Abstract
With the goal of solving the whole-cell problem with Escherichia coli K-12 as a model cell, highly accurate genomes were determined for two closely related K-12 strains, MG1655 and W3110. Completion of the W3110 genome and comparison with the MG1655 genome revealed differences at 267 sites, including 251 sites with short, mostly single-nucleotide, insertions or deletions (indels) or base substitutions (totaling 358 nucleotides), in addition to 13 sites with an insertion sequence element or defective prophage in only one strain and two sites for the W3110 inversion. Direct DNA sequencing of PCR products for the 251 regions with short indel and base disparities revealed that only eight sites are true differences. The other 243 discrepancies were due to errors in the original MG1655 sequence, including 79 frameshifts, one amino-acid residue deletion, five amino-acid residue insertions, 73 missense, and 17 silent changes within coding regions. Errors in the original MG1655 sequence (<1 per 13 000 bases) were mostly within portions sequenced with out-dated technology based on radioactive chemistry.
Keywords: crp mutation, E. coli K-12 genome, E. coli K-12 pedigree, genome corrections, rpoS mutations
Introduction
From the dawn of modern biology, the intestinal bacterium Escherichia coli has been the most intensively studied organism. Many basic molecular processes, best understood in E. coli, are universal throughout the natural world. The wealth of information on E. coli makes it an ideal test bed for pushing forward the limits of our ability to understand a cell through computational modeling (Wanner et al, 2005). As a first step of an E. coli systems biology project in Japan (Mori, 2004), we undertook the task of determination of highly accurate E. coli K-12 genomes, which are key for precisely defining the cell parts.
We present back-to-back manuscripts on more accurate E. coli K-12 genomes (this paper) and new resources (Baba et al, 2006) of value for both basic biology and systems-level research on E. coli K-12. A key tenet of postgenomics sciences requires an accurate appraisal of the cell parts. Here, we describe determination of highly accurate genome sequences of two common ‘wild-type' K-12 strains. Knowledge of E. coli gene sequences, products, and functions is of value not only to E. coli cell biologists but also to others who rely on E. coli information for understanding of processes in diverse cells having conserved genes, proteins, RNAs, or motifs. Elsewhere, we describe a community effort for re-annotation of these more accurate genomes (Riley et al, 2006). Postgenomic sciences can be accelerated by development and sharing of biological resources. In the accompanying paper, we describe construction of mutants that have in-frame, single-gene knockouts of nearly all nonessential E. coli protein-encoding genes (Baba et al, 2006) by use of a now standard method for direct modification of chromosomal genes (Datsenko and Wanner, 2000).
Systematic determination of the complete E. coli K-12 genome was among the first targets for whole-genome sequencing. From 1989 to 1997, projects led by T Yura and A Ishihama, by K Mizobuchi, and by T Horiuchi and H Mori in Japan and by F Blattner, by G Church, and by R Davis in the USA reported many long continuous sequence segments (contigs) of the E. coli K-12 genome (Daniels et al, 1992; Yura et al, 1992; Burland et al, 1993, 1995; Plunkett et al, 1993; Fujita et al, 1994; Sofia et al, 1994; Aiba et al, 1996; Itoh et al, 1996; Oshima et al, 1996; Yamamoto et al, 1997). While long contigs from the Church, Davis, and Mizobuchi projects were also deposited to GenBank™ or DNA Data Bank of Japan (DDBJ) over this period, results are unpublished. The complete E. coli genome sequence (Blattner et al, 1997) has provided a wealth of information on the gene products, gene organization, and chromosome structure.
All groups had chosen E. coli K-12 for whole-genome sequencing because more was known about it than any organism. The ancestral strain had been isolated from the stool of a convalescent diphtheria patient in 1922 and given the designation ‘K-12' when deposited in a strain collection at Stanford in 1925 (Bachmann, 1996). In the early 1940s, EL Tatum, who was then at Stanford, acquired E. coli K-12. Because it was prototrophic, easy to grow in a defined medium, and had a short generation time, he used it in his seminal studies of biochemical genetics (Tatum, 1959). In 1946, J Lederberg and EL Tatum demonstrated sexual recombination in E. coli K-12 (Lederberg and Tatum, 1946), a property requiring the F+ ‘fertility factor', which was later found to be rare among E. coli isolates from nature. Mating occurred between different K-12 derivatives because particular descendents had lost the F+ factor, which otherwise leads to incompatibility. In 1950, E Lederberg reported that the original Lederberg and Tatum K-12 strain was lysogenic for phage λ (Lederberg, 1950). Derivatives that had lost λ acted as sensitive hosts for λ released from lysogenic E. coli K-12 (Lederberg and Lederberg, 1953). Shortly thereafter, phage P1 (Bertani, 1951) was shown to carry out generalized transduction in E. coli (Lennox, 1955). Largely because of these early studies, E. coli K-12 became the primary source of basic information on innumerable biochemical and molecular processes over the past 60 years.
