Abstract
We present a genome-scale metabolic model for the archaeal methanogen Methanosarcina barkeri. We characterize the metabolic network and compare it to reconstructions from the prokaryotic, eukaryotic and archaeal domains. Using the model in conjunction with constraint-based methods, we simulate the metabolic fluxes and resulting phenotypes induced by different environmental and genetic conditions. This represents the first large-scale simulation of either a methanogen or an archaeal species. Model predictions are validated by comparison to experimental growth measurements and phenotypes of M. barkeri on different substrates. The predicted growth phenotypes for wild type and mutants of the methanogenic pathway have a high level of agreement with experimental findings. We further examine the efficiency of the energy-conserving reactions in the methanogenic pathway, specifically the Ech hydrogenase reaction, and determine a stoichiometry for the nitrogenase reaction. This work demonstrates that a reconstructed metabolic network can serve as an analysis platform to predict cellular phenotypes, characterize methanogenic growth, improve the genome annotation and further uncover the metabolic characteristics of methanogenesis.
Keywords: archaeal metabolism, metabolic modeling, methanogenesis, Methanosarcina barkeri, network reconstruction
Introduction
Metabolic reconstruction is a process through which the genes, proteins, reactions and metabolites that participate in the metabolic activity of a biological system are identified, categorized and interconnected to form a network. Most often, the system is a single cell of interest and, by using the genomic sequence as a scaffold, reconstructions can incorporate hundreds of reactions that approximate the entire metabolic activity of a cell. With the growing availability of genome sequences for eukaryotic, prokaryotic and archaeal species, genome-scale metabolic reconstructions have been performed for organisms across all three of these domains (for a review, see Reed et al, 2006).
Within the eukaryotic and prokaryotic domains in particular, metabolic reconstructions have been analyzed using constraint-based methods, which simulate our current understanding of metabolism in an organism and drive experiments to verify modeling predictions (Covert et al, 2004). Constraint-based methods enforce cellular limitations on biological networks such as physico-chemical constraints, spatial or topological constraints, environmental constraints or gene regulatory constraints (Price et al, 2004). One specific example of metabolic modeling using a constraint-based approach is flux balance analysis (FBA). FBA uses linear optimization to determine the steady-state reaction flux distribution in a metabolic network by maximizing an objective function, such as ATP production or growth rate (Kauffman et al, 2003). The effectiveness of metabolic modeling using constraint-based methods has been demonstrated in predicting the outcomes of gene deletions (Duarte et al, 2004), identifying potential drug targets (Yeh et al, 2004), engineering optimal production strains for bioprocessing (Burgard et al, 2003) and elucidating cellular regulatory networks (Covert et al, 2004). Analytical methods are continually being developed to understand additional emergent properties of metabolic models and to expand their application; for a review see Price et al (2004).
Surprisingly, constraint-based analysis has not yet been applied to study the metabolism of methanogenesis or archaeal organisms. Although high-quality organism-specific metabolic pathway databases are available for several archaea (Tsoka et al, 2004; see also BioCyc website, http://biocyc.org/), such databases have not yet been curated for constraint-based analysis which requires that the network (i) has been evaluated to produce biomass constituents, such as amino acids, nucleotides and lipids, (ii) has sufficient representation of how metabolites enter and leave the cell, and (iii) contains explicit substrates, products and reversibility for all reactions.
Among archaea, methanogens are an attractive model because of their utilization of low carbon substrates, metabolic diversity and the availability of detailed information on their metabolism. Methanogens also have major environmental and economic importance. They serve as a key component of the carbon cycle by degrading low carbon molecules in anaerobic environments to generate methane. Because of this, methanogens have been used for processing of industrial, agricultural and toxic wastes rich in organic matter (Zinder, 1993). Methanogens contribute to the greenhouse effect and are a potential source of renewable energy (Garcia et al, 2000). Moreover, a number of methanogenic archaea can form syntrophic relationships with eubacteria, allowing for the study of metabolite and energy coupling across species (Schink, 1997). Although many pieces of methanogenic metabolism are understood, there are still many questions to be answered about the biochemistry of methanogenesis and how these pieces work together in the context of the whole organism. Reconstruction and analysis at a genome scale would better determine the biochemical properties of key components and analyze methanogenic metabolism as a whole in its cellular context.
To better understand the general metabolic capabilities of the archaeal domain, and methanogenesis in particular, we reconstructed the metabolic network of the archaeal methanogen Methanosarcina barkeri and performed constraint-based analysis on the genome-scale model. M. barkeri is one of the most versatile methanogens and is capable of growing on all three of the major methanogenic substrates: methanol, acetate and H2/CO2 (Zinder, 1993). An isolated strain was also shown to utilize the uncommon methanogenic substrate, pyruvate (Bock et al, 1994). Our metabolic reconstruction, labeled iAF692 following a previously established naming convention (Reed et al, 2003), represents the first curated genome-scale model of an archaea generated specifically for constraint-based modeling. We use this model to determine the growth capabilities for M. barkeri for both wild type (WT) and mutant strains. We also examine the maintenance energy requirements for growth, minimal media requirements and the stoichiometry of energy-conserving proton and ion translocating reactions in the methanogenic process.
Results and discussion
Reconstructing the M. barkeri model
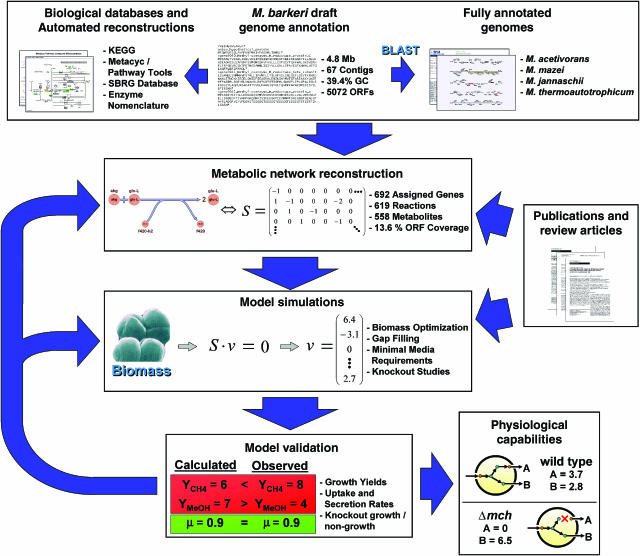
The metabolic reconstruction of M. barkeri, iAF692, was generated and refined using an iterative model building procedure (see Materials and methods and Figure 1). The model contains 692 metabolic genes associated with 509 reactions and 558 distinct metabolites (see Table I). An additional 110 reactions were included because they have been reported in prior literature, or because they were required to fill a gap in the reconstructed network (see Materials and methods). However, these are currently unassociated with any gene product in the annotation. iAF692 and constraint-based optimization results are available as Systems Biology Markup Language (SBML) files (level 2, version 1, http://sbml.org/, Supplementary information 4, 5, 6 and 7) or in spreadsheet form (Supplementary information 1).
Figure 1.
The iterative model building procedure used to generate iAF692. The draft genome annotation was used as a scaffold, on which GPR assignments were made. The reactions added to the model were taken from both biochemical databases and published data. Once a reaction was found to be in the network, it was manually curated and either associated to a potential ORF or added with no gene assignment. A biomass objective function was formulated to perform model simulations based on cellular composition. Modeling simulations were run under steady-state conditions to determine the reaction flux distribution in the network. The results from the simulations were interpreted and compared to experimental data. From the comparison, physiological capabilities of the cell were confirmed or the network was further refined or updated.
Table 1.
Properties of the archaeal metabolic reconstructions of M. barkeri and M. jannaschii
| iAF692 M. barkeri | Tsoka et al (2004) M. jannaschii | |
|---|---|---|
| Genome size | 4.8 Mb | 1.7 Mb |
| ORFs | 5072 | 1792 |
| Included genes | 692 | 436 |
| Unique proteins | 542 | 266 |
| Multiple gene associations | 96 | 67 |
| Enzyme complexes | 65 | NR |
| Instances of isozymes | 31 | NR |
| Reactions | 619 | 609 |
| Gene associated | 509 (82%) | 297 (49%) |
| No gene association | 110 (18%) | 312 (51%) |
| Transport reactions | 88 | 1 |
| Metabolites | 558 | 510 |
| Transported metabolites | 56 | 1 |
| Freely diffusible | 14 | NR |
NR, not reported.
