002; Li et al, 2004a) and takes a bow-tie structure (Csete and Doyle, 2004; Kitano, 2004) in which a highly conserved and robust core network ensures efficient processing of diverse inputs and generation of diverse outputs. Such a network structure is also observed in bacterial metabolic networks (Ma and Zeng, 2003; Tanaka, 2005). In the bacterial flora network, highly redundant clusters of species and strains under selected divisions form a conserved robust core network, and diverse bacterial species interface with this core network. In this regard, the bacterial flora network may resemble a bow-tie structure, but is also affected by competition among bacterial species and the nature of pressures imposed. If the network actually has a bow-tie structure, the metabolic network of bacterial flora and host tissue can be highly robust against elimination of bacterial species. This is in contrast to the scale-free network, where selective removal of hubs significantly degrades its performance. In addition, a bow-tie metabolic network shall be highly efficient, whereas the scale-free network may suffer from the bottleneck of a hub node capacity. The excessive use of antibiotics impedes bacterial flora regardless of network topology as their effects are nonspecific and eliminates a significant portion of bacterial flora. Although bow-tie networks with HOT properties are robust, efficient, and scale rich, they can be fragile when specific pathogenic bacterial species take over the flora, while maintaining its metabolic functions, and decoy possible molecular interactions to fake non-pathogenic bacteria (Li et al, 2004a).
Conclusion
Our assumption in this article is that commensal bacterial flora and the immune system create a state of symbiosis that effectively provides host defense against various pathogens, and that the involvement of commensal bacterial flora is pivotal to better accomplish this function through potential adaptability to environmental changes, thus increasing the robustness of the host–symbiont system.
This notation may have an interesting implication for the function of the immune system, namely, what is the immune system optimized for? The logical consequence of our assumption is that the immune system is optimized to provide robustness to the host–symbiont system, instead of providing defense to the host organism alone, against a variety of pathogenic threats from the environment. This means that the immune system enhances the robustness of the organisms by enabling them to harbor a broader range of symbionts, yet rejecting pathogenic microorganisms. The premise of this hypothesis is that harboring a broader range of symbionts increases the robustness of the host organism by enabling a spectrum of adaptive responses against changes in dietary and nutrient contents and exposure to pathogenic microorganisms.
Owing to inherent trade-offs of the host immune system, it is critically important that bacterial flora homeostasis is properly maintained to prevent and mitigate immunological disorders. Although there have been sporadic reports on the possible impacts of controlling bacterial flora to counter autoimmune diseases and allergies, no convincing result has yet been reported. This is perhaps owing to our lack of understanding of this highly complex ecosystem of bacterial flora that also interacts with the host and the environment. The future lies in a better understanding of this biological continuum of the internal and external ecosystems. Interestingly, recent developments in metagenome (or systems ecology) research (Venter et al, 2004; Delong, 2005) may provide us with tools and concepts to answer the question.
Acknowledgments
This research was supported in part by the Exploratory Research for Advanced Technology (ERATO) and the Solution-Oriented Research for Science and Technology (SORST) programs (Japan Science and Technology Organization), a NEDO (New Energy and Industrial Technology Development Organization) grant of the Japanese Ministry of Economy, Trade and Industry (METI), the Special Coordination Funds for Promoting Science and Technology and the Center of Excellence Program for Keio University (Ministry of Education, Culture, Sports, Science, and Technology), and The Genome Network Project by the Ministry of Education, Culture, Sports, Science, and Technology.
Conflict of interest:The authors declare no financial conflicts.
