Abstract
Neuronal synapses play fundamental roles in information processing, behaviour and disease. Neurotransmitter receptor complexes, such as the mammalian N-methyl-D-aspartate receptor complex (NRC/MASC) comprising 186 proteins, are major components of the synapse proteome. Here we investigate the organisation and function of NRC/MASC using a systems biology approach. Systematic annotation showed that the complex contained proteins implicated in a wide range of cognitive processes, synaptic plasticity and psychiatric diseases. Protein domains were evolutionarily conserved from yeast, but enriched with signalling domains associated with the emergence of multicellularity. Mapping of protein–protein interactions to create a network representation of the complex revealed that simple principles underlie the functional organisation of both proteins and their clusters, with modularity reflecting functional specialisation. The known functional roles of NRC/MASC proteins suggest the complex co-ordinates signalling to diverse effector pathways underlying neuronal plasticity. Importantly, using quantitative data from synaptic plasticity experiments, our model correctly predicts robustness to mutations and drug interference. These studies of synapse proteome organisation suggest that molecular networks with simple design principles underpin synaptic signalling properties with important roles in physiology, behaviour and disease.
Keywords: LTP, NMDA receptor, protein complex, synaptic plasticity
Introduction
In the last 5 years, proteomic studies of brain synapses have increased the number of known synaptic proteins by a factor of 5–10 revealing a surprisingly high molecular complexity (Husi et al, 2000; Collins et al, 2005). Comprising over 1000 proteins, the macromolecular complexes of neurotransmitter receptors connected with the postsynaptic density (PSD) are perhaps the most complex molecular structures known in mammals. Since many of these proteins participate in information processing in the brain, and also play roles in disease, it is of fundamental importance to ask if there is a molecular logic or organisation of the synapse proteome.
Synapses not only transmit information between neurons, but also process information by detecting patterns of neural activity that activate intracellular biochemical pathways, changing the properties of the neuron (Greengard, 2001; Kandel, 2001). Current molecular models focus on the excitatory neurotransmitter glutamate, which activates postsynaptic receptors that can be broadly categorised into those that transmit the electrical depolarisation (α-amino-3-hydroxy-5-methylisoxazole-4-proprionic acid (AMPA) receptors) and those that activate signalling and plasticity mechanisms (N-methyl-D-aspartate receptor (NMDA) and metabotropic receptors (mGluR)). Proteomic profiling of glutamate receptors isolated from brain reveal that NMDA and mGluR are assembled into large complexes of 186 proteins measuring 2–3 MDa and AMPA receptors into much smaller complexes of ∼10 proteins (Husi et al, 2000; Husi and Grant, 2001; Farr et al, 2004; Collins et al, 2005). These neurotransmitter receptor complexes are embedded within the PSD, which is visible with electron microscopy and comprises 1124 identified proteins (Collins et al, 2005). Studies of binary protein–protein interactions show that the NMDA–mGluR receptors are linked via adaptor proteins that also link the receptors to signalling enzymes and structural proteins (Tu et al, 1999). The adaptor proteins include Membrane-Associated Guanylate Kinase (MAGUK) proteins such as PSD-95 and SAP 102, which directly bind the cytoplasmic domains of NMDA receptors. These 186 protein complexes of NMDA receptors, mGluR receptors, MAGUK proteins and associated molecules, referred to as the NMDA receptor complex/MAGUK-associated signalling complex (NRC/MASC), are the subject of our analysis.
Functional studies of synaptic signalling have centred on the cellular mechanisms and behavioural roles of synaptic plasticity. Electrophysiological studies show that particular patterns of neuronal activity can induce changes in synaptic strength (e.g. long-term potentiation (LTP)) and other neuronal properties, which are currently thought to contribute to learning (mediated in the hippocampus), fear conditioning (mediated in the amygdala) and other forms of behavioural plasticity. Early pharmacological studies of synaptic plasticity revealed a role for glutamate receptors and subsequent knockout studies of MAGUK proteins revealed that these receptors require the assembly of signalling complexes for LTP and learning (Migaud et al, 1998). However, the progression of this field of research has been confused by the high number of molecules that are essential for normal forms of LTP (Sanes and Lichtman, 1999; A Howell et al, unpublished). In excess of 100 mouse gene knockouts show impairments in LTP and a similarly high number are involved with forms of behaviour involving glutamate receptors (Howell et al, in preparation). A second, and perhaps not unrelated problem, is that these single-gene perturbations do not usually completely block LTP (Howell et al, in preparation). The apparent robustness of synaptic plasticity may reflect its intrinsically important role as a necessary biological process. How this complexity and robustness can be related to the physiology of plasticity and learning is an important problem requiring new approaches.
In this paper, we utilise synapse proteomic data and present a detailed analysis of the NRC/MASC complex using annotation, network and statistical approaches. Using information on the function, interactions and phylogeny of individual proteins, their roles in synaptic plasticity and behavioural plasticity in the hippocampus and amygdala, and their roles in human diseases, we develop a model that explains many of the features of synaptic signalling. We find that the model explains the structural and functional aspects of synapse molecular complexity and why mutations in many genes have only partial effects on synapse plasticity. We suggest general principles of functional organisation that should provide a basis for new functional genomic approaches to synapse function and behaviour and be applicable to other cellular models of signal transduction.
Results
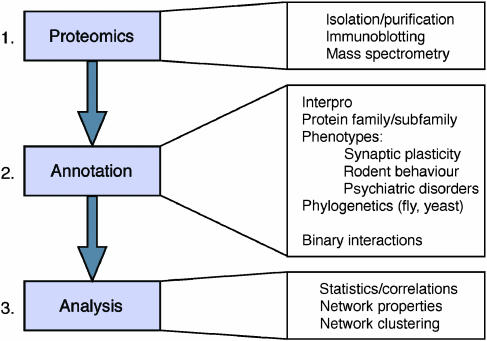
We adopted a three-step strategy of proteomics, annotation and analysis of synapse proteins (Figure 1). The proteomic step consisted of profiling of protein components of the synapse proteome, and is reported elsewhere (Husi et al, 2000; Collins et al, 2005). Step 2 involved the annotation of protein structure and function, including physiological and disease roles. Here we classify proteins according to function and molecular features (e.g. kinases, phosphatases) and known binding partners, as this informs on biochemical pathways. The physiological data were obtained from rodent experimental systems, where mutations or drugs that specifically interfere with a given protein have been tested for their effects on synapse physiology and animal behaviour. Finally, we have searched the literature for information on the involvement of specific molecules in human diseases. Step 3 comprised statistical and network analyses, where we ask if there are correlations and connections between proteins and functions. Integration of these data allows us to search for underlying principles of organisation and generate new models.
Figure 1.
A three-step strategy for analysis of synapse proteome organisation. Step 1 (Proteomics) was the collection of proteomic data identifying specific proteins. Step 2 (Annotation) was the collection of specific structural and functional data on individual proteins from Step 1, which was followed by Step 3 (Analysis) using statistical and network approaches.
Functional annotation of MASC
MASC proteins were assigned to functional families/subfamilies (Table I and Supplementary Table 1). Membrane-spanning channels, receptors and adhesion proteins, together with their associated signal transduction machinery, including adaptors and enzymes, account for 83% of the complex. Interpro annotations were retrieved via SwissProt (Supplementary Table 2). The protein domains most commonly found in MASC proteins (Table II) were highly enriched (3–12-fold) when compared to their frequency in the proteome as a whole. These top 10 domains represent key functionality associated with synaptic signalling: calcium binding (calcium-binding EF hand, C2, IQ calmodulin-binding region), G-protein-coupled signal transduction (small GTP-binding protein domain), phosphorylation (protein kinases, serine/threonine protein kinase), scaffolding (SH3, PDZ/DHR/GLGF) and membrane localisation (Pleckstrin homology type, Pleckstrin type, C2). These functional family and domain annotations clearly reflect specialisation for intercellular signalling.
Table 1.