Owing to its widespread use, a huge number of E. coli K-12 derivatives now exist (Bachmann, 1996). In an effort to get away from the early heavily mutagenized Stanford strains, E. coli K-12 W3110 (λ−, F−) was extensively used as an ancestral stock (Bachmann, 1972). The first physical map of the whole E. coli chromosome was created using a W3110 genomic library (Kohara et al, 1987). Subsequently, groups in Japan chose W3110 for whole-genome sequencing (Yura et al, 1992), while the Blattner group chose MG1655 (Guyer et al, 1981), which is more closely related to ancestral E. coli K-12 (EMG2 or WG1), except for loss of the F+ factor and λ prophage (Figure 1).
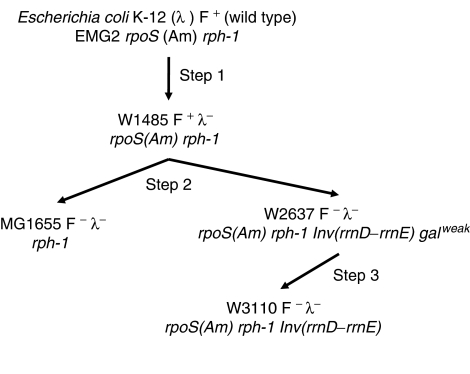
Figure 1.
E. coli K-12 pedigree. The relationships of E. coli K-12 MG1655 and W3110 with wild-type E. coli K-12 (EMG2 or WG1) have been described (Bachmann, 1972, 1996). Wild-type K-12 was cured of phage λ to make W1485 prior to 1954 (Step 1), which in turn was cured of the F+ factor to make W2637 (Step 2), from which W3110 was selected for a strongly galactose-fermenting strain in 1956 (Step 3). More recently, W1485 was cured of the F+ factor to make MG1655 (Guyer et al, 1981). E. coli K-12 EMG2, W1485, W2637, and W3110 have the same rpoS396(Am) allele (codon 33, TAG (Am); Rod et al, 1988; Atlung et al, 2002; KA Datsenko and BL Wanner, unpublished data), while MG1655 has the pseudorevertant Q33 allele (Atlung et al, 2002).
Results and discussion
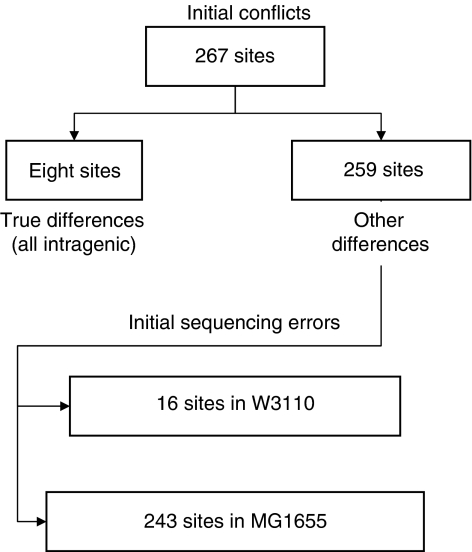
Determination of the complete W3110 genome and comparison with that of MG1655 (GenBank™ U00096, 1998 submission) revealed differences at 282 locations. These included 13 sites where an insertion sequence (IS) or defective phage exists in only one strain, two sites due to the W3110 inversion (Hill and Harnish, 1981), and 267 sites with sequence conflicts (Figure 2). To determine how many of the latter are true differences, these regions were PCR amplified from both strains and directly sequenced. Only eight are true differences. In all, 16 of the 267 sites with conflicts were due to errors in the W3110 sequence. These differences (totaling 17 nucleotides (nt); Supplementary Table 1) were due to errors in cloning (5 nt), sequencing (6 nt), or assembly (6 nt).
Figure 2.
Resolution of E. coli K-12 W3110 and MG1655 sequence differences. See text.