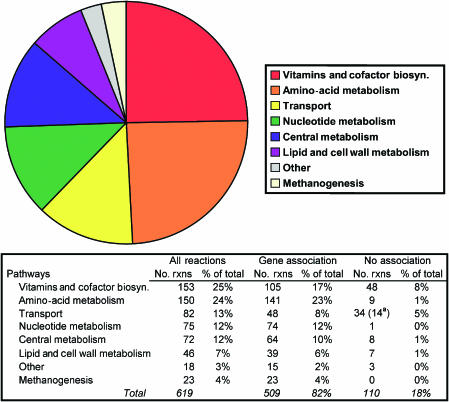
The reactions in iAF692 were subdivided into eight high-level functional categories based on the major metabolic roles of the cell (Figure 2). The largest number of reactions (153) was involved in the biosynthesis of vitamins and cofactors, probably because M. barkeri synthesizes many large molecular weight cofactors that require multiple enzymatic steps (Graham and White, 2002). M. barkeri contains all of the de novo pathways required to synthesize the 20 common amino acids (Zinder, 1993) and these pathways, containing 141 gene-associated reactions, are well characterized in methanogenic archaea (Peregrin-Alvarez et al, 2003). Transport reactions were another major functional class. Of the 88 transport reactions, 54 were included from the annotation, 31 were included from physiological data alone, whereas three were added from growth simulation requirements. The high number of transport reactions with no gene assignment in M. barkeri points to the fact that further work is needed to characterize the mechanisms and machinery involved in the transport of molecules in archaea.
Figure 2.
The distribution of reactions in iAF692. The table gives the pathway distribution for the total, gene-associated and non gene-associated reactions. Non gene-associated reactions were added on the basis of biochemical, physiological or modeling evidence. aDenotes that 14 reactions are diffusion reactions and would not require a gene association.
Reconstruction as an annotation tool
The iAF692 model suggests 55 new functional annotations for predicted open reading frames (ORFs) in the M. barkeri genome. These ORFs were either uncharacterized (30 genes) or likely misannotated in the draft annotation (25 genes). The model assists with functional annotation in cases in which a gene has multiple strong BLAST hits versus other species, or has only weak sequence homologies to other genes. The model acts to filter these lists of ambiguous matches, by indicating which homologous genes fulfill a metabolic requirement of the cell or bridge a gap between metabolites in the network. A list of the potential ORFs annotated during the reconstruction is given in Supplementary information 1.
One example of a functional prediction made during reconstruction is the case of 7,8-didemethyl-8-hydroxy-5-deazariboflavin (FO) synthase. FO is a chromophore that comprises part of the methanogenic cofactor coenzyme F420 (Graham et al, 2003). M. barkeri has been verified to produce coenzyme F420 for use in the methanogenic process (de Poorter et al, 2005). Although there are no genes annotated as FO synthase in M. barkeri or in any of the other Methanosarcina species, the enzyme has been characterized in Methanococcus jannaschii and is catalyzed by two different subunits, CofG and CofH (Graham et al, 2003). Three sequential genes from contig 187 (gene 838, 839, 840) of the M. barkeri draft annotation were identified as orthologs to the biochemically verified genes CofG and CofH from M. jannaschii using BLAST. Gene 838 is a predicted ortholog to the CofG gene, whereas genes 839 and 840 are two predicted paralogs that are orthologous to the CofH gene of M. jannaschii. The sequential chromosomal location of the three genes on contig 187 also supports the gene-protein-reaction (GPR) assignments in the model.
Comparison of iAF692 with previous metabolic reconstructions
The major differences between iAF692 and previous archaeal reconstructions (Tsoka et al, 2004; see also BioCyc website, http://biocyc.org/) are as follows: (i) the number of gene-associated reactions, (ii) the organism specificity of the reactions, (iii) defined reversibility and assurance of elementally and charge balanced reactions, (iv) the inclusion of sufficient transport reactions to support growth, (v) incorporation of physiological information and (vi) further curation of the model after comparison of flux simulations with experimental data. For instance, in the reconstruction of M. jannaschii by Tsoka et al (2004), 49% of the reactions in the network were gene-associated after curation in comparison to 82% in iAF692 (Table I). It is surprising that the metabolic model of M. jannaschii had roughly the same number of reactions as iAF692, but a much smaller genome. Although this result could indicate that many M. barkeri genes encode for functions that are nonmetabolic, it was at least partially because of the fact that reactions involving DNA, proteins and unspecified products/substrates were included in the M. jannaschii reconstruction, and that some predicted ORFs from the M. barkeri draft annotation may not be real genes.
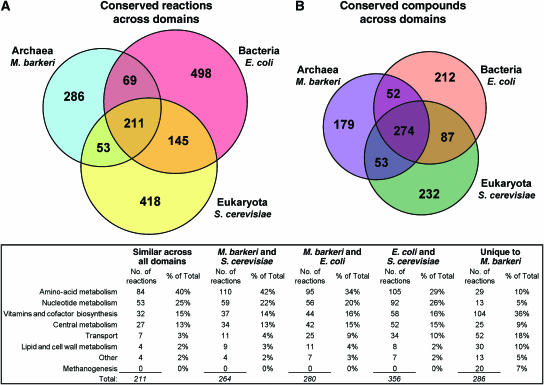
We also systematically compared iAF692 to previous metabolic models from the prokaryotic and eukaryotic domains, the other two domains of life. Figure 3 compares the content (reactions and metabolites) of the M. barkeri model with that of Escherichia coli, iJR904 (Reed et al, 2003), and Saccharomyces cerevisiae, iND750 (Duarte et al, 2004). All of these models shared a core set of 211 reactions (12.6% overall) and 274 metabolites (25.2% overall), indicating that the metabolites contained in the models are more highly conserved than the biochemical conversions between them (the reactions). This core set of reactions predominately involved biosynthesis and degradation of amino acids and nucleotides; a full list of conserved reactions is available in Supplementary information 2. Several pathways encoded by the model were primarily found only in or are specific to archaea. For instance, iAF692 contains all of the reactions in the methanogenic pathway necessary for growth on all known M. barkeri substrates (23 reactions associated with 125 distinct genes) and the biosynthetic pathways to generate all of the specific Methanosarcina species cofactors. Included are the biosynthetic pathways for coenzyme M, coenzyme B, tetrahydrosarcinapterin (H4SPT), coenzyme F420, coenzyme F430, coenzyme F390 and the anaerobic pathway for the synthesis of a vitamin B12 derivative (see Supplementary information 1 for references). iAF692 also contains the biosynthesis pathways for the unique archaeal ether-linked lipids (46 reactions generating nine distinct lipids; Nishihara and Koga, 1995). Fifty-two transport reactions were unique to M. barkeri, probably because of the specialized nature of many of the methanogenic substrates (Hippe et al, 1979). A map of the complete metabolic network of iAF692 including the methanogenic pathway, the lipid biosynthesis pathway, vitamin and cofactor biosynthesis, amino-acid metabolism and nucleotide metabolism is available in Supplementary Figure 1.
Figure 3.
(A) The conserved reactions and (B) the conserved compounds among reconstructed metabolic models from the three phylogenetic domains. The models of the archaea M. barkeri, iAF692, the bacteria E. coli, iJR904, and the eukaryote S. cerevisiae, iND750, were compared to determine identical reactions and compounds contained in the models. All of the models were decompartmentalized so that only the reactions and cytosolic transporters were compared and not their location inside of the cell. The table gives information on the distribution of reactions in their respective pathways.
A comparison of global topological properties of the metabolic networks is given in Table II. Comparing M. barkeri to other models generated specifically for constraint-based analysis (see Table II), M. barkeri and S. cerevisiae were more similar to each other than to E. coli. For instance, M. barkeri and S. cerevisiae had a longer average path length than E. coli, and also had a smaller average degree and network diameter. These findings show that the E. coli metabolic network is more connected than those of M. barkeri and S. cerevisiae and suggest that these latter models have less redundancy in their network structure. All three networks followed a power law degree distribution implying that the models are scale-free networks (see Supplementary Figure 2) and also contained one large connected component of reactions (the giant strong component (GSC), see Ma and Zeng, 2003) along with several isolated subnetworks composed of linear and significantly smaller connected pathways. As argued by Ma and Zeng (2003), the GSC contains most of the core metabolites. The number of metabolites in each subnetwork is given in Table II and the metabolites present in each subnetwork for each model are given in Supplementary information 3. iAF692 contained fewer links (reactions) and nodes (metabolites) than iJR904 or iND750. This was not surprising given the level of genetic characterization of both E. coli and S. cerevisiae (Janssen et al, 2005). Table II also shows that constraint-based models are more connected than those generated from biochemical databases and reversibility rules (Ma and Zeng, 2003).
Table 2.