Functional families present in MASC
| Family | n MASC a | % MASC b |
|---|---|---|
| Channels and receptors | 12 | 6.4 |
| Cell adhesion and cytoskeletal | 35 | 18.8 |
| G-proteins and modulators | 19 | 10.2 |
| Kinases | 22 | 11.8 |
| MAGUKs/adaptors/scaffolders | 20 | 10.8 |
| Protein phosphatases | 8 | 4.3 |
| Signalling molecules and enzymes | 39 | 21.0 |
| Synaptic vesicles/protein transport | 23 | 12.4 |
| Transcription and translation | 5 | 2.7 |
| Uncharacterised/novel | 3 | 1.6 |
aNumber of MASC proteins assigned to family.
bPercentage of MASC proteins assigned to family.
Table 2.
Domain profile of MASC—10 most common
| Domain | Accession | % MASC a | % Proteome b | Ratio c |
|---|---|---|---|---|
| Protein kinase | IPR000719 | 11.8 | 3.75 | 3.16 |
| Serine/threonine protein kinase | IPR002290 | 10.2 | 1.69 | 6.05 |
| SH3 | IPR001452 | 8.06 | 1.51 | 5.33 |
| Pleckstrin-like | IPR001849 | 5.91 | 1.25 | 4.72 |
| PDZ/DHR/GLGF | IPR001478 | 5.91 | 0.74 | 8.04 |
| Small GTP-binding protein domain | IPR005225 | 5.38 | 1.49 | 3.62 |
| Pleckstrin homology-type | IPR011993 | 4.84 | 1.08 | 4.49 |
| Calcium-binding EF-hand | IPR002048 | 4.84 | 1.65 | 2.93 |
| C2 | IPR000008 | 4.84 | 0.82 | 5.92 |
| IQ calmodulin-binding region | IPR000048 | 3.76 | 0.31 | 12.0 |
aPercentage of MASC proteins possessing domain annotation.
bPercentage of mouse proteome possessing domain annotation.
cRatio of MASC to proteome (measure of enrichment in MASC).
We next searched the literature for evidence that MASC proteins were involved in synaptic physiology and rodent behaviour. The systematic text searching and manual curation of the published literature that was utilised is described in detail elsewhere (Grant et al, 2005; Howell et al, in preparation). We specifically searched for genetic and pharmacological evidence that disruption of MASC proteins interferes with LTP and long-term depression (LTD), forms of synaptic plasticity found at most central nervous system synapses (Supplementary Table 3). In total, 44 (24%) proteins represented in MASC were known to be essential: without the function of these proteins, synaptic plasticity was impaired. We also searched for studies reporting the involvement of specific proteins/genes in behavioural plasticity, focusing on rodent learning and conditioning paradigms, as these represent the largest body of molecular data (Supplementary Table 3). We annotated papers into those affecting spatial learning (primarily mediated by the hippocampus), cue/context conditioning (primarily mediated by the amygdala) and other behavioural paradigms (Kandel et al, 2000). Overall there were 48 (26%) MASC proteins involved with behaviour, of which 42 (23%) were important for learning. Of those involved in learning, 32 (17%) were involved with spatial learning and 25 (13.5%) with cue/contextual conditioning.
Although it is generally accepted on the basis of anatomical homology and lesion data that cognitive mechanisms are conserved between mice and humans, it is unclear to what degree the rodent molecular studies map onto human psychiatric conditions. We therefore examined the possibility that MASC proteins may be involved in human psychiatric and neurological disorders. We identified 54 (29%) MASC proteins implicated in mental illness (Supplementary Table 3). Although we searched all mental disorders, we found 33 (18%) in schizophrenia, 23 (12%) in mental retardation, 12 (6.5%) in bipolar disorder and 14 (7.5%) in depressive illness. This apparent bias toward schizophrenia and mental retardation may be biologically relevant as they both have a major cognitive component to their primary symptoms. In total, 49 (26%) proteins were linked to cognitive disorders (schizophrenia, mental retardation), compared to only 22 (12%) implicated in affective disorders (bipolar, depression).
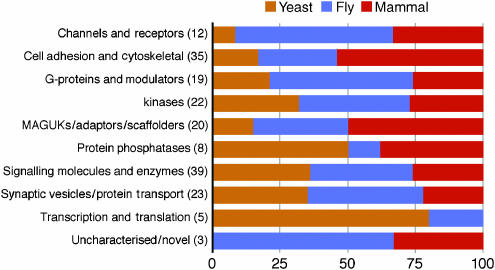
To investigate the evolutionary conservation of MASC proteins in invertebrates and unicellular eukaryotes, we searched for orthologues in the genomic databases of yeast (Saccharomyces cerevisae) and fruit fly (Drosophila melanogaster) (see Materials and methods). In total, 117 (63%) MASC proteins were identified in fly and 51 (27.5%) in yeast (Supplementary Table 4). Overall, 63 (34%) were found only in mouse (i.e. have no identified orthologues in yeast or fly), 135 (72.5%) were metazoan (not found in yeast) and 45 (24%) conserved (present in both fly and yeast). While transcription/translation proteins were generally conserved from yeast (Figure 2), those protein families involved in intercellular signalling were primarily metazoan, consistent with previous observations (Manning et al, 2002). Although all functional families were found in yeast, and thus predate the evolution of the nervous system, there were distinct patterns among these families relevant to synapse specialisation. For example, significant expansion of most families arises at the metazoan transition (yeast to fly) and there is additional expansion from fly to mammals in specific classes (Cell Adhesion and Cytoskeletal, MAGUKs/Adaptors/Scaffolders). This is supported by more detailed statistical analysis described in the following section. These expansions toward the mammalian lineage may be relevant to the more complex range of behaviours and neuroanatomy of the mammalian nervous system and the physiological role of MASC proteins.
Figure 2.
Evolutionary expansion of protein families in MASC. Functional classes of proteins are represented as a horizontal line and the number of these proteins found in mouse MASC is shown in parenthesis. The percentage of each functional family making its earliest appearance in yeast (brown), fly (blue) or mammal (mouse, rat, human) (red) is indicated using the species-specific colour scheme. For example, only one out of 12 (8.5%) mammalian channels and receptors were found in yeast, in contrast to transcription and translation regulators, where four out of five (80%) were found in yeast. It is interesting to note that the novel/uncharacterised proteins arise in metazoans.
Statistical analysis of annotations
The entire set of functional, phenotypic and phylogenetic annotations (Supplementary Tables 1–6) were subjected to a statistical analysis, looking at their distribution and overlap (see Materials and methods). For each pair of annotations (e.g. glutamate receptors and schizophrenia), we identified the number of common proteins, calculating the probability of an overlap as or less likely occurring by chance. As an example, of the 32 MASC proteins implicated in schizophrenia, five are glutamate receptor proteins. In total, there are only six glutamate receptor proteins in MASC—if 32 MASC proteins are selected at random, it is most likely that only one of them will be a glutamate receptor. The probability of finding five or more glutamate receptors in a random sample of 32 taken from MASC is ∼0.0007. This suggests that the overlap between glutamate receptors and schizophrenia is of biological relevance. Only probabilities <0.01 were considered as potentially significant (Supplementary Table 7). In both this and the following section, references to annotations are indicated in italics, brief descriptions of which are collected below.
Glutamate receptors showed a striking correspondence with all studies of synaptic plasticity (drug P<10−4, mutation P<10−3, synaptic plasticity P<10−3), all behavioural assays (spatial learning P<10−2, cue/contextual conditioning P<10−3, behavioural (other) P<10−3, behavioural plasticity P<10−2), cognitive disorders in general (P<10−2) and schizophrenia in particular (P<10−3). The only proteins to occur in all of these categories were the NMDA receptor subunits NR1, NR2A and NR2B: evidence of a strong link between NMDA receptor signalling and cognitive function. Notable overlaps were also found between phosphatases and synaptic plasticity (drug P<10−2, synaptic plasticity P<10−2), between G-α proteins and affective disorders (affective P<10−2, bipolar P<10−2, depressive P<10−2) and between C2 (Ca-dependent membrane-targeting) domain proteins and behavioural phenotypes (behavioural plasticity P<10−3, cue/contextual conditioning P<10−3, spatial learning P<10−2). While the functional subfamily of other enzymes had a significant overlap with mental retardation, four of the five proteins responsible for this overlap (a lactate dehydrogenase, phosphofructokinase, pyruvate kinase and triosephosphate isomerase) are metabolic enzymes involved in glycolysis. As such, the phenotypes of these enzymes are more likely to be due to widespread disruption of electrophysiological properties than the perturbation of specific signalling pathways.