The remaining 243 (totaling 358 nt; Supplementary Table 2) were errors in the original MG1655 GenBank™ deposit. These included 104 sites with 1-, 2-, or multiple (short) nt substitutions, 134 sites with 1-, 2-, or 4-nt indels (Table I). MG1655 segments sequenced were deposited to DDBJ in January 2004 (Accession numbers AG613214–AG613378) and incorporated into a new MG1655 GenBank™ release (U00096.2; June 2004 version).
Table 1.
Summary of E. coli K-12 MG1655 genome correctionsa
| Change | Locationb | No. sitesc |
|---|---|---|
| 1-nt substitution | Intergenic | 12 |
| Coding | 56 | |
| 2-nt substitution | Intergenic | 5 |
| Coding | 26 | |
| Multiple nt substitution | Intergenic | 2 |
| Codingd | 2 | |
| RNA | 1 | |
| 1-nt indel | Intergenic | 48 (27) |
| Coding | 75 (50) | |
| RNA | 1 (0) | |
| 2-nt indel | Intergenic | 6 (5) |
| Coding | 3 (2) | |
| 3-nt indele | Coding | 4 (3) |
| 4-nt indel | Coding | 1 (1) |
| 6-nt indelf | Coding | 1 (1) |
| Total | 243 (193) |
aThe actual sequence corrections are in Supplementary Table 2.
bMost genes affected have only single corrections. Exceptions had five (yfjP, alx, ppiC), six (yhdZ), seven (yieP, yjgN), 11 (yigL), and 14 (yibJ) corrections.
cTotals are given with the number of insertions in parentheses. Indels changed not only the length of particular gene products but also the number of gene products, for example, corrections resulting in gene fusion event(s), or conversion from one to two genes.
dOne multiple nt substitution changes coding of ebgA (CAAG to AGCA at nt 3 222 944); the other results in a frameshift in gntT due to a 2-nt addition (C to GCG at nt 3 544 358).
eA 1-codon deletion lies in yghG and 1-codon insertions lie in rffE, yiaY, and yieP.
fA 2-codon insertion lies in arcB.
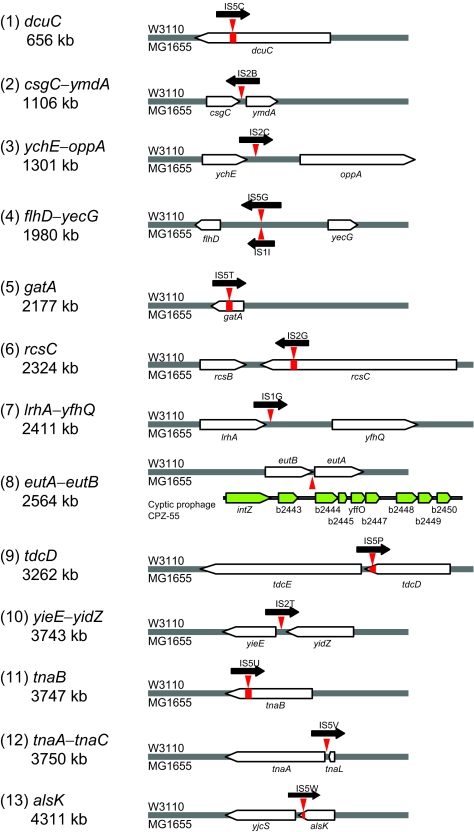
In total, 13 sites have an IS element or defective phage in only E. coli K-12 W3110 or MG1655 (Figure 3). Of these, 11 sites have an IS element only in W3110. One defective phage (CPZ-55) is only in MG1655. One site has an IS5 element in W3110 and an IS1 element in MG1655. Locations of all IS elements and defective phages in MG1655 and W3110 and the W3110 inversion are shown in Supplementary Figure 1.
Figure 3.
IS element and defective phage differences. Locus names and genome locations on the left side are based on the MG1655 genome. IS1A, IS1B, IS1C, etc. are named alphabetically to distinguish individual insertions (Supplementary Figure 1). IS elements, black arrows; sites, red arrowheads; six ISs disrupt orfs, red bars (alsK, dcuA, gatA, rcsC, tdcD, and tnaB); and phage genes, green arrows.