References
- Abbas AK, Murphy KM, Sher A (1996) Functional diversity of helper T lymphocytes. Nature 383: 787–793 [DOI] [PubMed] [Google Scholar]
- Adachi O, Kawai T, Takeda K, Matsumoto M, Tsutsui H, Sakagami M, Nakanishi K, Akira S (1998) Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity 9: 143–150 [DOI] [PubMed] [Google Scholar]
- Akira S, Takeda K, Kaisho T (2001) Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol 2: 675–680 [DOI] [PubMed] [Google Scholar]
- Bach JF (2002) The effect of infections on susceptibility to autoimmune and allergic diseases. N Engl J Med 347: 911–920 [DOI] [PubMed] [Google Scholar]
- Bach JF (2005) The protective effect of infections on immune disorders. J Pediatr Gastroenterol Nutr 40 (Suppl 1): S8 [DOI] [PubMed] [Google Scholar]
- Backhed F, Ley R, Sonnenburg J, Peterson D, Gordon JI (2005) Host–bacterial mutualism in the human intestine. Science 307: 1915–1920 [DOI] [PubMed] [Google Scholar]
- Barabasi AL, Oltvai ZN (2004) Network biology: understanding the cell's functional organization. Nat Rev Genet 5: 101–113 [DOI] [PubMed] [Google Scholar]
- Baum H, Davies H, Peakman M (1996) Molecular mimicry in the MHC: hidden clues to autoimmunity? Immunol Today 17: 64–70 [DOI] [PubMed] [Google Scholar]
- Bergogne-Berezin E (2000) Treatment and prevention of antibiotic associated diarrhea. Int J Antimicrob Agents 16: 521–526 [DOI] [PubMed] [Google Scholar]
- Beutler B (2004) Inferences, questions and possibilities in Toll-like receptor signalling. Nature 430: 257–263 [DOI] [PubMed] [Google Scholar]
- Bracke JW, Cruden DL, Markovetz AJ (1979) Intestinal microbial flora of the of the American cockroach, Periplaneta americana L. Appl Environ Microbiol 38: 945–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JH, Jardetzky TS, Gorga JC, Stern LJ, Urban RG, Strominger JL, Wiley DC (1993) Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature 364: 33–39 [DOI] [PubMed] [Google Scholar]
- Carlson JM, Doyle J (2000) Highly optimized tolerance: robustness and design in complex systems. Phys Rev Lett 84: 2529–2532 [DOI] [PubMed] [Google Scholar]
- Carlson JM, Doyle J (2002) Complexity and robustness. Proc Natl Acad Sci USA 99 (Suppl 1): 2538–2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csete M, Doyle J (2004) Bow ties, metabolism and disease. Trends Biotechnol 22: 446–450 [DOI] [PubMed] [Google Scholar]
- Day R (2002) Adverse reactions to TNF-alpha inhibitors in rheumatoid arthritis. Lancet 359: 540–541 [DOI] [PubMed] [Google Scholar]
- Delong EF (2005) Microbial community genomics in the ocean. Nat Rev Microbiol 3: 459–469 [DOI] [PubMed] [Google Scholar]
- Dunne JA, Williams RJ, Martinez ND (2002) Food-web structure and network theory: the role of connectance and size. Proc Natl Acad Sci USA 99: 12917–12922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson U, Ricci R, Hunziker L, Kurrer MO, Oudit GY, Watts TH, Sonderegger I, Bachmaier K, Kopf M, Penninger JM (2003) Dendritic cell-induced autoimmune heart failure requires cooperation between adaptive and innate immunity. Nat Med 9: 1484–1490 [DOI] [PubMed] [Google Scholar]
- Feldmann M, Brennan FM, Paleolog E, Cope A, Taylor P, Williams R, Woody J, Maini RN (2004) Anti-TNFalpha therapy of rheumatoid arthritis: what can we learn about chronic disease? Novartis Found Symp 256: 53–69; discussion 69–73, 106–111, 266–109 [PubMed] [Google Scholar]
- Feldmann M, Maini RN (2002) Discovery of TNF-alpha as a therapeutic target in rheumatoid arthritis: preclinical and clinical studies. Joint Bone Spine 69: 12–18 [DOI] [PubMed] [Google Scholar]
- Gilroy DW, Lawrence T, Perretti M, Rossi AG (2004) Inflammatory resolution: new opportunities for drug discovery. Nat Rev Drug Discov 3: 401–416 [DOI] [PubMed] [Google Scholar]