Synaptic plasticity and behavioural plasticity were intimately connected within MASC (P<10−11), which strongly supports the model that the overall complex is a molecular machine underpinning cellular and behavioural plasticity. The close correspondence found between spatial learning and cue/contextual conditioning (P<10−6) indicates a common molecular basis for learning in the amygdala and hippocampus. Synaptic plasticity showed a high degree of overlap with studies of cognitive and to a lesser extent affective disorders (cognitive P<10−8, schizophrenia P<10−6, mental retardation P<10−2, affective P<10−2, bipolar P<10−2). In contrast, behavioural plasticity displayed a high degree of overlap with both (cognitive P<10−9, affective P<10−6, bipolar P<10−4, depressive P<10−3). In addition, cognitive and affective disorders showed a significant overlap (P<10−6). Within the cognitive disorders, schizophrenia showed a significant overlap with both affective disorders (depressive P<10−7, bipolar P<10−4) and mental retardation with bipolar (P<10−2). However, the overlap between schizophrenia and mental retardation was not significant (P>0.1). In general, the degree of commonality between mental retardation and other annotations tended to be among the least significant of all phenotypes. Together, these results validate the use of rodent models of human mental illness, particularly schizophrenia, and promote the perturbation of MASC and its effect on synaptic plasticity as a major underlying factor.
The physiological and behavioural annotations did not have significant overlap with the phylogenetic annotations. Nevertheless, consistent with Figure 2 and earlier results, cell adhesion and cytoskeletal proteins were under-represented in fly (P<10−2) and significantly expanded in mammals (mammalian P<10−2). All ATP synthases were conserved (P<10−2) and all Ser/Thr-specific phosphatases were found in yeast. All MASC proteins containing L27 domains (found in receptor-targeting proteins) appeared to be mammalian specific (P<10−2).
Drawing these analyses together, MASC appears to be a highly specialised signal transduction complex, whose evolutionary expansion reflects a role in neural information processing unique to higher organisms. The composition of MASC suggests that diverse cell-biological responses are co-ordinated within the complex. The coupling of these responses to glutamatergic signalling induces synaptic plasticity, which results in behavioural learning. Disruption of the complex perturbs the orchestration of responses, causing altered synaptic plasticity. This manifests as impaired behavioural plasticity in rodents and psychiatric disorders in humans. To the limited extent to which they can be separated, cognitive disorders appear intimately linked to signal transduction via the NMDA receptor, while affective disorders show closer correspondence to G-protein-coupled signalling and mGluRs. On balance, information processing within MASC appears primarily related to cognitive function. The relatively weak correlation between mental retardation and other annotations (i.e. essentially random overlap) suggests that it involves random disruption within MASC, and that the major cognitive component to its symptoms simply reflects the primary role of the complex.
Protein interaction network analysis
The annotation studies presented above only consider the list of components and do not take into consideration their organisation or assembly into a complex through protein–protein interactions. We therefore obtained high-quality interaction data, curated from the literature, in order to generate a network representation of the complex (see Materials and methods) and analyse its features.
We identified 248 binary interactions between 105 of the MASC proteins (Supplementary Table 8). This number excludes self-interactions, which were not used in network construction. No interaction data were found for the remaining 77 proteins (apart from self-interactions). When represented as an undirected graph, the largest network component consisted of 101 proteins linked by 246 interactions. This connected component constitutes core functional elements of the complex. It links together all glutamate receptors and a high proportion of the signal transduction machinery responsible for the reception and integration of calcium and G-protein-coupled synaptic signalling (Table IIIa). That the component captures key functional processes is supported by the fact that it contains the majority of all phenotypically linked proteins (Table IIIb). The sole exception to this is mental retardation, whose representation within the connected component (65%) is close to that of MASC proteins as a whole. This supports the hypothesis that mental retardation entails general disruption of the complex, and argues against enrichment of the component with other phenotypic annotations being solely due to bias in the literature. All further analysis concerns this 101-protein component.
Table 3.
Composition of the largest network component
| Annotation | n Ccpt b | % Ccpt b | +/− c | Probability d |
|---|---|---|---|---|
| (a) Functional family a | ||||
| Channels and receptors (12) | 8 | 67 | ∼ | 0.6 |
| Cell adhesion and cytoskeletal (35) | 19 | 54 | ∼ | 1. |
| G-proteins and modulators (20) | 17 | 85 | + | 0.007 |
| Kinases (21) | 19 | 90 | + | 0.0007 |
| MAGUKs/adaptors/scaffolders (17) | 15 | 88 | + | 0.004 |
| Protein phosphatases (7) | 6 | 86 | ∼ | 0.1 |
| Signalling molecules and enzymes (39) | 10 | 26 | − | 0.00004 |
| Synaptic vesicles/protein transport (23) | 7 | 30 | − | 0.01 |
| Transcription and translation (5) | 0 | 0 | − | 0.02 |
| Uncharacterised/novel (3) | 0 | 0 | − | 0.09 |
| Glutamate receptors (6) | 6 | 100 | + | 0.03 |
| (b) Phenotype a | ||||
| Synaptic plasticity (44) | 38 | 86 | + | 10−6 |
| Behavioural plasticity (41) | 35 | 85 | + | 10−5 |
| Psychiatric disorder (53) | 40 | 75 | + | 0.0005 |
| Schizophrenia (32) | 27 | 84 | + | 0.0003 |
| Mental retardation (23) | 15 | 65 | ∼ | 0.4 |
| Bipolar (13) | 11 | 85 | + | 0.04 |
| Depressive (13) | 11 | 85 | + | 0.04 |
aNumber of MASC proteins contained in each family or implicated in each phenotype indicated in brackets. Also shown in (a) is the representation of glutamate receptors, a subfamily of channels and receptors.
bNumber and percentage of each annotation present in the largest connected component of network.
cAnnotations over/under-represented in the connected component denoted by (+/−). The representation of an annotation is considered to be indistinguishable from a random sample (∼) if the probability given in the final column is >0.1.
dProbability of this overlap (n Ccpt) between annotation and network component using the statistical method described in the text.
Summarised are the representation of (a) functional family and (b) phenotype annotations within the largest network component. The number of MASC proteins contained in each family (or implicated in each phenotype) is indicated in brackets. Subsequent columns show the number (n Ccpt) and percentage (% Ccpt) of these present in the largest connected component, whether this is more or less than expected by chance (+/−) and the probability of this overlap between annotation and connected component using the statistical method described in the text. The representation of an annotation is considered to be indistinguishable from a random sample (∼) if this probability is >0.1. Also shown in (a) is the representation of glutamate receptors, a subfamily of channels and receptors.
The average number of interactions separating any pair of proteins was very low (average shortest path length=3.3), implying a high level of crosstalk between signal transduction pathways. While this suggests an ability to rapidly integrate disparate sources of information and orchestrate coherent responses, it does not sit comfortably with a model of well-defined linear pathways of limited overlap. It suggests instead that functional roles are distributed over sets, or clusters of proteins within MASC. We therefore sought to identify and evaluate any clustering inherent in the network (see Materials and methods).