Network properties for selected metabolic reconstructions
| Organism | Reconstruction reference | Linksa (irr/rev) | Metsb | APL | D | 〈k〉 | SC | Network structure subsets | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| GSC | S | P | IS | ||||||||
| M. barkeri | This study | 636/253 | 592 | 8.00 | 24 | 3.03 | 7 | 322 | 54 | 131 | 85 |
| E. coli | Reed et al (2003) | 1076/333 | 725 | 6.75 | 19 | 3.89 | 8 | 468 | 145 | 65 | 47 |
| S. cerevisiae | Duarte et al (2004) | 1073/536 | 972 | 8.00 | 31 | 3.36 | 11 | 629 | 98 | 125 | 120 |
| E. coli | Ma and Zeng (2003) | NR | 811 | 8.20 | 23 | NR | 29 | 274 | 93 | 161 | 283 |
| S. cerevisiae | Ma and Zeng (2003) | NR | 679 | 9.71 | NR | NR | NR | 206 | 54 | 164 | 255 |
| NR, not reported; irr, irreversible; rev, reversible; Mets, metabolites; APL, average path length—average over the shortest paths between all metabolites; D, network diameter—shortest path length between the most distant metabolites in a network; 〈k〉, average degree—average number of links per metabolite; SC, strong components—fully connected subnetworks; GSC, giant strong component—the largest strong component; S, substrate subset—metabolites that can be converted to metabolites in the GSC; P, product subset—metabolites that can be produced from metabolites in the GSC; IS, isolated subset—metabolites that can not be converted to or produced from metabolites in the GSC. | |||||||||||
| The network properties for the metabolic reconstructions generated by Reed et al (2003), Duarte et al (2004), and those of the M. barkeri model were calculated in this study. These models were built specifically for use with constraint-based methods. The additional network properties were reported for reconstructions generated by Ma and Zeng (2003). | |||||||||||
aModel compartmentalization was conserved (Duarte et al, 2004).
bCurrency metabolites were removed from each network.
Computational analysis of minimal media for M. barkeri
Minimal media conditions were determined which could produce all of the biomass constituents found in the biomass objective function (BOF) for M. barkeri. The BOF is a linear equation consisting of the molar amounts of metabolites that comprise the dry weight content of the cell (Table III) along with a growth maintenance reaction (see Supplementary information 1). Optimization of the network to maximize the reaction flux through the BOF simulates a cell which strives to maximize the generation of biomass constituents from available media substrates. Beyond the primary source (methanol, acetate, CO2 or pyruvate), two additional carbon-containing compounds were needed to generate the metabolites present in the BOF: p-aminobenzoic acid (pABA) and nicotinic acid. pABA is needed for the biosynthesis of folic acid (Buchenau and Thauer, 2004) and H4SPT (Graham and White, 2002), whereas nicotinic acid is used in the generation of nicotinaminde coenzymes.
Table 3.
Estimated biomass composition of M. barkeri
| Metabolite | mmol gDW−1 | Metabolite | mmol gDW−1 | Metabolite | mmol gDW−1 |
|---|---|---|---|---|---|
| Protein (63%) a | RNA (24%) a | Soluble pools (4%) | |||
| Alanine | 0.5621 | ATP | 0.1846 | Putrescine | 0.0262 |
| Arginine | 0.3237 | CTP | 0.1379 | Homospermidine | 0.0047 |
| Asparagine | 0.2638 | GTP | 0.2222 | Acetyl-Coenzyme A | 0.0001 |
| Aspartate | 0.2638 | UTP | 0.1489 | Coenzyme A | <0.0001 |
| Cysteine | 0.1002 | DNA (4%) a | NAD+ | 0.0022 | |
| Glutamine | 0.2880 | dATP | 0.0331 | NADH | 0.0001 |
| Glutamate | 0.2880 | dCTP | 0.0215 | NADP+ | 0.0001 |
| Glycine | 0.6704 | dGTP | 0.0215 | NADPH | 0.0004 |
| Histidine | 0.1037 | dTTP | 0.0331 | Succinyl-Coenzyme A | <0.0001 |
| Isoleucine | 0.3179 | Lipid (5%) | Coenzyme M | 0.0206 | |
| Leucine | 0.4930 | Archaetidylglycerol | 0.0007 | Coenzyme F420 | 0.0008 |
| Lysine | 0.3755 | Hydroxyarchaetidylglycerol | 0.0101 | Tetrahydrosarcinapterin | 0.0236 |
| Methionine | 0.1682 | Archaetidylinositol | 0.0027 | Adenosylcobalamin-HBI | 0.0047 |
| Phenylalanine | 0.2027 | Hydroxyarchaetidylinositol | 0.0135 | Coenzyme F430 | 0.0020 |
| Proline | 0.2419 | Archaetidylethanolamine | 0.0007 | Coenzyme B | 0.0005 |
| Serine | 0.2361 | Hydroxyarchaetidylethanolamine | 0.0027 | 5,6,7,8-Tetrahydrofolate | 0.0001 |
| Threonine | 0.2776 | Archaetidylserine | 0.0013 | Coenzyme F390 | <0.0001 |
| Tryptophan | 0.0622 | Hydroxyarchaetidylserine | 0.0115 | ATP | 0.0040 |
| Tyrosine | 0.1509 | Glucosaminyl archaetidylinositol | 0.0162 | ADP | 0.0020 |
| Valine | 0.4631 | Carbohydrates (<1%) | AMP | 0.0010 | |
| Glycogen | 0.0154 |
aRelative ratios of wt% of protein, RNA and DNA were estimated from the composition of a typical prokaryotic cell (Neidhardt et al, 1990). The values were taken from published data and converted to mmol gDW−1 and the wt% of each category is given (for references, see Supplementary information 1).
However, Buchenau and Thauer (2004) reported that pABA may not be necessary for the biosynthesis of H4SPT. A possible alternate pathway in M. barkeri is discussed below. Other carbon-containing compounds commonly found in M. barkeri media (Wolin et al, 1963) such as biotin, folic acid (from pABA), thiamine, pantothenate and vitamin B12 can be synthesized by the network and thus were not essential for simulated growth. There is corroborating experimental evidence that M. barkeri is not dependent on these compounds for optimal growth (Scherer and Sahm, 1981). On the other hand, Scherer and Sahm (1981) stated that riboflavin (found to be nonessential using iAF692) was required for optimal growth. This finding suggests that the de novo pathway to synthesize riboflavin in M. barkeri (http://genome.ornl.gov/microbial/mbar/, described for similar archaea by Fischer et al, 2004) may not be sufficient for optimal growth. In addition to carbon-containing compounds, the required compounds are metals, phosphate, sulfur and nitrogen. The stoichiometry of the nitrogenase reaction in M. barkeri was computationally determined (see below) and further details for the other media requirements are provided in Supplementary text.
Estimation of the proton translocation efficiency of the Ech hydrogenase reaction
To investigate the predictive power of iAF692, we examined three different uncharacterized areas of M. barkeri metabolism using a model-driven approach: (i) the stoichiometry of the Ech hydrogenase reaction, (ii) the stoichiometry of the nitrogenase reaction and (iii) an alternate pathway for the biosynthesis of H4SPT.
Although the methanogenic process is well defined and has been considerably reviewed (see Supplementary information 1 for references), several aspects are still poorly understood. One of these aspects is the efficiency of the energy-conserving ion translocating reactions of the methanogenic pathway, specifically the Ech hydrogenase catalyzed reaction. These reactions couple conversion of metabolites with the transport of ions across compartmental membranes to create an electrochemical potential (Thauer et al, 1977). This electrochemical potential is used to generate ATP through ATP synthase (ATPS) and to drive reactions that are otherwise energetically unfavorable (Deppenmeier, 2004). Ech hydrogenase is one of the six energy-conserving ion translocating enzymes of the methanogenic pathway in M. barkeri, and the only one for which the stoichiometry is unknown (i.e., the number of protons translocated per electrons transferred) (Deppenmeier, 2004; Muller, 2004).
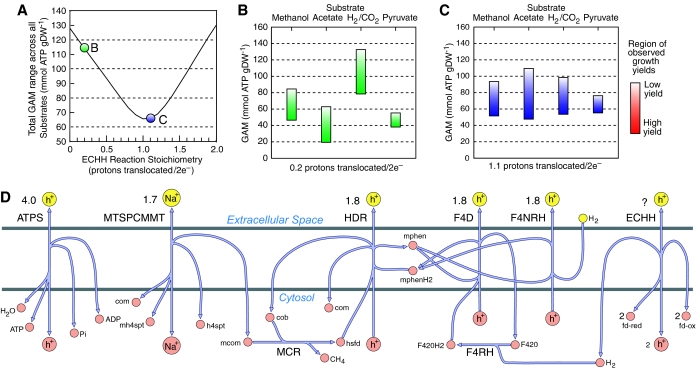
We examined the effect of the Ech hydrogenase reaction stoichiometry on the flux distribution and resulting growth yields for M. barkeri using iAF692. In this approach, we used FBA and BOF optimization to determine which choices of Ech hydrogenase stoichiometry resulted in the experimentally observed growth yields for various substrates (see Materials and methods). However, to calculate a flux distribution, additional parameters were needed. Two of these parameters, the growth-associated maintenance (GAM) and non-growth-associated (NGAM) maintenance, were also unknown because of the lack of experimental data. To reduce the number of unknown variables, the NGAM was set as 2.5% of the GAM based on previous analyses (see Materials and methods). This left the GAM and the stoichiometry of the Ech hydrogenase reaction as the remaining unknowns. A constraint on the Ech hydrogenase reaction stoichiometry was that it cannot exceed 2 protons translocated/1e− because of thermodynamic reasons (Hedderich, 2004). Given that two electrons are transferred in the Ech hydrogenase reaction, this provided a possible range of 0–4 protons translocated/2e−. However, the value probably lies closer to that of another hydrogenase from the same family, hydrogenase 3 from E. coli, which has an apparent stoichiometry of 1.3 protons translocated/2e− (Hedderich, 2004; Hakobyan et al, 2005).