The finding that the complete genome sequences of MG1655 and W3110 are nearly alike gives high confidence in the assembly. Resolution of discrepancies showed that the original MG1655 genome sequence was highly accurate (<1 error per 13 000 nt). Independent cloning and sequencing and reconciliation of differences have provided a pair of highly accurate E. coli K-12 genomes.
Most (ca. 88%) of the E. coli K-12 genome encodes proteins. As expected, the majority of the 1-, 2-, and 4-nt indel corrections (79 of 134) lie within coding regions; these 79 corrections resulted in frame shifting of 77 different open reading frames (orfs) (Table I). One multiple nt substitution changed adjacent residues; another changed the reading frame. Five indel corrections resulted in one 1-codon deletion, three 1-codon insertions, and one 2-codon insertion. Accordingly, 84 corrections dramatically alter protein coding regions by frame shifting or otherwise changing lengths of orfs. Of the 78 frameshifts, 23 resulted in fusing adjacent or overlapping orfs into a single orf, two led to fission of orfs into two, and one led to recognition of a conserved coding sequence on the opposite strand to that previously predicted, that is, an inversion with respect to the predicted coding region. Examples are illustrated elsewhere (Riley et al, 2006). Other corrections in coding regions included 73 amino-acid switches and 17 silent changes. It is more difficult to assess effects of corrections in intergenic regions (73 corrections) or RNA genes (two corrections).
E. coli K-12 W3110 has been widely used as a wild-type strain in Japan, the USA, and elsewhere from 1956. Because both MG1655 and W3110 are descendents of W1485 (Figure 1), they diverged more than 50 years ago. Yet, they have few differences. Further, only two of the 12 W3110-specific IS insertions are in common among stocks of W3110 from nine different laboratories in Japan. Two others are in the majority of these stocks. Eight are only in the Kohara stock that was used for genome sequencing (unpublished data). Because transposition of IS elements occurs in resting E. coli K-12 (Naas et al, 1994), the additional IS copies in W3110 Kohara probably arose during storage in stabs. The finding of so few differences is consistent with these strains having been stored as lyophilized or frozen cultures during much of the interim (Barratt and Tatum, 1950). Presumably, the defective CPZ-55 phage in MG1655 is in ancestral K-12 and was lost in the line leading to W3110.
The eight site (9 nt) differences between MG1655 and W3110 include seven in orfs and one in an rRNA gene (Table II). Two (rpoS and dcuA) are nonfunctional alleles in W3110. Because progenitor E. coli probably has the E33 (GAG) allele, and W3110 (like ancestral EMG2) has the Stop33 (TAG) allele (Figure 1), pseudoreversion to Q33 (CAG) apparently arose in MG1655 (Table II). Different stocks of W3110 have also been shown to carry different rpoS alleles (Jishage and Ishihama, 1997).
Table 2.
Confirmed sequence differences between E. coli K-12 W3110 and MG1655
| Gene | b numa | JW id | Function | Changes | Ancestral typea | |
|---|---|---|---|---|---|---|
| W3110 | MG1655 | |||||
| ycdT | b1025 | JW5143 | Conserved membrane protein | V130 (GTA) | A130(GCA) | MG1655 |
| acnA | b1276 | JW1268 | Aconitate hydratase 1 | G522 (GGC) | A522 (AGC) | W3110 |
| intQ | b1579 | JW1571 | Qin prophage; predicted defective integrase | L274 (CTC) | F274 (TTC) | W3110 |
| yedJ | b1942 | JW1926 | Conserved protein | V219 (GTT) | V219 (GTC) | MG1655 |
| rpoS | b2741 | JW5437 | RNA polymerase, sigma S (sigma38) factor | Stop33 (TAG) | Q33 (CAG) | W3110 |
| crp | b3357 | JW5702 | DNA-binding transcriptional dual regulator; cyclic AMP receptor protein | K29 (AAG) | T29(ACG) | MG1655 |
| rrlE | b4009 | JWR109 | 23S ribosomal RNA (rrlE) | A2256 | G2256 | ND |
| dcuA | b4138 | JW5735 | C4-dicarboxylate antiporter; anaerobic | Frameshiftc | MG1655 | |
| ND=not determined. | ||||||
aThe b number (b num) and JW identifier (JW id) are the locus tags in the MG1655 (GenBank™ U00096.2) and W3110 (DDBJ AP009048) genomes.
bAncestral K-12 is EMG2 (Figure 1). EMG2 and W1485 were shown to be alike and to have the same allele as MG1655 or W3110, as indicated. For reasons given elsewhere (Atlung et al, 2002), progenitor E. coli likely had the rpoS Q33 (CAG) allele, while a number of E. coli K-12 derivatives have a true reversion to Q33 or a mutant (E33, Y33, S33, or L33) allele. ND, not determined.
c dcuA has a 2-nt insertion (TT) after nt 182 of its coding region.