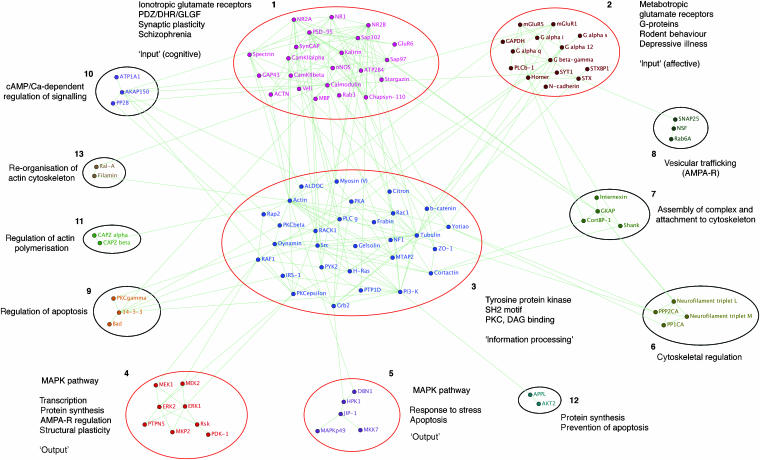
The connected component was found to possess a clearly modular structure (Figure 3 and Supplementary Table 9), with ∼75% of its proteins contained in the five largest clusters. To evaluate the functional significance of these clusters, their overlap with each functional and phenotypic annotation (such as those shown in Figure 4) was analysed using the statistical method introduced earlier (Supplementary Table 10).
Figure 3.
Network cluster analysis. Clustering of the largest connected component of the MASC network identified 13 clusters. Significant overlap with functional and phenotypic annotations is indicated for the largest clusters, 1–3. Clusters 4 and 5 both correspond to MAPK signalling pathways regulating various functional processes (briefly summarised). All five are followed by a brief descriptive phrase indicating their general functional role. Functional roles suggested by composition and interactions are indicated for the remaining clusters. Graphical representation of network produced using BioLayout (Enright and Ouzounis, 2001).
Figure 4.
Network clustering—phenotypic overlay. Proteins with various phenotypes (electrophysiological, behavioural and human psychiatric) are highlighted within the MASC network. The layout of proteins, identical to that of Figure 3, reflects network clustering.
Cluster 1 contains all ionotropic glutamate receptor proteins (P<10−3) and a large number of PDZ/DHR/GLGF scaffolding molecules (P<10−3), particularly MAGUKs. In total, ∼50% of its proteins are essential to normal synaptic plasticity (P<10−2) and ∼40% are implicated in schizophrenia (P<10−2). Within MASC, these features have a strong association with cognitive function.
Cluster 2 appears specialised for metabotropic/G-protein-coupled signalling (G-proteins P<10−2, metabotropic glutamate receptor P<10−2). Half of its proteins have known behavioural phenotypes (behavioural (other) P<10−2) and it contains a third of all MASC proteins implicated in depressive illness (P<10−2). With Homer coupling mGluRs to IP3 receptors in the endoplasmic reticulum and PLC β localised to the membrane via PH and C2 domains, the cluster is capable of directly regulating Ca2+ release from internal stores. Also present are several proteins closely associated with vesicular release. These link to cluster 8, which contains proteins implicated in the postsynaptic trafficking of AMPA receptors, for example, NSF (Nishimune et al, 1998).
Cluster 3, the largest, is strongly connected to clusters 1 and 2. Its size and centrality within the network (see Figure 3) suggest that it assimilates signals from various sources and co-ordinates common effector mechanisms. This seems to be borne out by its composition. The well-studied Ser/Thr kinase PKA, known as an integrator of signals in synaptic plasticity, is found within the cluster. The cluster also contains a concentration of tyrosine kinases (P<10−3) and SH2 motif proteins (P<10−3). SH2 domains bind specifically to phosphotyrosine in a wide range of substrates, interactions known to regulate diverse signal transduction pathways (Pawson, 2004). The tyrosine kinases are themselves a point of convergence for multiple signalling pathways regulating NMDA receptor activity (Salter and Kalia, 2004). These data suggest that cluster 3 integrates the ionotropic and metabotropic signals of clusters 1 and 2 with modulatory sources external to MASC. This is supported by the concentration of Ser/Thr kinases sensitive to the second messenger diacylglycerol (DAG), another route for external modulation (PKC, phorbol ester/DAG binding P<10−2). These processes are closely interconnected: Citron, a dual-specificity kinase, contains a DAG-binding motif, while PLC γ hydrolyses PI(4,5)P2 to form DAG and IP3 (another link to Ca2+ signalling) when activated by tyrosine phosphorylation.
Cluster 4 encapsulates the well-studied MAPK–ERK signalling pathway (Ser/Thr kinases P<10−4, Erk1/2 MAP kinase P<10−2, tyrosine & dual-specificity phosphatase P<10−2). ERK activation has been linked to transcription, protein synthesis, regulation of AMPA receptors and structural plasticity (Thomas and Huganir, 2004). Cluster 5 is another MAPK pathway (Ser/Thr kinases P<10−2), mediating response to stress through JNK3 (MAPKp49). It may be of note that cluster 4 interacts with cluster 3 via proteins containing DAG-binding motifs (RAF1 and PKC ɛ), while cluster 5 interacts through Grb2—an SH2 domain adapter protein. With reference to clusters 4 and 5, it is interesting to note the existence of the small cluster 12 linking AKT2 (PKB β) to PI3-K via the scaffolding protein APPL. Interplay between PI3-K and MAPK signalling has a complex effect on LTP, the mechanisms of which are still unclear (Opazo et al, 2003). PI3-K has been implicated in vesicular trafficking and cytoskeletal rearrangement, while PI3-K-dependent activation of PKB contributes to the control of protein synthesis and the prevention of apoptosis (Rogers and Theibert, 2002).
For the smaller clusters still to be discussed, we indicate functional roles suggested by their composition and interactions (Supplementary Tables 8 and 9). Clusters 6, 7, 11 and 13 mediate interactions with the cytoskeleton. Three of these regulate cytoskeletal structure and its rearrangement: cluster 6 via neurofilaments, clusters 11 and 13 via actin. Assembly and localisation of the complex are influenced by cluster 7, which mediates colocalisation of receptor subcomplexes (clusters 1 and 2) and their attachment to the cytoskeleton. Cluster 10 controls channel and kinase activity through the integration of cAMP and Ca signals. Regulation of Na/K channel ATP1A1 alters the postsynaptic resting potential, while control of PKA and PKC isoforms strongly modulates MASC signalling. Cluster 9 regulates the induction of apoptosis through the phosphorylation and sequestration of Bad. These clusters reflect the role of the NMDA receptor in structural plasticity, cell death and the synaptic localisation of proteins.
In general terms, the network seems to be organised into a few large, highly connected clusters directing MASC function (clusters 1–3) and a greater number of smaller, more sparsely connected clusters dedicated to specific functional processes (clusters 6, 7, 8, 9, 11 and 13). Other clusters are intermediate between the two (clusters 4, 5, 10 and 12).
Topology and functional robustness of the MASC network
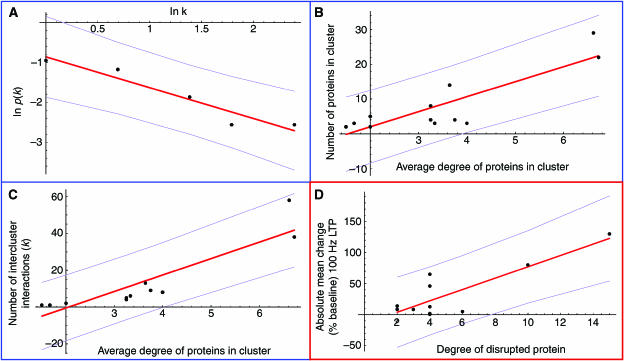
Synaptic plasticity is surprisingly robust, with disruption by mutation or drugs only partially impairing rather than completely abolishing plasticity in most cases (e.g, see Grant et al, 1992; Watabe et al, 2000; Komiyama et al, 2002; Opazo et al, 2003; Yasuda et al, 2003). A potential source of robustness lies in the pattern of connectivity within the network. The degree distribution of the network (the probability ρ(k) of a protein being involved in k interactions) was found to be well fitted by a power law, with ρ(k)∼k −1.2 (P<10−5). This reflects the presence of a few highly connected proteins mediating interaction between the more sparsely connected proteins that constitute the bulk of the network. It provides a level of structural robustness whereby the degree of a protein (the number of interactions it is involved in) correlates with the structural disruption caused by its removal (Albert et al, 2000). In yeast this has been shown to translate into functional robustness—while highly connected proteins form a small percentage of the proteome, their disruption is more likely to prove lethal (Jeong et al, 2001). Of the eight MASC proteins with >10 interactions, there are five (62.5%) with phenotypes in all three major categories (synaptic plasticity, behavioural plasticity and psychiatric disorder), while of the 61 MASC proteins with <5 interactions, there are only nine (14.8%).