For proton translocation efficiencies in the feasible range (0–4 protons translocated/2e−), FBA simulations were used to find the corresponding ranges of GAM values that were consistent with observed growth yields. The variability in these ranges is shown in Figure 4A as a function of the Ech hydrogenase reaction stoichiometry (0–2.0 protons translocated/2e−). Any stoichiometry >2.0 protons translocated/2e− created an increasingly larger variability in the GAM values consistent with observed yields across all substrates (see Figure 4 and Supplementary Figure 3). Figure 4B and C show the regions of experimentally observed growth yields for each substrate calculated at a given stoichiometry. A probable stoichiometry for the Ech hydrogenase reaction would be approximately 1.1 protons translocated/2e− if similar GAM values were found for growth on different substrates (Figure 4C). Using this data set, a stoichiometry ⩾2.0 or ⩽0.2 protons translocated/2e− appears unlikely because (i) GAM values typically vary less than 2.5-fold across all substrates for an extreme case (Russell and Cook, 1995) and (ii) the lowest value of GAM which produced a consistent yield is approaching the minimum theoretical cost for the polymerization of cellular macromolecules, 26 mmol ATP gDW−1 (see Figure 4B and Supplementary Figure 3). The overall rates of end-product formation and product/substrate yields produced during FBA simulations were determined to be unique for each substrate and consistent with experimental data (Supplementary Table).
Figure 4.
The effect of the Ech hydrogenase reaction stoichiometry on growth yields. (A) Variability in the growth-associated maintenance (GAM) that will produce growth yields consistent with experimental data. This variability is shown as a function of Ech hydrogenase reaction stoichiometry. Yields were calculated using iAF692 for four different substrates (methanol, acetate, H2/CO2 and pyruvate). Panel A shows the total GAM variability across all substrates. (B, C) GAM variability for each substrate at a constant Ech hydrogenase reaction stoichiometry (0.2 protons translocated/2e− (B, green) and 1.1 protons translocated/2e− (C, purple)). The ranges calculated were constrained by experimental values. (D) A diagram of all energy-conserving ion translocating reactions in M. barkeri, each labeled with the stoichiometry of the translocated ion. Any value greater than 2 protons translocated/2e− for Ech hydrogenase created a larger total GAM range across all substrates. ATPS, ATP synthase; MTSPCMMT, methyl-H4SPT:coenzyme M methyltransferase; HDR, heterodisulfide reductase; F4NRH, F420-nonreducing hydrogenase; F4D, F420H2 dehydrogenase; ECHH, Ech hydrogenase; MCR, methyl-coenzyme M reductase; F4RH, F420-reducing hydrogenase; Pi, pyrophosphate; com, coenzyme m, mh4spt, methyl-tetrahydrosarcinapterin, h4spt, tetrahydrosarcinapterin; mcom, methyl-coenzyme M; cob, coenzyme B; hsfd, heterodisulfide; mphen, oxidized methanophenazine; mphenH2, reduced methanophenazine; F420H2, reduced coenzyme F420; F420, oxidized coenzyme F420; fd-red, reduced ferredoxin; fd-ox, oxidized ferredoxin.
Determination of the stoichiometry for the nitrogenase reaction in M. barkeri
M. barkeri was found to contain two different clusters of genes potentially encoding nitrogenases (Chien et al, 2000) and can fix molecular nitrogen at a lower yield per substrate than when supplied with ammonium (Bomar et al, 1985). The stoichiometry (efficiency of energy coupling) of the nitrogenase reaction in M. barkeri is unknown and we estimated this value using iAF692 and experimental data. The data used were growth rates, total substrate consumption and growth yields on methanol and either N2 (which requires nitrogenase to convert N2 to NH4) or NH4 (a control, as nitrogenase is not needed) (Bomar et al, 1985). Again, we were faced with a number of unknown variables in our system (see previous analysis on Ech hydrogenase reaction stoichiometry and Materials and methods). For this analysis, the Ech hydrogenase translocation efficiency was approximated at 1 proton translocated/2e−. For growth on methanol, the overall growth rate is less dependent on the translocation efficiency of Ech hydrogenase compared to acetate or H2/CO2 (at most ±10% in the range of 0.2–2.0 protons translocated/2e−, see Materials and methods). The value of the GAM was determined from BOF optimization using the constraints from the control data (NH4 as a nitrogen source). With all other variables defined for our system, the stoichiometry of the nitrogenase reaction was determined using BOF optimization constrained by the data from diazotrophic growth (growth using N2 as a nitrogen source) and is given in the following balanced equation:
![]()
This nitrogenase stoichiometry shows that energy coupling between ATP and the nitrogenase enzyme(s) was greater than the theoretical limit of ATP hydrolyzed per electron transferred (Rees and Howard, 2000). This finding was not uncommon (Rees and Howard, 2000) and quantifies the energy needed for M. barkeri to grow diazotrophically.
This represents an improvement over the classical approximations used in Bomar et al (1985), in which overall growth yields and amounts of methanol assimilated were approximated using a standardized constant. iAF692 determines these values directly by computing the network flux through the physiological reactions available to the cell. The percentage of methanol assimilated for diazotrophic growth compared to growth on NH4 indicates how much additional methanol is needed to generate ATP for fixing N2 (43 and 56%, respectively). These percentages were significantly different (6 and 14%, respectively) to those computed by Bomar et al (1985). It is also worth mentioning that cells grown diazotrophically and those grown on NH4 had similar nitrogen content (0.069 and 0.066 gN gDW−1, respectively; Bomar et al, 1985), which provides confidence in the use of the same BOF under both growth conditions.
Examination of a possible alternate pathway for the biosynthesis of H4SPT
The possibility of an alternate pathway for the biosynthesis of H4SPT in M. barkeri was proposed by Buchenau and Thauer (2004) when they found that pABA was not required for H4SPT synthesis. Unfortunately, the only pathway characterized for the synthesis of H4SPT involved pABA and was characterized for a H4SPT derivative in M. jannaschii (Graham and White, 2002; Scott and Rasche, 2002). This prevented a computational comparison of opposed pathways using iAF692 and experimental measurements. However, the model was used to identify a possible precursor metabolite, which may aid in the discovery of an alternate pathway (Chunhui et al, 2004). In review of biochemical databases and metabolites present in iAF692, compounds were analyzed to determine their structural similarity to pABA (see Materials and methods). Chorismate was found to be the closest structurally related compound that could be synthesized from basic media substrates in simulations using iAF692. Chorismate can also be converted to pABA in some microbes (Nichols et al, 1989), but this is unlikely in M. barkeri as this would contradict the finding that growth is dependent on either pABA or folic acid (Buchenau and Thauer, 2004). Another possible lead for an alternative pathway was found when a homology search indicated two genes in M. barkeri with strong sequence similarity to the single gene in M. jannaschii responsible for synthesis of the H4SPT derivative from pABA (Scott and Rasche, 2002). A search of the M. jannaschii genome found no probable paralogs to this gene, indicating that M. barkeri may have an additional metabolic capability similar to the function of the pABA utilizing gene. Characterization of the substrates for these probable enzymes in M. barkeri (gene5036 and gene4403) could lead to evidence supporting or refuting a possible alternate pathway.
Gene deletion analysis for the methanogenic pathways in M. barkeri
The essential genes and reactions in the methanogenic pathway needed for growth of M. barkeri on different methanogenic substrates were computationally determined (Figure 5). By using FBA with BOF optimization, it was possible to determine the active gene-encoded reactions and their flux values (Figure 6) that are essential for generating sufficient energy (or any at all) for growth under given substrate conditions (see Materials and methods). Each reaction in turn was deleted from the model, simulating a loss-of-function mutation of any single gene or group of genes associated with the reaction. Through interpretation of the computational results, it was possible to determine why certain mutation states fail to grow whereas others are still viable. The results were categorized into three different conditions: methanogenic growth, acetogenic growth, and no growth (see Materials and methods and Figure 5).
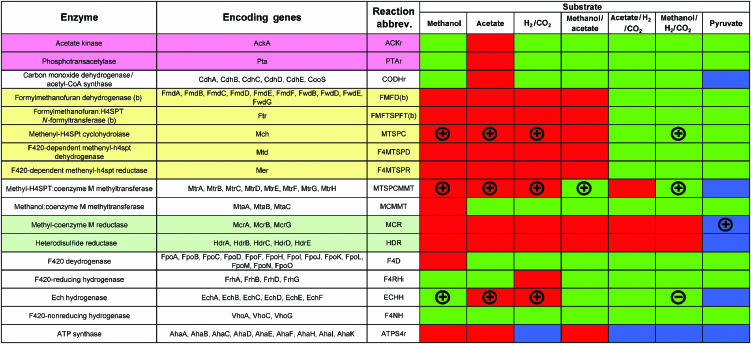
Figure 5.