- Gionchetti P, Rizzello F, Helwig U, Venturi A, Lammers KM, Brigidi P, Vitali B, Poggioli G, Miglioli M, Campieri M (2003) Prophylaxis of pouchitis onset with probiotic therapy: a double-blind, placebo-controlled trial. Gastroenterology 124: 1202–1209 [DOI] [PubMed] [Google Scholar]
- Gionchetti P, Rizzello F, Venturi A, Brigidi P, Matteuzzi D, Bazzocchi G, Poggioli G, Miglioli M, Campieri M (2000) Oral bacteriotherapy as maintenance treatment in patients with chronic pouchitis: a double-blind, placebo-controlled trial. Gastroenterology 119: 305–309 [DOI] [PubMed] [Google Scholar]
- Granucci F, Vizzardelli C, Pavelka N, Feau S, Persico M, Virzi E, Rescigno M, Moro G, Ricciardi-Castagnoli P (2001) Inducible IL-2 production by dendritic cells revealed by global gene expression analysis. Nat Immunol 2: 882–888 [DOI] [PubMed] [Google Scholar]
- Harmsen HJ, Wildeboer-Veloo AC, Raangs GC, Wagendorp AA, Klijn N, Bindels JG, Welling GW (2000) Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J Pediatr Gastroenterol Nutr 30: 61–67 [DOI] [PubMed] [Google Scholar]
- Hooper LV, Midtvedt T, Gordon JI (2002) How host–microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr 22: 283–307 [DOI] [PubMed] [Google Scholar]
- Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI (2001) Molecular analysis of commensal host–microbial relationships in the intestine. Science 291: 881–884 [DOI] [PubMed] [Google Scholar]
- Horak I, Lohler J, Ma A, Smith KA (1995) Interleukin-2 deficient mice: a new model to study autoimmunity and self-tolerance. Immunol Rev 148: 35–44 [DOI] [PubMed] [Google Scholar]
- Hornef MW, Wick MJ, Rhen M, Normark S (2002) Bacterial strategies for overcoming host innate and adaptive immune responses. Nat Immunol 3: 1033–1040 [DOI] [PubMed] [Google Scholar]
- Jordan MS, Singer AL, Koretzky GA (2003) Adaptors as central mediators of signal transduction in immune cells. Nat Immunol 4: 110–116 [DOI] [PubMed] [Google Scholar]
- Kapsenberg ML (2003) Dendritic-cell control of pathogen-driven T-cell polarization. Nat Rev Immunol 3: 984–993 [DOI] [PubMed] [Google Scholar]
- Kawai T, Adachi O, Ogawa T, Takeda K, Akira S (1999) Unresponsiveness of MyD88-deficient mice to endotoxin. Immunity 11: 115–122 [DOI] [PubMed] [Google Scholar]
- Kitano H (2004) Biological robustness. Nat Rev Genet 5: 826–837 [DOI] [PubMed] [Google Scholar]
- Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, Nunez G, Flavell RA (2005) Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science 307: 731–734 [DOI] [PubMed] [Google Scholar]
- Li L, Alderson D, Tanaka R, Doyle J, Willinger W (2004a) Towards a Theory of Scale-free Graphs: Definition, Properties, and Implications (Extended Version). Pasadena, CA: California Institute of Technology [Google Scholar]
- Li L, Alderson D, Willinger W, Doyle J (2004b) A first-principle approach to understand the Internet's Router-level topology. Proceedings of ACM Sigcomm. Portland, OR: ACM [Google Scholar]
- Li Q, Verma IM (2002) NF-kappaB regulation in the immune system. Nat Rev Immunol 2: 725–734 [DOI] [PubMed] [Google Scholar]
- Liew FY (2002) T(H)1 and T(H)2 cells: a historical perspective. Nat Rev Immunol 2: 55–60 [DOI] [PubMed] [Google Scholar]
- Luther SA, Cyster JG (2001) Chemokines as regulators of T cell differentiation. Nat Immunol 2: 102–107 [DOI] [PubMed] [Google Scholar]
- Ma HW, Zeng AP (2003) The connectivity structure, giant strong component and centrality of metabolic networks. Bioinformatics 19: 1423–1430 [DOI] [PubMed] [Google Scholar]
- Macpherson AJ, Hunziker L, McCoy K, Lamarre A (2001) IgA responses in the intestinal mucosa against pathogenic and non-pathogenic microorganisms. Microbes Infect 3: 1021–1035 [DOI] [PubMed] [Google Scholar]