Given that the more interactions a protein has the more likely it is to influence multiple processes (e.g. effector mechanisms), correlation between protein connectivity and severity of effect on disruption naturally arises. When combined with a power-law degree distribution, this suggests a loose functional hierarchy composed of a few highly interacting proteins, largely responsible for overall functional co-ordination, a broad range of lower degree proteins influencing various aspects of functionality and a large number of low interacting proteins specific to individual functional processes. This has clear parallels with the modular organisation of MASC, discussed above. Our analysis up to this point led us to propose the following model for the structural and functional organisation of MASC.
Each functional process is the net result of complex interactions within a subset of MASC proteins. These subsets overlap, with some proteins being involved in multiple functions: there is a positive correlation between the degree of a protein and the number of functional processes it influences. This correlation is intimately connected with the emergence of a power-law degree distribution and low average path length. Physical interactions cluster MASC proteins into functional modules. The network formed by these modules is subject to the same organisational principles: the mean shortest path length between modules is low; the size and intercluster connectivity (degree of interaction with other clusters) of each module correlates with the extent of its functional influence; and module–module interactions possess a roughly power-law degree distribution. These common principles facilitate co-ordination and impart robustness to functional processes at both levels. Individual modules are specialised for functional roles including signal reception, signal integration and processing and the regulation of effector mechanisms. Through the dynamic balance of interactions within and between modules, MASC integrates multiple streams of information and co-ordinates diverse cell-biological processes in response, regulating the induction of synaptic plasticity.
While more data are needed to properly evaluate this model, some preliminary observations can be made. The low average path length separating proteins and the power-law distribution of their interactions have already been demonstrated. The mean shortest path length between modules was 1.86. Limited by the low number of modules, the probability ρ(k) of a module interacting with k others showed a marginally significant fit to a power law: ρ(k)∼k −0.77 (P=0.010, see Figure 5A). Functional influence is positively correlated with properties at two levels: the degree of individual proteins and the size and connectivity of clusters of proteins. Compatibility between the two requires the degree of a protein to be correlated with the size/intercluster connectivity of the module in which it is found. The average degree of proteins belonging to each cluster of the MASC network was found to have a significant correlation with both cluster size (linear fit: P<10−3, Pearson correlation=0.87; see Figure 5B) and intercluster connectivity (linear fit: P<10−4, Pearson correlation=0.90; see Figure 5C). Correlation was also found between cluster size and intercluster connectivity (linear fit: P<10−6, Pearson correlation=0.96). These correlations persist when data are restricted to the five largest, most clearly defined clusters, and also when connections between clusters are taken to be binary (data not shown). The relationship with functional influence is reflected in the concentration of highly interacting, influential proteins (e.g. NR2A/B, PSD-95, calmodulin, CamKII α, PI3-K, actin) within the largest, most highly connected clusters, 1 and 3.
Figure 5.
Support for model of MASC organisation. The model of MASC functional organisation makes several predictions: interconnectivity between modules follows a power law; protein degree is correlated with the size and interconnectivity of the enclosing module; and the degree of a protein is correlated with the perturbation of synaptic plasticity caused by its disruption. In all plots, the best linear fit is shown in red, with magenta lines at one standard deviation. (A) The probability p(k) of a cluster being connected to k other clusters is shown as a function of k in a log–log plot, a power-law distribution being characterised by a straight-line plot. (B) The number of proteins contained in a cluster is plotted as a function of their average degree. (C) The number of interactions with proteins external to a cluster is shown as a function of the average degree of proteins within the cluster. (D) The absolute change (% baseline) in 100 Hz LTP is shown as a function of the degree (number of interaction partners) of the protein disrupted. Where multiple sets of experimental data were available for a single protein, the absolute value of the mean experimentally recorded change was used (a significant linear fit was also evident for the full data set of 36 points).
Most significantly, the model makes the following prediction: if MASC controls the induction of synaptic plasticity, then a correlation between protein degree and extent of functional influence entails a correlation between protein degree and quantitative perturbation of LTP/LTD on disruption.
Quantitative data on the perturbation of LTP/LTD caused by disruption of individual proteins were available for a subset of MASC proteins with synaptic plasticity phenotypes. As LTP varies with the frequency of presynaptic stimulation, we considered only experimental data obtained using 100 Hz stimulation protocols, by far the most common. Such data are available for 36 experiments covering 11 MASC proteins, all of which are present in the connected component (Supplementary Table 11). Despite the inherent variability of this data due to differences in experimental protocols, protein degree and quantitative perturbation of LTP on disruption were found to be strongly correlated (linear fit: P<10−3, Pearson correlation=0.85, see Figure 5D).
Discussion
Here we present an integrated analysis of molecular organisation, signal transduction, physiology and diseases of a neurotransmitter receptor signalling complex as a step toward synapse systems biology. We also present a new model for understanding the molecular complexity of the synapse proteome and its relationship to synapse physiology (Figure 6). In addition to its specific features relevant to neurobiology, this model has some general properties applicable to other areas of signal transduction and receptor biology. Below we discuss the elements of this model progressing from signal transduction, to physiology and finally to behaviour and disease. We then compare our model of network organisation to other descriptions of network topology, before placing our analysis in perspective with some general observations.
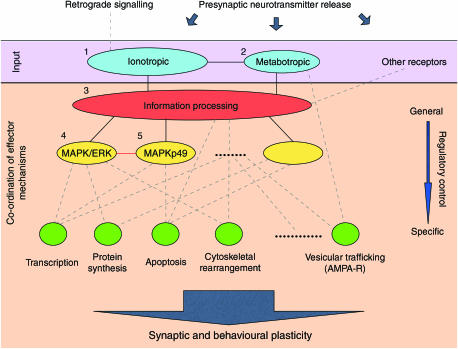
Figure 6.
Modular structure and functional organisation within MASC. MASC proteins are clustered into modules with well-defined functional roles. Primary signal reception modules (blue) are formed around ionotropic and metabotropic receptors. These inputs are integrated within a large signal-processing module (red) responsible for overall co-ordination of functional processes. Other sources of input (‘other receptors') may feed into this module directly, or through smaller input/processing modules (such as cluster 10, Figure 3). Note that, within this general structure, individual modules may play multiple functional roles (e.g. regulation of effector mechanisms by input modules 1 and 2). In this way, information processing and regulation of effector pathways are distributed over multiple modules. The general principles underlying functional organisation within MASC are apparent in the co-ordinated regulation of common downstream effector pathways: a single, large module (red) is responsible for overall co-ordination; several intermediate modules (yellow) regulate overlapping sets of pathways, while numerous small modules (green) are specific to individual effector responses. Note that this is not a simple feed-forward mechanism, rather a dynamical balance between multiple functional processes. The resulting synchronisation of multiple cell-biological processes induces synaptic plasticity, manifest at a higher level of neurological function through behavioural learning. Numbering of the five largest clusters reflects that of Figure 3, as do the interactions between them (solid black lines). Internal/external modulation of MASC function and the regulation of effector mechanisms are denoted by dashed lines. The red line between clusters 4 and 5 denotes the fact that other interactions (e.g. phosphorylation) play an important role in MASC function.