In addition to nonfunctional rpoS and dcuA alleles, W3110 has ISs disrupting four genes of known function (gatA for galactitol PTS enzyme II; dcuC for aerobic and anaerobic C4-dicarboxylate transporters; rcsC for a hybrid sensory kinase controlling capsule biosynthesis; and tnaB for a low-affinity tryptophan permease in the tryptophanase operon). These are likely to affect metabolism such as growth on galactitol (gatA) or succinate (dcuA and dcuC), polysaccharide biosynthesis (rcsC), or use of tryptophan as a carbon and nitrogen source (tnaB). These illustrate the breadth of phenotypic differences that can arise among isolates of a single species maintained separately for several decades.
Five true differences between MG1655 and W3110 are missense changes; one is silent. Whether the missense ones are functional has not been determined. The T29K change in crp affects a surface-exposed residue not involved in interactions with cAMP, DNA, or RNA polymerase. Substitutions of this residue are likely to be neutral (RH Ebright, personal communication). MG1655 has the ancestral (EMG2 and W1485) allele for four; W3110 has the ancestral allele for three (Figure 1; Table II).
The creation of highly accurate E. coli K-12 genome sequences provided the impetus for a cooperative re-annotation of both MG1655 and W3110 (Riley et al, 2006). The complete W3110 genome with the latest annotation has the Accession number DDBJ AP009048. These highly accurate E. coli K-12 genomes were used in the design of a collection of in-frame, gene knockout mutants (the Keio collection), whose construction is described in the accompanying manuscript (Baba et al, 2006).
Materials and methods
In all, 60% (2.6 Mb) of the E. coli K-12 W3110 genome had been previously completely determined and deposited in DDBJ (Yura et al, 1992; Aiba et al, 1996; Itoh et al, 1996; Oshima et al, 1996; Yamamoto et al, 1997). Most of the remainder and uncertain regions were completely determined in this work by using a set of λ clones (Kohara et al, 1987). Initially, each chromosomal segment was amplified by long-range PCR, fragmented by sonication, cloned into an M13 vector and sequenced (Aiba et al, 1996; Itoh et al, 1996; Oshima et al, 1996; Yamamoto et al, 1997). Later, 20 continuous λ clones were separately amplified, mixed, fragmented, cloned, and sequenced, and the sequences were assembled into 100–200 kbp continuous regions. The remaining 10% was determined by insertion of two rare I-SceI restriction sites into the genome within the fadB-yicN and hflX-thrA intervals. The intervening regions were recovered by digestion, fragmented, cloned, and sequenced as described (Blattner et al, 1997). Ancestral alleles were determined by sequencing the respective PCR-amplified regions from EMG2 and its immediate descendent W1485. Chromosomal DNAs for W3110 and MG1655 were from Yuji Kohara and the National Institute of Genetics (Shizuoka Japan), respectively. Strains EMG2 and W1485F+ were from Mary Berlyn. Automated sequencing was carried out with an ABI 3100 sequencer.
Supplementary Material
Supplementary Table 1
Supplementary Figure 1
Supplementary Table 2
Supplementary Information
Acknowledgments
We thank Yuji Kohara for strain W3110, Yukiko Yamazaki for sequence analysis, Naomi Ishine, Masami Inagaki, Kayo Shirai, and Mineko Shimizu for technical assistance, Nicole Perna and Guy Plunkett III for helpful discussions and sharing unpublished data, Mary Berlyn for information on K-12 pedigrees, and our many collaborators for helpful discussions at the E. coli re-annotation meetings. This work was supported by CREST, JST (Japan Science and Technology) to TH and HM. and BLW is supported by NIH GM62662.