- Macpherson AJ, Martinic MM, Harris N (2002) The functions of mucosal T cells in containing the indigenous commensal flora of the intestine. Cell Mol Life Sci 59: 2088–2096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen K, Cornish A, Soper P, McKaigney C, Jijon H, Yachimec C, Doyle J, Jewell L, De Simone C (2001) Probiotic bacteria enhance murine and human intestinal epithelial barrier function. Gastroenterology 121: 580–591 [DOI] [PubMed] [Google Scholar]
- Maeda S, Hsu LC, Liu H, Bankston LA, Iimura M, Kagnoff MF, Eckmann L, Karin M (2005) Nod2 mutation in Crohn's disease potentiates NF-kappaB activity and IL-1beta processing. Science 307: 734–738 [DOI] [PubMed] [Google Scholar]
- Mahnke K, Schmitt E, Bonifaz L, Enk AH, Jonuleit H (2002) Immature, but not inactive: the tolerogenic function of immature dendritic cells. Immunol Cell Biol 80: 477–483 [DOI] [PubMed] [Google Scholar]
- Mai V, Morris JG Jr (2004) Colonic bacterial flora: changing understandings in the molecular age. J Nutr 134: 459–464 [DOI] [PubMed] [Google Scholar]
- Malek TR, Bayer AL (2004) Tolerance, not immunity, crucially depends on IL-2. Nat Rev Immunol 4: 665–674 [DOI] [PubMed] [Google Scholar]
- McCune JM (2001) The dynamics of CD4+ T-cell depletion in HIV disease. Nature 410: 974–979 [DOI] [PubMed] [Google Scholar]
- Medzhitov R (2001) Toll-like receptors and innate immunity. Nat Rev Immunol 1: 135–145 [DOI] [PubMed] [Google Scholar]
- Mills KH (2004) Regulatory T cells: friend or foe in immunity to infection? Nat Rev Immunol 4: 841–855 [DOI] [PubMed] [Google Scholar]
- Moser M, Murphy KM (2000) Dendritic cell regulation of TH1–TH2 development. Nat Immunol 1: 199–205 [DOI] [PubMed] [Google Scholar]
- Murphy KM, Reiner SL (2002) The lineage decisions of helper T cells. Nat Rev Immunol 2: 933–944 [DOI] [PubMed] [Google Scholar]
- Nathan C (2002) Points of control in inflammation. Nature 420: 846–852 [DOI] [PubMed] [Google Scholar]
- Nicholson JK, Holmes E, Wilson ID (2005) Gut microorganisms, mammalian metabolism and personalized health care. Nat Rev Microbiol 3: 431–438 [DOI] [PubMed] [Google Scholar]
- Nicholson JK, Wilson ID (2003) Opinion: understanding ‘global' systems biology: metabonomics and the continuum of metabolism. Nat Rev Drug Discov 2: 668–676 [DOI] [PubMed] [Google Scholar]
- Ott SJ, Musfeldt M, Wenderoth DF, Hampe J, Brant O, Folsch UR, Timmis KN, Schreiber S (2004) Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut 53: 685–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxley AP, Shipton W, Owens L, McKay D (2002) Bacterial flora from the gut of the wild and cultured banana prawn, Penaeus merguiensis . J Appl Microbiol 93: 214–223 [DOI] [PubMed] [Google Scholar]
- Read S, Powrie F (2001) CD4(+) regulatory T cells. Curr Opin Immunol 13: 644–649 [DOI] [PubMed] [Google Scholar]
- Roncarolo MG, Bacchetta R, Bordignon C, Narula S, Levings MK (2001) Type 1T regulatory cells. Immunol Rev 182: 68–79 [DOI] [PubMed] [Google Scholar]
- Rudensky A, Preston-Hurlburt P, Hong SC, Barlow A, Janeway CA Jr (1991) Sequence analysis of peptides bound to MHC class II molecules. Nature 353: 622–627 [DOI] [PubMed] [Google Scholar]
- Sartor RB (2003) Targeting enteric bacteria in treatment of inflammatory bowel diseases: why, how, and when. Curr Opin Gastroenterol 19: 358–365 [DOI] [PubMed] [Google Scholar]
- Sartor RB (2005) Probiotic therapy of intestinal inflammation and infections. Curr Opin Gastroenterol 21: 44–50 [PubMed] [Google Scholar]
- Savage DC (1977) Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol 31: 107–133 [DOI] [PubMed] [Google Scholar]
- Schmitt-Wagner D, Friedrich MW, Wagner B, Brune A (2003) Phylogenetic diversity, abundance, and axial distribution of bacteria in the intestinal tract of two soil-feeding termites (Cubitermes spp.). Appl Environ Microbiol 69: 6007–6017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevach EM (2002) CD4+ CD25+ suppressor T cells: more questions than answers. Nat Rev Immunol 2: 389–400 [DOI] [PubMed] [Google Scholar]