Signal transduction
Analysis of protein interactions leads us to propose the following view of signal transduction within the complex, summarised in Figure 6. Clusters of proteins around ionotropic and metabotropic glutamate receptors (modules 1 and 2) form the primary sites for signal reception. The density of interactions surrounding NMDA receptor subunits and the extent of their phenotypic involvement suggest that ionotropic signalling dominates. Indeed, this is supported by electrophysiological studies using blockers of NMDA and metabotropic receptors, where the NMDA receptors have a severe phenotype (Bliss and Collingridge, 1993). These clusters may directly regulate effector mechanisms such as retrograde signalling (ionotropic) and release of calcium from internal stores (metabotropic). The latter generates a second calcium signal with a spatiotemporal profile quite distinct to that of the ionotropic current. The relative timing, strength and duration of these signals are likely to be important factors in the initiation of downstream signalling events. These signals are modulated by other sources of information, both internal and external to the complex. The main body of proteins (module 3) integrates these disparate sources, co-ordinating common effector pathways via a cascade of smaller clusters (modules 4, 5 and others). This integrated model fits well with classical reductionist studies of molecular plasticity, where, for example, these output clusters, such as the ERK pathway, are well known as important outputs of glutamate receptor-mediated synaptic plasticity (Sweatt, 2004).
Physiology
At the physiological level, our analysis suggests that MASC is central to the postsynaptic processing of information encoded in neural activity, orchestrating the cellular responses underlying synaptic plasticity. We propose that the complex as a whole is responsible for the induction of synaptic plasticity. The strongest case can be made for hippocampal LTP/LTD, with MASC containing a large number of proteins implicated through study of CA3–CA1 synapses. While it was obvious that NMDA receptors were involved in both plasticity and learning, it was not obvious that so many of the other proteins were involved with both synaptic plasticity and behaviour and could be ‘unified' in the complex. Thus, the fact that there was such an extremely high overlap (probability of an overlap as or more extreme occurring by chance <10−11) is indeed very striking. The physiological role of the complex is strongly reinforced by the correlation between protein connectivity within the complex and quantitative perturbation of LTP on disruption (Figure 5D).
Behaviour and disease
Extending from the physiology to behaviour and disease, we see marked evidence of a common molecular foundation to synaptic plasticity, rodent behaviour and human mental illness. Of the psychiatric disorders, the complex is most closely associated with schizophrenia, which itself shows the highest correlation to synaptic and behavioural plasticity. Mouse genetic studies have been used to dissect distinct cognitive subprocesses (such as strategy choice, perception and learning) and show that these processes can be separated by mutations in different genes in NRC/MASC (Migaud et al, 1998; Cuthbert et al, submitted). This is consistent with the modularity within MASC mapping onto these distinct cognitive processes. For example, signal reception modules (clusters 1 and 2) were clearly specialised for different streams of information transmitted by distinct mechanisms (ionotropic and metabotropic receptors) that map onto cognitive and affective disorders and processing, respectively. The fact that cognitive and affective disorders display contrasting associations at the level of input should not be taken as indicating a sharp separation, as they appear closely intertwined, with elements of ionotropic and metabotropic signalling implicated in both. On balance, the evidence argues for a primarily cognitive role to MASC function. It is interesting to note that although schizophrenia and mental retardation/learning disability are both cognitive disorders, mental retardation genes are found scattered throughout the genome in large numbers, whereas schizophrenia genes are found in a small number of regions, and that schizophrenia is likely to entail more specific disruption within the complex.
A potentially exciting feature of our model for the aetiology of human psychiatric diseases arises from our study of molecular complexity and robustness, which helps explain why many molecules participate in a phenotype. We speculate that the genetic complexity of schizophrenia or other diseases affecting this complex (Grant et al, 2005) may emerge from combinations of common polymorphisms in the many genes encoding MASC. These polymorphisms alone may have no clear phenotype, until they are in combination with others, which together have a cumulative effect on MASC function. Complementary to this aetiological model is the possibility of using the network to identify disease-modifying pharmaceuticals that target specific proteins.
Network organisation
We observed evidence for simple principles underlying the functional organisation of MASC at the level of both proteins and clusters: the characteristic path length (average shortest path length) separating elements (proteins/modules) is low; the connectivity of elements follows an approximately power-law distribution and the connectivity of each element is correlated with the extent of its functional influence. These imply a functional hierarchy ranging from a few highly connected elements responsible for overall coordination, to numerous sparsely connected elements specific to individual functional processes. In between these extremes lies a range of elements through which particular sets of functional processes are coordinated. We suggest that these principles extend to all levels of organisation within the postsynaptic proteome.
The properties of this model are best understood through comparison with other models of network topology—small-world, scale-free and bow-tie architectures—all of which have been identified in biological networks. The small-world models of Watts and Strogatz (1998) combined local clustering and a low characteristic path length. Both of these properties are present in our model, where we would expect them to reproduce the enhanced signal propagation and functional coordination with which they were associated. Many biological networks have been described as scale-free due to their approximately power-law degree distribution (Jeong et al, 2000), another component of our model. This reflects the presence of a few highly connected nodes (molecules/clusters) that mediate interaction within the less-well-connected bulk of the network. Such networks are structurally robust to random deletions, but fragile to targeted removal of highly interacting nodes (Albert et al, 2000). Given that the more interactions a node has the more likely it is to influence multiple functional processes (as in our model); this naturally extends to a correlation between node connectivity and severity of functional effect on disruption, a correlation also observed in the yeast proteome (Jeong et al, 2001). Higher-level structure has been modelled as a ‘bow-tie' (Ma and Zeng, 2003; Csete and Doyle, 2004) in which multiple inputs converge on a tightly integrated core of processes that drive an array of output pathways. While it is tempting to identify the MASC network (Figures 3 and 6) as a bow-tie structure—modules 1 and 2 as input, 3 as core functional processing, and 4, 5 and others as outputs—this would be overly simplistic. Input clusters 1 and 2 are directly linked to output pathways (retrograde signalling, vesicular trafficking), while cluster 10 (Figure 3) integrates second messenger signals, strongly modulates information processing in cluster 3 and directly regulates ion channel properties. In effect, the core functionality of information processing and integration is distributed rather than centralised. As we have shown, higher-level structure is better described by a network of modules with power-law degree distribution.
The proposal that all levels of organisation follow the same structural pattern is supported by evidence of self-similarity in biological networks (Song et al, 2005). By identifying this common pattern as a power-law distribution, each level is endowed with structural and functional robustness. The combination of modularity and distributed function also appears to be a novel feature of the model. Each cluster integrates a particular set of inputs (either external or internally processed signals) and influences a particular set of functional processes (e.g. other modules, effector pathways). Stated another way, each module reflects the correlation between a particular set of functions in response to a specific range of stimuli. This implies that different sets of signals (e.g. different patterns of action potentials) are processed by different sets of modules, and that the relative importance of each module varies according to the information being processed.
Predictions arising from the model have been confirmed, most significantly in the correlation between protein connectivity and quantitative perturbation of LTP. The fact that this correlation emerges with current interaction data suggests that the network used in our analysis and the organisation apparent within it accurately reflects the general structure of the complete network.
General observations
Although we have referred to the complex as if it were identical at all synapses, this is known not to be the case in all parts of the central nervous system (Porter et al, 2005). Ongoing systematic studies of MASC proteins using microarray data and protein localisation show a high degree of coexpression for most MASC proteins in forebrain structures, including hippocampus, cortex, striatum and amygdala (Zapala et al, 2005). Given the number of different proteins involved and the complexity of their interactions, it seems most likely that any given synapse will contain a distribution of complexes of varying composition. This implies that the coordination of signalling responses takes place both within individual complexes and as a function of the distribution as a whole. Dynamic interactions could be factored into future analysis both at the level of protein turnover and phosphorylation. We also recognise the incompleteness of some of our data sets that ideally would be obtained in systematic unbiased studies and such programs are underway (e.g, www.genes2cognition.org; www.gensat.org; www.brainatlas.org). Not only will the availability of this data refine our ‘draft' maps of synapse proteome organisation, but our maps can also be used to prioritise areas for data acquisition and development of analytical techniques relevant to the nervous system. Future directions will not only include refinements based on more systematic data availability, but also act as a starting point for systems biology approaches to the synapse. This presents an attractive approach within the overall complexity of the brain proteome and transcriptome, as it has defined functional roles amenable to genetic and pharmacological manipulation. This model of MASC can now be extended to the synapse proteome as a whole and encompass neurological and psychiatric disease genes. This model also presents an exciting new avenue for the integration of the molecular networks presented here with synapse models of neural networks.