Essential reactions and genes in the methanogenic pathway of M. barkeri. Listed are the enzymes of the methanogenic pathway, the protein encoding genes needed to produce the functional enzymes and the abbreviation for the reaction they perform. Each of the reactions catalyzed by the enzyme listed was removed from the network and growth phenotypes were determined. For each computational prediction, green indicates methanogenic growth, blue indicates acetogenic growth, and red indicates no growth (0 net flux through the BOF). A plus symbol for a given condition indicates an agreement between model predictions and experimental characterization. A negative symbol indicates a disagreement. The colored enzyme and encoding gene sets (pink, yellow and green) indicate equal flux correlated reaction sets that possess the same reaction flux value under all growth conditions since they belong to a linear pathway. The simulation results can be used to determine the growth phenotypes of mutant strains and interpret the active pathways under each given condition.
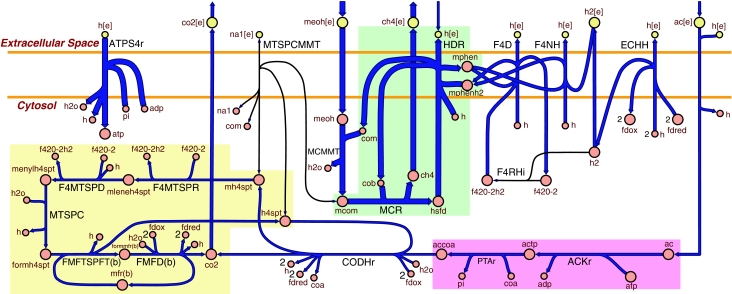
Figure 6.
A flux map of the methanogenic pathway for growth of an mtr mutant on methanol and acetate. Blue arrows indicate the direction of enzymatic activity and the arrow thicknesses are proportional to the flux through each reaction (a thicker arrow has a larger flux). The uptake of acetate (ac) is dependent on the uptake of methanol (meoh), which was constrained at 16 mmol gDW−1 h−1. The MTSPCMMT reaction encoded by the mtr operon is not available to the cell (because of the mutated state) and the F4RHi reaction is predicted not to be active for this optimal growth solution. All of the primary metabolites (large circles) are connected, except for ATP which participates in ATPS4r and ACKr. The secondary metabolites (small circles) that appear more than once in the map are adp, pi, h, h2o, fdred, fdox, f420-2h2, f420-2, coa and com. The different colored regions correspond to the equal flux correlated reaction sets that were determined during the gene deletion analysis and are listed in Figure 5. All the reactions in each set will contain the same flux value for a given condition. The map does not display the stoichiometry for the ion translocating reactions (see Figure 4). The reaction abbreviations are listed in Figure 4 and also in Supplementary information 1 along with the metabolite abbreviations.
Simulation results were compared to experimental measurements on M. barkeri mutants (Figure 5). Three single reaction loss-of-function mutations in the methanogenic pathway (the ech operon, mtr operon and mch gene) have been generated for M. barkeri and characterized for growth on different substrates (Meuer et al, 2002; Guss et al, 2005; Welander and Metcalf, 2005). Also, Bock and Schonheit (1995) characterized growth on pyruvate for an isolated M. barkeri strain when the methyl coenzyme M reductase reaction was completely inhibited (similar to a loss-of-function mutation). The predictions made using iAF692 fully agree with the findings for a M. barkeri mutant lacking the function of the mtr operon (Welander and Metcalf, 2005), the mch gene (Guss et al, 2005) or when the methyl coenzyme M reductase reaction was not available to the cell (Bock and Schonheit, 1995) (see Supplementary text for a discussion of selected substrate and genetic growth conditions).
Meuer et al (2002) characterized a mutant of M. barkeri lacking the functional ech operon, encoding genes involved in methanogenesis. The predictions made using iAF692 agree with the experimental observations on single substrate cultures for the ech mutant (see Figure 5). Conversely, the model does not reproduce the finding that the ech mutant does not grow on methanol/H2/CO2. This is surprising in that the mutant would grow on methanol alone, but not with the addition of H2/CO2 to the medium. One possibility that has been proposed is that the ech mutant did not grow because of repression of the oxidative branch of methanogenesis (mh4spt to co2, see Figure 6) when H2 was added to the medium (Meuer et al, 2002). Although an active oxidative pathway was determined to produce a higher growth rate, this pathway was not essential for simulated growth under these conditions. With only one false positive in this limited data set, the reason for this disagreement will likely become evident once additional genetic mutants can be analyzed using iAF692. It is worthwhile to mention that FBA will never predict reduced growth with only the addition of substrates to medium (like the addition of H2) unless the cell is forced to take them up.
Conclusions
We have reconstructed the metabolic network of M. barkeri and analyzed the metabolic network using constraint-based analysis. Sequence homology data were combined with flux simulations to assign probable functions for unknown genes. This method of ‘functional annotation' will become increasingly useful for interpreting ambiguous homology searches as more enzymes and their genomic sequences are characterized.
The level of agreement between iAF692 predictions and experimental findings, especially for growth phenotypes, shows promise in the use of the model as a high-throughput analysis tool for studying growth of M. barkeri. The model can not only correctly predict growth phenotypes, but it can also determine (i) active reactions and pathways, (ii) the flux distribution in the network reactions, (iii) the redundancy or robustness of reactions in the network to a particular objective function, (iv) product formation and additional substrates for growth under a given state, such as predicted WT growth solely on cysteine and, (v) areas of disagreement between current knowledge (the model content) and experimental findings, as was seen when minimal media conditions were examined. The findings that iAF692 can accurately predict phenotypes on mixed substrate conditions are also of interest because the pathways that are active during these conditions are poorly understood.
Although iAF692 is already a useful model, continual refinement and updating is necessary. If the incorrect growth prediction of the ech mutant on methanol/H2/CO2 was caused by regulatory events, iAF692 will be instrumental in the interpretation of experimental data and characterization of metabolic regulation. With the ability to examine all aspects of metabolism, an iterative modeling process can generate logical hypotheses and identify conditions (such as regulatory events) that would reconcile disagreements between experimental observations and simulation results. These hypotheses can then be further investigated. As the overall amount of data on M. barkeri increases (for instance, an updated annotation and specific growth maintenance values), iAF692 will continue to expand in its scope and accuracy to predict cellular phenotypes. In the future, iAF692 can serve as a starting point for additional archaeal reconstructions and as an analysis platform for the study of methanogenesis and microbial communities.
Materials and methods
Model reconstruction
The reconstruction software SimPheny™, version 1.7.1.1 (Genomatica Inc., San Diego, CA), was the software platform on which the model was built. The ORF draft annotations for M. barkeri Fusaro, downloaded from the ORNL website (http://genome.ornl.gov/microbial/mbar/, February 2004), were used as a framework on which translated metabolic proteins were assigned to form GPR assignments. The draft genome consisted of 67 contigs of length 4.8 Mb and 5072 predicted candidate ORFs. Most GPR assignments were made from the genome annotation and the model was constructed on a pathway basis manually. Biochemical databases such as KEGG (http://www.genome.jp/kegg/), the Enzyme Nomenclature Database (http://www.chem.qmul.ac.uk/iubmb/enzyme/) and the MetaCyc database (http://metacyc.org/) were used as general guides for pathways and sources for previous genome annotations. When a reaction was entered into the model, the participating metabolites were characterized according to their chemical formula and charge determined for a cytosolic pH of 7.2, a value consistent with the intracellular range determined for methanogens (von Felten and Bachofen, 2000; de Poorter et al, 2003, 2005). Metabolite charge was determined using its pK a value. When the metabolite pK a was not available, charge was determined using the pK a of ionizable groups present in a metabolite. It should be mentioned that the charge of almost all metabolites in the network will not change for a pH increase or decrease of greater than ∼1.5 pH units based on the pK a values of the ionizable groups (most frequently, carboxyl groups and amines, pK a ∼4 and ∼9, respectively). The BLAST algorithm (Altschul et al, 1997) was implemented to infer gene function for enzymes needed to form complete pathways where no gene could be found in the annotation (see Supplementary information 1 for detailed BLAST results). Operon structure was also considered when assigning function when multiple genes having identical annotations were found. GPR associations were also made directly from biochemical evidence presented in journal publications and reviews (see Supplementary information 1). The Pathway Tools software, version 8.5 (http://bioinformatics.ai.sri.com/ptools/), was used to generate an automated metabolic reconstruction and the pathways were analyzed and used to form or confirm GPR associations after manual inspection. Organism specificity of the reactions was achieved by including (i) the unique metabolites present in M. barkeri, such as H4SPT (Grahame and DeMoll, 1996), methanofuran-b (Bobik et al, 1987), and coenzyme F420 (Raemakers-Franken et al, 1991), (ii) specific physiological cofactors, such as ADP for phosphofructokinase (Verhees et al, 2003) and coenzyme F420 as an electron donor in glutamate synthase (Raemakers-Franken et al, 1991), (iii) the measured stoichiometric values for proton and ion translocation reactions in the electron transport chain of M. barkeri (Deppenmeier, 2004; Muller, 2004), and (iv) the necessary metabolic transport reactions for substrates and products of metabolism. Transport reactions were added to the network from the genome annotation or alternatively from physiological data (these were added when a metabolite was taken up into the cell or excreted into the media; Krzycki et al, 1985; Bock et al, 1994; Buchenau and Thauer, 2004). All of the reactions entered into the network were both elementally and charged balanced and were labeled either reversible or irreversible. Reversibility was determined first from primary literature if an enzyme was characterized and additionally from thermodynamic considerations, for example, reactions that consume high-energy metabolites (ATP, GTP, etc.) are generally irreversible.