- Stappenbeck TS, Hooper LV, Gordon JI (2002) Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci USA 99: 15451–15455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelling J, Sauer U, Szallasi Z, Doyle FJ III, Doyle J (2004) Robustness of cellular functions. Cell 118: 675–685 [DOI] [PubMed] [Google Scholar]
- Suau A, Bonnet R, Sutren M, Godon JJ, Gibson GR, Collins MD, Dore J (1999) Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl Environ Microbiol 65: 4799–4807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan A, Nord CE (2005) Probiotics and gastrointestinal diseases. J Intern Med 257: 78–92 [DOI] [PubMed] [Google Scholar]
- Swidsinski A, Ladhoff A, Pernthaler A, Swidsinski S, Loening-Baucke V, Ortner M, Weber J, Hoffmann U, Schreiber S, Dietel M, Lochs H (2002) Mucosal flora in inflammatory bowel disease. Gastroenterology 122: 44–54 [DOI] [PubMed] [Google Scholar]
- Tanaka R (2005) Scale-rich metabolic networks. Phys Rev Lett 94: 168101 [DOI] [PubMed] [Google Scholar]
- Tiveljung A, Soderholm JD, Olaison G, Jonasson J, Monstein HJ (1999) Presence of eubacteria in biopsies from Crohn's disease inflammatory lesions as determined by 16S rRNA gene-based PCR. J Med Microbiol 48: 263–268 [DOI] [PubMed] [Google Scholar]
- Ushijima T, Ozaki Y (1986) Potent antagonism of Escherichia coli, Bacteroides ovatus, Fusobacterium varium, and Enterococcus faecalis, alone or in combination, for enteropathogens in anaerobic continuous flow cultures. J Med Microbiol 22: 157–163 [DOI] [PubMed] [Google Scholar]
- Ushijima T, Ozaki Y (1988) Factors influencing potent antagonistic effects of Escherichia coli and Bacteroides ovatus on Staphylococcus aureus in anaerobic continuous flow cultures. Can J Microbiol 34: 645–650 [DOI] [PubMed] [Google Scholar]
- Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parsons R, Baden-Tillson H, Pfannkoch C, Rogers YH, Smith HO (2004) Environmental genome shotgun sequencing of the Sargasso Sea. Science 304: 66–74 [DOI] [PubMed] [Google Scholar]
- von Boehmer H (2005) Mechanisms of suppression by suppressor T cells. Nat Immunol 6: 338–344 [DOI] [PubMed] [Google Scholar]
- von Herrath MG, Fujinami RS, Whitton JL (2003) Microorganisms and autoimmunity: making the barren field fertile? Nat Rev Microbiol 1: 151–157 [DOI] [PubMed] [Google Scholar]
- Weiner HL (2001) The mucosal milieu creates tolerogenic dendritic cells and T(R)1 and T(H)3 regulatory cells. Nat Immunol 2: 671–672 [DOI] [PubMed] [Google Scholar]
- Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, Hooper LV, Gordon JI (2003) A genomic view of the human–Bacteroides thetaiotaomicron symbiosis. Science 299: 2074–2076 [DOI] [PubMed] [Google Scholar]
- Xu J, Gordon JI (2003) Inaugural article: honor thy symbionts. Proc Natl Acad Sci USA 100: 10452–10459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yachi S, Loreau M (1999) Biodiversity and ecosystem productivity in a fluctuating environment: the insurance hypothesis. Proc Natl Acad Sci USA 96: 1463–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yewdell JW, Bennink JR (2001) Cut and trim: generating MHC class I peptide ligands. Curr Opin Immunol 13: 13–18 [DOI] [PubMed] [Google Scholar]
- Zoetendal EG, Akkermanns AD, Akkermanns-van Vliet WM, Arjan G, de Visser GM, de Vos WM (2001) The host genotype affects the bacterial community in the human gastrointestinal trace. Microb Ecol Health Dis 13: 129–134 [Google Scholar]
- Zuany-Amorim C, Hastewell J, Walker C (2002) Toll-like receptors as potential therapeutic targets for multiple diseases. Nat Rev Drug Discov 1: 797–807 [DOI] [PubMed] [Google Scholar]