Materials and methods
Functional annotation
Throughout this study, we have stressed the use of high-quality, manually curated data sets. While bioinformatic tools are invaluable for extensive literature searches, subsequent manual curation is indispensable. The inadequacy of fully automated annotation methods is perhaps most acute when searching for complex disease associations. These human data include evidence of mendelian inheritance of polymorphisms and changes in protein or mRNA levels in brain tissue. It is generally unknown if any of these molecular changes alone produce mental illness. To compound this level of uncertainty with artefacts inevitably thrown up by automated methods is to risk losing all information to noise. The use of GO annotation has also been avoided, as its coverage of experimental data is generally incomplete. Given the partial nature of this data itself, comprehensive (automated and manual) literature searching and expert curation was felt to be the only means of obtaining a data set of minimal inherent bias.
Interpro annotations for all proteins were taken from SwissProt (Supplementary Table 2). To compare the frequency of domain occurrence within mouse, SwissProt identifiers for the NCBIM33 mouse gene data set were obtained from EnsMart. Out of 24 461 entries, identifiers were available for 7338 genes. Interpro annotations available through EnsMart were not used, as these were found to contain a large number of spurious entries (e.g. tyrosine kinases and nuclear localisation signals: IPR001245 and IPR001472). The annotation provided by SwissProt was found to contain significantly fewer such entries.
EnsMart was used to identify orthologues in the BDGP3.2.1 Drosophila and SGD1 yeast data sets (Supplementary Table 4). Human, mouse and rat Ensembl Gene Ids (Supplementary Table 1) were used for the search, and their results combined—a protein was deemed to possess an orthologue if at least one of its gene ids returned an orthologue.
In presenting the number and percentage of MASC proteins with a particular phenotypic annotation, a protein was counted as having that annotation if identified directly in the literature, or if a corresponding generic protein entity was referred to (e.g. a reference to ‘PKA' would be taken as implicating PRKACB and PKA-R2b).
Generic protein entities
The specificity with which interactions and phenotypes are reported varies considerably, and it is not uncommon to find references to classes of molecules (e.g. ‘G-α s'). In order to make use of such data, a number of generic proteins were defined (Supplementary Table 5). This was strictly limited to cases where isoforms were judged to be functionally identical, or where the level of resolution was most appropriate to the analysis. These were as follows: G-α proteins treated as four classes (s/i/q/12); G β/γ treated as a single entity; 14-3-3 isoforms treated as a single entity; PKA subunits fused into a single entity and PP2a isoforms fused into a single entity. Prior to this, the MASC set was supplemented with a minimal number of additional proteins: the catalytic subunit of PKA (the regulatory subunit having been found in MASC) and G-α i/q/12. While only G-α s was identified by proteomic studies, all classes are known to interact with MASC proteins, and all have been identified in the PSD. For a complex involved in signal transduction via ionotropic and metabotropic receptors, their inclusion seems natural. The manipulations described above, resulting in a final set of 182 proteins, were carried out prior to the statistical and network analyses described in the text.
Functional correlation
The significance of the overlap between a pair of annotations was evaluated by calculating its probability of occurrence under a random distribution. Suppose that within a set of N proteins, n a and n b possess annotations a and b, respectively. If both annotations are distributed randomly throughout the set, the probability of n ab proteins possessing both annotations is given by the function:
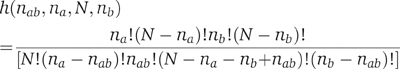
Given the actual number of proteins possessing both annotations, μ ab , we estimate its significance by calculating the probability P(μ ab ) of an overlap as or less likely under the random distribution:
![]()
These values were used without any further adjustment to account for the number of comparisons made. This was done for several reasons. Annotations are not independent, with many correlated to a significant degree (see, e.g, the Interpro annotations of Table I). It was also felt that, due to the incomplete and potentially uneven nature of much of the data, it was better policy to retain sensitivity. As a consequence, we have tried to avoid placing undue stress on isolated scores unsupported by other evidence.
Network analysis
Protein–protein binding data were mined from studies describing protein interactions in any relevant mammalian source (cell or specific organ) and has been described elsewhere (Husi and Grant, 2002). Protein sequence alignment was used to identify splice variants and orthologues across the mouse, rat and human genomes and synonyms were collected for each protein entity. The synonym list was used to search PubMed for scientific reports that may describe protein interactions. Interactions described in BIND, GRID and the commercial database NetPro (https://www.molecularconnections.com/home.html) were also identified. All interaction data were manually curated: evidence for binary interactions between protein pairs was expertly annotated and relevant PMID numbers stored with interactions (Supplementary tables). No high-throughput yeast 2 hybrid data was included unless confirmed by other techniques. A second curator re-checked all interactions used in our analysis.
Power-law analysis was performed as a linear regression fit of ln p(k) to ln k, where k is the number of interactions and p(k) the probability that a protein has k interactions.
Clustering was performed with the algorithm of Newman and Girvan (2004), using edge (rather than the shortest path) betweenness. The modularity score Q that they define was used to identify the best clustering of the network. Interaction data incorporated while this manuscript was in preparation was found to alter the clustering identified by the algorithm. With a modularity score of Q=0.56 (calculated with updated interaction data), the original clustering of the network appeared to better reflect its structure than the altered clustering (Q=0.53). To look at the variance in these scores, we calculated Q for all neighbouring clusterings that differ by moving a single protein into another cluster in which it has an interaction partner. For the neighbourhood containing the original clustering Q=0.5502±0.0074 (mean±s.d.), while for the altered clustering Q=0.5233±0.0073. The original clustering seems to be a markedly better reflection of network structure, and is the version presented here. Note that the majority of the significant overlaps between clusters and annotations discussed in the text were found to be valid for analogous clusters in the alternative network clustering.
The % change in 100 Hz LTP (as a percentage of baseline EPSP) was calculated as: 100 × (mutant−wild type)/(wild type−100), where wild type and mutant are the mean changes in amplitudes (% baseline) of excitatory postsynaptic potentials (evoked by test stimulus) caused by a 100 Hz stimulation protocol (see Supplementary Table 11). Where multiple sets of experimental data were available for a single protein, the absolute value of the mean change was used.

Supplementary Material
Legends to Supplementary tables
Supplementary Tables
Acknowledgments
We thank Dr B Webber, Dr D Barber, Mr T Theodosiou, Ms S Sarmento, Ms A Delaney, Mr MO Collins, Dr H Husi, Mr M Marshall for bioinformatics assistance and Ms JV Turner for editorial assistance. We also thank Dr D Blackwood, Dr P Brophy, Dr E Hawrot, Dr N Komiyama, Dr P Skehel and Dr J Choudhary for comments on an earlier version of the manuscript. SGNG, MC and JDA were supported by the Wellcome Trust Genes to Cognition programme. AJP was supported by the Medical Research Council (UK) through a Special Research Training Fellowship in Bioinformatics.