ORFs in the draft genome annotation that were determined to be previously unannotated were genes assigned functionality in the model which contained the words ‘hypothetical' or similar. Genes that were deemed misannotated were determined to be assigned a function considerably different or more specific than what was given in the draft annotation.
Network comparison
For the comparison of model content, both iJR904 and iND750 were decompartmentalized so that only the fundamental reaction and metabolite names were compared and not their location inside the cells (transport reactions across the cytosolic membrane were compared). Reversibility was not considered in this comparison. Three reactions from the methanogenic pathway in iAF692 were changed to different high-level functional categories according to their role in iJR904 and iND750 (PTA, ACK and ATPS).
For the comparison of network properties, currency metabolites (Supplementary information 3) were removed from each network as they participate in several reactions and form links that do not represent real metabolic pathways (Ma and Zeng, 2003) and model compartmentalization was conserved to maintain network structure. Irreversible and reversible links that appeared twice or more in a network were considered as one link. All of the network properties were calculated using the pajek software package (Batagelj and Mrvar, 1998).
Modeling simulations
A stoichiometric matrix, S (m × n), was constructed for the M. barkeri metabolic network where m is the number of metabolites and n is the number of reactions. The corresponding entry in the stoichiometric matrix, S ij , represents the stoichiometric coefficient for the participation of the ith metabolite in the jth reaction. FBA was then used to solve the linear programming problem under steady-state criteria (Varma and Palsson, 1994; Kauffman et al, 2003; Price et al, 2004). The linear steady-state problem can be represented by the equation:
![]()
where v (n × 1) is a vector of reaction fluxes. Because the linear problem is normally an underdetermined system for genome-scale metabolic models, there exists multiple solutions for v that satisfy equation (1). To find a solution for v , the cellular objective of producing the maximal amount of biomass constituents, represented by the ratio of metabolites in the BOF, is optimized in the linear system. This is achieved by adding an additional column vector to S , S i,BOF, containing the stoichiometric coefficients for the metabolites in the BOF and then subsequently maximizing the reaction flux through the corresponding element in v , v BOF, under the steady-state criteria. Additionally, constraints that are imposed on the system are in the form of:
![]()
where α i and β i are the lower and upper limits placed on each reaction flux, v i , respectively. For reversible reactions, −∞⩽v i ⩽∞, and for irreversible reactions, 0⩽v i ⩽∞.
The constraints on the reactions that allow metabolites entry to the extracellular space were set to 0⩽v i ⩽∞ if the metabolite was not present in the medium, meaning that the compounds could leave, but not enter the system. For the metabolites that were in the medium, the constraints were set to −∞⩽v i ⩽∞ for all except the limiting substrate and cysteine. When cysteine was a media component, it was allowed only for use as a source of sulfur by restricting hydrogen sulfide from exiting the system. Artificial transhydrogenase cycles in the network (Reed et al, 2006) were avoided by only allowing the net flux through a set of potential NAD(H)/NADP(H) cycling reactions in one direction. The reaction flux through the BOF was constrained from 0⩽v BOF⩽∞ and the BOF was generated as a linear equation consisting of the molar amounts of metabolic constituents that make up the dry weight content of the cell (Table III) and a GAM (mmol ATP gDW−1) reaction to account for nonmetabolic growth activity,
![]()
The full BOF is included in Supplementary information 1.
Aside from the BOF, an NGAM (mmol ATP gDW−1 h−1) value was used as an energy ‘drain' on the system during the linear programming calculations and accounts for nongrowth cellular activities (Pirt, 1965). The NGAM was represented as a set flux in the reaction flux vector, v NGAM. The corresponding reaction vector in the stoichiometric matrix, S i,NGAM, was in the form of an ATP maintenance reaction identical to equation (3).
Linear programming calculations were performed using the previously mentioned SimPheny™ software platform and the MATLAB®, version 7.0.0.19920 (The MathWorks Inc., Natick, MA) software platform on which the linear programming package LINDO (Lindo Systems Inc., Chicago, IL) was used as a solver.
Gap filling and determination of minimal media
To fill gaps in the network, biomass components were sequentially added to the BOF individually and FBA was used for BOF optimization under steady-state criteria. When a simulation resulted in a positive net flux through the BOF, a subsequent component was added to the BOF and the simulation was rerun. When a biomass component added to the BOF resulted in no flux through the BOF, the network was manually updated. This process was continued until all of the biomass constituents in Table III were included, and FBA optimization produced a positive flux in the BOF. The initial gap filling procedure was performed with common media conditions (Wolin et al, 1963) for all of the three major substrates: methanol, acetate and H2/CO2. Subsequent procedures were performed removing common media substrates to identify additional gaps that may have been overseen by using the complex common media.
After the gap filling procedure, nonessential compounds were individually removed from the common media conditions until a minimal set of compounds remained that produced a nonzero positive flux through the BOF for a simulation. When two compounds were found to fulfill the same metabolic requirement, the lesser carbon-containing compound was taken as a minimal component. All of the minimal media metabolites were manually analyzed to determine their necessity in producing the BOF constituents. The main substrate uptake rates used were the maximum uptake rates observed for each substrate (see following section). An approximate value of 50 mmol ATP gDW−1 was used for the GAM and the NGAM was not considered for the gap filling procedure and determination of minimal media.
Estimation of the proton translocation efficiency of the Ech hydrogenase reaction
The proton translocation stoichiometry of the Ech hydrogenase reaction was varied in S for different simulations. For each S generated, there were two unknown variables needed for BOF optimization using FBA: the GAM and the NGAM. For growth simulations using FBA and BOF optimization on microbial cells, the NGAM has ranged from 1.0 to 7.6 mmol ATP gDW−1 h−1 when calculated from experimental data (Varma and Palsson, 1994; Famili et al, 2003; Borodina et al, 2005). It was shown that the NGAM and GAM will vary proportionally (Borodina et al, 2005). Therefore, the NGAM was estimated as 2.5% of the GAM as it produced values similar to those from the previous studies. Because no data were available to directly calculate the GAM in this analysis, a wide range of GAM values were considered. Minimal media conditions were used with the addition of cysteine aside from the main growth substrate. The main growth substrate uptake rates were set as the maximum uptake rates for each growth substrate and were taken directly or approximated using biomass yields (gDW mol−1 substrate) and the maximal specific growth rates (h−1) from the cited studies: 16 mmol methanol gDW−1 h−1 and 8 mmol acetate gDW−1 h−1 (Elferink et al, 1994), 41 mmol H2 gDW−1 h−1 (Smith and Mah, 1978) (the amount of CO2 was not constrained) and 5 mmol pyruvate gDW−1 h−1 (Bock et al, 1994). The maximal growth yields (gDW mmol−1 CH4) determined using linear optimization were calculated from the BOF reaction flux (h−1) and the reaction flux of methane (mmol CH4 gDW−1 h−1) leaving the system. The substrate yields and molar yields of CO2 and CH4 were determined with a similar method. The observed growth yields that were used for comparison to simulation results were compiled from growth studies of M. barkeri on methanol (5–7 gDW mol−1 CH4) (Smith and Mah, 1978; Weimer and Zeikus, 1978; Hippe et al, 1979), acetate (2–4 gDW mol−1 CH4) (Smith and Mah, 1978; Bock et al, 1994), H2/CO2 (80:20 v/v) (6–9 gDW mol−1 CH4) (Smith and Mah, 1978; Weimer and Zeikus, 1978) and pyruvate (12–15 gDW mol−1 CH4) (Bock et al, 1994). Data from growth on different substrates were used as the Ech hydrogenase reaction operates in different directions depending on the substrate(s) (Meuer et al, 2002). The cost of the polymerization of the macromolecules was calculated using the cellular composition of M. barkeri (Table III) and the common polymerization and processing costs for a typical prokaryotic cell (Neidhardt et al, 1990). Supplementary information 4, 5, 6, 7 are the reaction flux distributions in iAF692 for optimized growth with the primary substrate of methanol, acetate, H2/CO2 and pyruvate, respectively. The flux distributions in the Supplementary information were generated using minimal media conditions with the addition of cysteine, the primary substrate uptake rates listed above, a GAM of 70 mmol ATP gDW−1, a NGAM of 1.75 mmol ATP gDW−1 h−1 and a stoichiometry of 1 proton translocated/2e− for the Ech hydrogenase reaction. These distributions are also provided as a Microsoft Excel worksheet in Supplementary information 1.