References
- Albert R, Jeong H, Barabasi AL (2000) Error and attack tolerance of complex networks. Nature 406: 378–382 [DOI] [PubMed] [Google Scholar]
- Bliss TV, Collingridge GL (1993) A synaptic model of memory: long-term potentiation in the hippocampus. Nature 361: 31–39 [DOI] [PubMed] [Google Scholar]
- Collins MO, Husi H, Yu L, Brandon JM, Anderson CNG, Blackstock WP, Choudhary J, Grant SGN (2005) Molecular characterization and comparison of the components and multi-protein complexes in the postsynaptic density. J Neurochemistry. Online Early, doi:10.1111/j.1471-4159.2005.03507.x [DOI] [PubMed] [Google Scholar]
- Csete M, Doyle J (2004) Bow ties, metabolism and disease. Trends Biotechnol 22: 446–450 [DOI] [PubMed] [Google Scholar]
- Enright AJ, Ouzounis CA (2001) BioLayout—an automatic graph layout algorithm for similarity visualization. Bioinformatics 17: 853–854 [DOI] [PubMed] [Google Scholar]
- Farr CD, Gafken PR, Norbeck AD, Doneanu CE, Stapels MD, Barofsky DF, Minami M, Saugstad JA (2004) Proteomic analysis of native metabotropic glutamate receptor 5 protein complexes reveals novel molecular constituents. J Neurochem 91: 438–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant SG, Marshall MC, Page KL, Cumiskey MA, Armstrong JD (2005) Synapse proteomics of multiprotein complexes: en route from genes to nervous system diseases. Hum Mol Genet 14 (suppl 2): R225–R234; doi:10.1093/hmg/ddi330 [DOI] [PubMed] [Google Scholar]
- Grant SG, O'Dell TJ, Karl KA, Stein PL, Soriano P, Kandel ER (1992) Impaired long-term potentiation, spatial learning, and hippocampal development in fyn mutant mice. Science 258: 1903–1910 [DOI] [PubMed] [Google Scholar]
- Greengard P (2001) The neurobiology of slow synaptic transmission. Science 294: 1024–1030 [DOI] [PubMed] [Google Scholar]
- Husi H, Grant SG (2001) Isolation of 2000-kDa complexes of N-methyl-D-aspartate receptor and postsynaptic density 95 from mouse brain. J Neurochem 77: 281–291 [DOI] [PubMed] [Google Scholar]
- Husi H, Grant SG (2002) Neuroscience Databases: A Practical Guide. Boston/Dordrecht/London: Kluwer Academic Publishers, pp 51–62 [Google Scholar]
- Husi H, Ward MA, Choudhary JS, Blackstock WP, Grant SG (2000) Proteomic analysis of NMDA receptor-adhesion protein signaling complexes. Nat Neurosci 3: 661–669 [DOI] [PubMed] [Google Scholar]
- Jeong H, Mason SP, Barabasi AL, Oltvai ZN (2001) Lethality and centrality in protein networks. Nature 411: 41–42 [DOI] [PubMed] [Google Scholar]
- Jeong H, Tombor B, Albert R, Oltvai ZN, Barabasi AL (2000) The large-scale organization of metabolic networks. Nature 407: 651–654 [DOI] [PubMed] [Google Scholar]
- Kandel ER (2001) The molecular biology of memory storage: a dialogue between genes and synapses. Science 294: 1030–1038 [DOI] [PubMed] [Google Scholar]
- Kandel ER, Schwartz JH, Jessell TM (2000) Principles of Neural Science. New York, USA: McGraw-Hill Health Professions Division [Google Scholar]
- Komiyama NH, Watabe AM, Carlisle HJ, Porter K, Charlesworth P, Monti J, Strathdee DJ, O'Carroll CM, Martin SJ, Morris RG, O'Dell TJ, Grant SG (2002) SynGAP regulates ERK/MAPK signaling, synaptic plasticity, and learning in the complex with postsynaptic density 95 and NMDA receptor. J Neurosci 22: 9721–9732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma HW, Zeng AP (2003) The connectivity structure, giant strong component and centrality of metabolic networks. Bioinformatics 19: 1423–1430 [DOI] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S (2002) The protein kinase complement of the human genome. Science 298: 1912–1934 [DOI] [PubMed] [Google Scholar]
- Migaud M, Charlesworth P, Dempster M, Webster LC, Watabe AM, Makhinson M, He Y, Ramsay MF, Morris RG, Morrison JH, O'Dell TJ, Grant SG (1998) Enhanced long-term potentiation and impaired learning in mice with mutant postsynaptic density-95 protein. Nature 396: 433–439 [DOI] [PubMed] [Google Scholar]
- Newman MEJ, Girvan M (2004) Finding and evaluating community structure in networks. Phys Rev E 69: 026113 [DOI] [PubMed] [Google Scholar]
- Nishimune A, Isaac JT, Molnar E, Noel J, Nash SR, Tagaya M, Collingridge GL, Nakanishi S, Henley JM (1998) NSF binding to GluR2 regulates synaptic transmission. Neuron 21: 87–97 [DOI] [PubMed] [Google Scholar]
- Opazo P, Watabe AM, Grant SG, O'Dell TJ (2003) Phosphatidylinositol 3-kinase regulates the induction of long-term potentiation through extracellular signal-related kinase-independent mechanisms. J Neurosci 23: 3679–3688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson T (2004) Specificity in signal transduction: from phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell 116: 191–203 [DOI] [PubMed] [Google Scholar]
- Porter K, Komiyama NH, Vitalis T, Kind PC, Grant SG (2005) Differential expression of two NMDA receptor interacting proteins, PSD-95 and SynGAP during mouse development. Eur J Neurosci 21: 351–362 [DOI] [PubMed] [Google Scholar]
- Rogers EE, Theibert AB (2002) Functions of PI 3-kinase in development of the nervous system. Int J Dev Neurosci 20: 187–197 [DOI] [PubMed] [Google Scholar]
- Salter MW, Kalia LV (2004) Src kinases: a hub for NMDA receptor regulation. Nat Rev Neurosci 5: 317–328 [DOI] [PubMed] [Google Scholar]
- Sanes JR, Lichtman JW (1999) Can molecules explain long-term potentiation? Nat Neurosci 2: 597–604 [DOI] [PubMed] [Google Scholar]
- Song C, Havlin S, Makse HA (2005) Self-similarity of complex networks. Nature 433: 392–395 [DOI] [PubMed] [Google Scholar]
- Sweatt JD (2004) Mitogen-activated protein kinases in synaptic plasticity and memory. Curr Opin Neurobiol 14: 311–317 [DOI] [PubMed] [Google Scholar]
- Thomas GM, Huganir RL (2004) MAPK cascade signalling and synaptic plasticity. Nat Rev Neurosci 5: 173–183 [DOI] [PubMed] [Google Scholar]
- Tu JC, Xiao B, Naisbitt S, Yuan JP, Petralia RS, Brakeman P, Doan A, Aakalu VK, Lanahan AA, Sheng M, Worley PF (1999) Coupling of mGluR/Homer and PSD-95 complexes by the Shank family of postsynaptic density proteins. Neuron 23: 583–592 [DOI] [PubMed] [Google Scholar]
- Watabe AM, Zaki PA, O'Dell TJ (2000) Coactivation of beta-adrenergic and cholinergic receptors enhances the induction of long-term potentiation and synergistically activates mitogen-activated protein kinase in the hippocampal CA1 region. J Neurosci 20: 5924–5931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts DJ, Strogatz SH (1998) Collective dynamics of ‘small world' networks. Nature 393: 440–442 [DOI] [PubMed] [Google Scholar]
- Yasuda H, Barth AL, Stellwagen D, Malenka RC (2003) A developmental switch in the signaling cascades for LTP induction. Nat Neurosci 6: 15–16 [DOI] [PubMed] [Google Scholar]
- Zapala MA, Hovatta I, Ellison JA, Wodicka L, Del Rio JA, Tennant R, Tynan W, Broide RS, Helton R, Stoveken BS, Winrow C, Lockhart DJ, Reilly JF, Young WG, Bloom FE, Lockhart DJ, Barlow C (2005) Adult mouse brain gene expression patterns bear an embryologic imprint. Proc Natl Acad Sci USA 102: 10357–10362 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Legends to Supplementary tables
Supplementary Tables