Determination of the stoichiometry for the nitrogenase reaction in M. barkeri
The stoichiometry of the nitrogenase reaction was determined using the growth rates, total amounts of methanol consumed and growth yields for growth on NH4 and diazotrophic growth (Bomar et al, 1985). Minimal media conditions were used in addition to methanol as the main substrate (Bomar et al, 1985). The value of 1 proton translocated/2e− was used for the Ech hydrogenase reaction stoichiometry as this was the approximate stoichiometry for a similar hydrogenase (Hakobyan et al, 2005). Using the NH4 growth conditions, the stoichiometry of the Ech hydrogenase reaction was found to affect the predicted growth rate, at most, ±10% in the range of 0.2–2.0 protons translocated/2e−. The GAM used in the BOF was calculated by finding the value that produced the observed growth rate from the given methanol uptake rate (Bomar et al, 1985) using BOF optimization when NH4 was the nitrogen source in the simulation (the nitrogenase reaction was not needed under these conditions). The value determined was 30 mmol ATP gDW−1, the NGAM was set to 2.5% of this value and these values were used in all subsequent simulations. The stoichiometry of the nitrogenase reaction was then determined by finding the value of p in equation (4) that produced the observed growth rate from the given methanol uptake rate (Bomar et al, 1985) using BOF optimization when N2 was the nitrogen source in the simulation (the nitrogenase reaction was needed under these conditions). Equation (4) is the balanced overall enzymatic reaction for nitrogenase presented by Rees and Howard (2000):
![]()
Examination of a possible alternate pathway for the biosynthesis of H4SPT
When searching for an alternate pathway for the synthesis of H4SPT, M. jannaschii gene MJ1427 (Scott and Rasche, 2002) was used as a query sequence for BLAST (M. barkeri genes gene5036 and gene4403 had e-values of 2e-63 and 4e-62, respectively). Structural similarity of metabolites was determined by finding compounds that could be converted to pABA in the smallest number of known enzymatic steps.
Gene essentiality
To determine the effect of a gene deletion, the reaction associated with each gene in the methanogenic pathway was individually deleted from S and FBA was used to predict the mutation growth phenotype. Using the same maximum uptake rates for each growth substrate as indicated in the examination of Ech hydrogenase reaction, the flux through the BOF was optimized in the mutated network, S ′ , for each substrate. The criteria used to determine growth was a positive flux through the BOF (v BOF>0) using S ′ under steady state (equation (1)). When v BOF>0 for a given S ′ , a positive growth phenotype was categorized under two different types of growth: methanogenic growth or acetogenic growth. For methanogenic growth, methane was formed as a major end product (green boxes, Figure 5) and for acetogenic growth, acetate was formed as a major end product (blue boxes, Figure 5). When an optimization of the BOF resulted in v BOF=0, this was determined to be a no-growth phenotype (red boxes, Figure 5). All of the values of v BOF for the positive mutation growth phenotypes were greater than 10% of the WT value under the same substrate conditions. The GAM was varied from 30 to 110 mmol ATP gDW−1 and the NGAM was 2.5% of the GAM for the gene deletion study. Growth phenotypes were also determined with a variable stoichiometry of 0.2–2.0 protons translocated/2e− for the Ech hydrogenase reaction. The GAM, NGAM or choice of Ech hydrogenase reaction stoichiometry did not have an effect on the predicted growth phenotypes in the range considered. Acetogenic growth was reported if it was the only means for growth and was possible under all Ech hydrogenase reaction stoichiometries. The gene deletion analysis was performed using the SimPheny™ software platform.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Text
Supplementary Table 1
Legend to Supplementary Material
Supplementary Data 1
Supplementary Data 2
Supplementary Data 3
Supplementary Data 4
Supplementary Data 5
Supplementary Data 6
Supplementary Data 7
Acknowledgments
We thank Jennifer Reed, Thuy Vo, Natalie Duarte, Sharon Wiback, Iman Famili, Radhakrishnan Mahadevan and Chris Workman for their invaluable insight. We also thank the National Institutes of Health for a Fellowship that provided training during the project. The M. barkeri sequence data were produced by the US Department of Energy Joint Genome Institute, http://www.jgi.doe.gov/. Work on this project was funded by the Laboratory Directed Research and Development (LDRD) Program at the Pacific Northwest National Laboratory, a multi-program national laboratory operated by Battelle for the US Department of Energy under Contract DE-AC056-76RLO1830.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batagelj V, Mrvar A (1998) Pajek: Program for large network analysis. Connections 21: 47–57 [Google Scholar]
- Bobik TA, Donnelly MI, Rinehart KL Jr, Wolfe RS (1987) Structure of a methanofuran derivative found in cell extracts of Methanosarcina barkeri . Arch Biochem Biophys 254: 430–436 [DOI] [PubMed] [Google Scholar]
- Bock AK, Prieger-Kraft A, Schoenheit P (1994) Pyruvate—a novel substrate for growth and methane formation in Methanosarcina barkeri . Arch Microbiol 161: 33–46 [Google Scholar]
- Bock AK, Schonheit P (1995) Growth of Methanosarcina barkeri (Fusaro) under nonmethanogenic conditions by the fermentation of pyruvate to acetate: ATP synthesis via the mechanism of substrate level phosphorylation. J Bacteriol 177: 2002–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomar M, Knoll K, Widdel F (1985) Fixation of molecular nitrogen by Methanosarcina barkeri . FEMS Microbiol Ecol 31: 47–55 [Google Scholar]
- Borodina I, Krabben P, Nielsen J (2005) Genome-scale analysis of Streptomyces coelicolor A3(2) metabolism. Genome Res 15: 820–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchenau B, Thauer RK (2004) Tetrahydrofolate-specific enzymes in Methanosarcina barkeri and growth dependence of this methanogenic archaeon on folic acid or p-aminobenzoic acid. Arch Microbiol 182: 313–325 [DOI] [PubMed] [Google Scholar]
- Burgard AP, Pharkya P, Maranas CD (2003) Optknock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol Bioeng 84: 647–657 [DOI] [PubMed] [Google Scholar]
- Chien YT, Auerbuch V, Brabban AD, Zinder SH (2000) Analysis of genes encoding an alternative nitrogenase in the archaeon Methanosarcina barkeri 227. J Bacteriol 182: 3247–3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chunhui L, Henry CS, Jankowski MD, Ionita JA, Hatzimanikatis V, Broadbelt LJ (2004) Computational discovery of biochemical routes to specialty chemicals. Chem Eng Sci 59: 5051–5060 [Google Scholar]
- Covert MW, Knight EM, Reed JL, Herrgard MJ, Palsson BO (2004) Integrating high-throughput and computational data elucidates bacterial networks. Nature 429: 92–96 [DOI] [PubMed] [Google Scholar]
- de Poorter LM, Geerts WG, Theuvenet AP, Keltjens JT (2003) Bioenergetics of the formyl-methanofuran dehydrogenase and heterodisulfide reductase reactions in Methanothermobacter thermautotrophicus . Eur J Biochem 270: 66–75 [DOI] [PubMed] [Google Scholar]
- de Poorter LM, Geerts WJ, Keltjens JT (2005) Hydrogen concentrations in methane-forming cells probed by the ratios of reduced and oxidized coenzyme F420. Microbiology 151: 1697–1705 [DOI] [PubMed] [Google Scholar]
- Deppenmeier U (2004) The membrane-bound electron transport system of Methanosarcina species. J Bioenerg Biomembr 36: 55–64 [DOI] [PubMed] [Google Scholar]
- Duarte NC, Herrgard MJ, Palsson BO (2004) Reconstruction and validation of Saccharomyces cerevisiae iND750, a fully compartmentalized genome-scale metabolic model. Genome Res 14: 1298–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elferink O, H SJW, Visser A, Hulshoff Pol LW, Stams AJM (1994) Sulfate reduction in methanogenic bioreactors. FEMS Microbiol Rev 15: 119–136 [Google Scholar]
- Famili I, Forster J, Nielsen J, Palsson BO (2003) Saccharomyces cerevisiae phenotypes can be predicted by using constraint-based analysis of a genome-scale reconstructed metabolic network. Proc Natl Acad Sci USA 100: 13134–13139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M, Schott AK, Romisch W, Ramsperger A, Augustin M, Fidler A, Bacher A, Richter G, Huber R, Eisenreich W (2004) Evolution of vitamin B2 biosynthesis. A novel class of riboflavin synthase in archaea. J Mol Biol 343: 267–278 [DOI] [PubMed] [Google Scholar]
- Garcia J-L, Patel BKC, Ollivier B (2000) Taxonomic, phylogenetic, and ecological diversity of methanogenic archaea. Anaerobe 6: 205–226 [DOI] [PubMed] [Google Scholar]
- Graham DE, White RH (2002) Elucidation of methanogenic coenzyme biosyntheses: from spectroscopy to genomics. Nat Prod Rep 19: 133–147 [DOI] [PubMed] [Google Scholar]
- Graham DE, Xu H, White RH (2003) Identification of the 7,8-didemethyl-8-hydroxy-5-deazariboflavin synthase required for coenzyme F(420) biosynthesis. Arch Microbiol 180: 455–464 [DOI] [PubMed] [Google Scholar]
- Grahame DE, DeMoll E (1996) Partial reactions catalyzed by protein components of the acetyl-CoA decarbonylase synthase enzyme complex from Methanosarcina barkeri . J Biol Chem 271: 8352–8358 [DOI] [PubMed] [Google Scholar]
- Guss AM, Mukhopadhyay B, Zhang JK, Metcalf WW (2005) Genetic analysis of mch mutants in two Methanosarcina species demonstrates multiple roles for the methanopterin-dependent C-1 oxidation/reduction pathway and differences in H metabolism between closely related species. Mol Microbiol 55: 1671–1680 [DOI] [PubMed] [Google Scholar]
- Hakobyan M, Sargsyan H, Bagramyan K (2005) Proton translocation coupled to formate oxidation in anaerobically grown fermenting Escherichia coli . Biophys Chem 115: 55–61 [DOI] [PubMed] [Google Scholar]
- Hedderich R (2004) Energy-converting [NiFe] hydrogenases from archaea and extremophiles: ancestors of complex I. J Bioenerg Biomembr 36: 65–75 [DOI] [PubMed] [Google Scholar]
- Hippe H, Caspari D, Fiebig K, Gottschalk G (1979) Utilization of trimethylamine and other N-methyl compounds for growth and methane formation by Methanosarcina barkeri . Proc Natl Acad Sci USA 76: 494–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen P, Goldovsky L, Kunin V, Darzentas N, Ouzounis CA (2005) Genome coverage, literally speaking. The challenge of annotating 200 genomes with 4 million publications. EMBO Rep 6: 397–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman KJ, Prakash P, Edwards JS (2003) Advances in flux balance analysis. Curr Opin Biotechnol 14: 491–496 [DOI] [PubMed] [Google Scholar]
- Krzycki JA, Lehman LJ, Zeikus JG (1985) Acetate catabolism by Methanosarcina barkeri: evidence for involvement of carbon monoxide dehydrogenase, methyl coenzyme M, and methylreductase. J Bacteriol 163: 1000–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma HW, Zeng AP (2003) The connectivity structure, giant strong component and centrality of metabolic networks. Bioinformatics 19: 1423–1430 [DOI] [PubMed] [Google Scholar]
- Meuer J, Kuettner HC, Zhang JK, Hedderich R, Metcalf WW (2002) Genetic analysis of the archaeon Methanosarcina barkeri Fusaro reveals a central role for Ech hydrogenase and ferredoxin in methanogenesis and carbon fixation. Proc Natl Acad Sci USA 99: 5632–5637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller V (2004) An exceptional variability in the motor of archaeal A1A0 ATPases: from multimeric to monomeric rotors comprising 6–13 ion binding sites. J Bioenerg Biomembr 36: 115–125 [DOI] [PubMed] [Google Scholar]
- Neidhardt FC, Ingraham JL, Schaechter M (1990) Physiology of the Bacterial Cell: A Molecular Approach. Sunderland, MA: Sinauer Associates [Google Scholar]
- Nichols BP, Seibold AM, Doktor SZ (1989) para-aminobenzoate synthesis from chorismate occurs in two steps. J Biol Chem 264: 8597–8601 [PubMed] [Google Scholar]
- Nishihara M, Koga Y (1995) Two new phospholipids, hydroxyarchaetidylglycerol and hydroxyarchaetidylethanolamine, from the Archaea Methanosarcina barkeri . Biochim Biophys Acta 1254: 155–160 [DOI] [PubMed] [Google Scholar]
- Peregrin-Alvarez JM, Tsoka S, Ouzounis CA (2003) The phylogenetic extent of metabolic enzymes and pathways. Genome Res 13: 422–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirt SJ (1965) The maintenance energy of bacteria in growing cultures. Proc R Soc London B Biol Sci 163: 224–231 [DOI] [PubMed] [Google Scholar]
- Price ND, Reed JL, Palsson BO (2004) Genome-scale models of microbial cells: evaluating the consequences of constraints. Nat Rev Microbiol 2: 886–897 [DOI] [PubMed] [Google Scholar]
- Raemakers-Franken PC, Brand RJ, Kortstee AJ, Van der Drift C, Vogels GD (1991) Ammonia assimilation and glutamate incorporation in coenzyme F420 derivatives of Methanosarcina barkeri . Antonie Van Leeuwenhoek 59: 243–248 [DOI] [PubMed] [Google Scholar]
- Reed JL, Famili I, Thiele I, Palsson BO (2006) Towards multidimensional genome annotation. Nat Rev Genet 7: 130–141 [DOI] [PubMed] [Google Scholar]
- Reed JL, Vo TD, Schilling CH, Palsson BO (2003) An expanded genome-scale model of Escherichia coli K-12 (iJR904 GSM/GPR). Genome Biol 4: R54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees DC, Howard JB (2000) Nitrogenase: standing at the crossroads. Curr Opin Chem Biol 4: 559–566 [DOI] [PubMed] [Google Scholar]
- Russell JB, Cook GM (1995) Energetics of bacterial growth: balance of anabolic and catabolic reactions. Microbiol Rev 59: 48–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer P, Sahm H (1981) Effect of trace elements and vitamins on the growth of Methanosarcina barkeri . Acta Biotechnol 1: 57–65 [Google Scholar]
- Schink B (1997) Energetics of syntrophic cooperation in methanogenic degradation. Microbiol Mol Biol Rev 61: 262–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JW, Rasche ME (2002) Purification, overproduction, and partial characterization of beta-RFAP synthase, a key enzyme in the methanopterin biosynthesis pathway. J Bacteriol 184: 4442–4448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MR, Mah RA (1978) Growth and methanogenesis by Methanosarcina strain 227 on acetate and methanol. Appl Environ Microbiol 36: 870–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thauer RK, Jungermann K, Decker K (1977) Energy conservation in chemotrophic anaerobic bacteria. Bacteriol Rev 41: 100–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoka S, Simon D, Ouzounis CA (2004) Automated metabolic reconstruction for Methanococcus jannaschii . Archaea 1: 223–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma A, Palsson BO (1994) Stoichiometric flux balance models quantitatively predict growth and metabolic by-product secretion in wild-type Escherichia coli W3110. Appl Environ Microbiol 60: 3724–3731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhees CH, Kengen SW, Tuininga JE, Schut GJ, Adams MW, De Vos WM, Van Der Oost J (2003) The unique features of glycolytic pathways in Archaea. Biochem J 375: 231–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Felten P, Bachofen R (2000) Continuous monitoring of the cytoplasmic pH in Methanobacterium thermoautotrophicum using the intracellular factor F(420) as indicator. Microbiology 146 (Part 12): 3245–3250 [DOI] [PubMed] [Google Scholar]
- Weimer PJ, Zeikus JG (1978) One carbon metabolism in methanogenic bacteria. Cellular characterization and growth of Methanosarcina barkeri . Arch Microbiol 119: 49–57 [DOI] [PubMed] [Google Scholar]
- Welander PV, Metcalf WW (2005) Loss of the mtr operon in Methanosarcina blocks growth on methanol, but not methanogenesis, and reveals an unknown methanogenic pathway. Proc Natl Acad Sci USA 102: 10664–10669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin EA, Wolin MJ, Wolfe RS (1963) Formation of methane by bacterial extracts. J Biol Chem 238: 2882–2886 [PubMed] [Google Scholar]
- Yeh I, Hanekamp T, Tsoka S, Karp PD, Altman RB (2004) Computational analysis of Plasmodium falciparum metabolism: organizing genomic information to facilitate drug discovery. Genome Res 14: 917–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinder SH (1993) Physiological ecology of methanogens. In Methanogenesis, Ferry JG (ed), pp 128–206. Routledge, New York: Chapman & Hall [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Text
Supplementary Table 1
Legend to Supplementary Material
Supplementary Data 1
Supplementary Data 2
Supplementary Data 3
Supplementary Data 4
Supplementary Data 5
Supplementary Data 6
Supplementary Data 7