References
- Aiba H, Baba T, Hayashi K, Inada T, Isono K, Itoh T, Kasai H, Kashimoto K, Kimura S, Kitakawa M, Kitagawa M, Makino K, Miki T, Mizobuchi K, Mori H, Mori T, Motomura K, Nakade S, Nakamura Y, Nashimoto H, Nishio Y, Oshima T, Saito N, Sampei G, Seki Y, Sivasundaram S, Tagami H, Takeda J, Takemoto K, Takeuchi Y, Wada C, Yamamoto Y, Horiuchi T (1996) A 570-kb DNA sequence of the Escherichia coli K-12 genome corresponding to the 28.0–40.1 min region on the linkage map. DNA Res 3: 363–377 [DOI] [PubMed] [Google Scholar]
- Atlung T, Nielsen HV, Hansen FG (2002) Characterisation of the allelic variation in the rpoS gene in thirteen K12 and six other non-pathogenic Escherichia coli strains. Mol Genet Genom 266: 873–881 [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Okumura Y, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H (2006) Construction of Escherichia coli K-12 in-frame, single-gene knock-out mutants—the Keio collection. Mol Syst Biol 2006.0008. doi:10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann BJ (1972) Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev 36: 525–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann BJ (1996) Derivations and genotypes of some mutant derivatives of Escherichia coli K-12. In Escherichia coli and Salmonella typhimurium Cellular and Molecular Biology, Neidhardt FC, Curtiss III R, Ingraham JL, Lin ECC, Low Jr KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (eds), 2 edn, pp 2460–2488. Washington, DC: ASM Press [Google Scholar]
- Barratt RW, Tatum EL (1950) A simplified method of lyophilizing microorganisms. Science 112: 122–123 [DOI] [PubMed] [Google Scholar]
- Bertani G (1951) Studies of lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli . J Bacteriol 62: 293–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y (1997) The complete genome sequence of Escherichia coli K-12. Science 277: 1453–1462 [DOI] [PubMed] [Google Scholar]
- Burland V, Plunkett G III, Daniels DL, Blattner FR (1993) DNA sequence and analysis of 136 kilobases of the Escherichia coli genome: organizational symmetry around the origin of replication. Genomics 16: 551–561 [DOI] [PubMed] [Google Scholar]
- Burland V, Plunkett G III, Sofia HJ, Daniels DL, Blattner FR (1995) Analysis of the Escherichia coli genome VI: DNA sequence of the region from 92.8 through 100 min. Nucleic Acids Res 23: 2105–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels DL, Plunkett G III, Burland V, Blattner FR (1992) Analysis of the Escherichia coli genome: DNA sequence of the region from 84.5 to 86.5 min. Science 257: 771–778 [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Mori H, Yura T, Ishihama A (1994) Systematic sequencing of the Escherichia coli genome: analysis of the 2.4–4.1 min (110 917–193 643 bp) region. Nucleic Acids Res 22: 1637–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyer MS, Reed RR, Steitz JA, Low KB (1981) Identification of a sex-factor-affinity site in E. coli as γδ. Cold Spring Harbor Symp Quant Biol 45: 135–140 [DOI] [PubMed] [Google Scholar]
- Hill CW, Harnish BW (1981) Inversions between ribosomal RNA genes of Escherichia coli . Proc Natl Acad Sci USA 78: 7069–7072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Aiba H, Baba T, Hayashi K, Inada T, Isono K, Kasai H, Kimura S, Kitakawa M, Kitagawa M, Makino K, Miki T, Mizobuchi K, Mori H, Mori T, Motomura K, Nakade S, Nakamura Y, Nashimoto H, Nishio Y, Oshima T, Saito N, Sampei G, Seki Y, Sivasundaram S, Tagami H, Takeda J, Takemoto K, Wada C, Yamamoto Y, Horiuchi T (1996) A 460-kb DNA sequence of the Escherichia coli K-12 genome corresponding to the 40.1–50.0 min region on the linkage map. DNA Res 3: 379–392 [DOI] [PubMed] [Google Scholar]
- Jishage M, Ishihama A (1997) Variation in RNA polymerase sigma subunit composition within different stocks of Escherichia coli W3110. J Bacteriol 179: 959–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohara Y, Akiyama K, Isono K (1987) The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell 50: 495–508 [DOI] [PubMed] [Google Scholar]
- Lederberg EM (1950) Lysogenicity of Escherichia coli strain K-12. Microb Genet Bull 1: 5–7 [Google Scholar]
- Lederberg EM, Lederberg J (1953) Genetic studies of lysogenicity in E. coli . Genetics 38: 51–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederberg J, Tatum EL (1946) Gene recombination in Escherichia coli . Nature 158: 558 [DOI] [PubMed] [Google Scholar]
- Lennox ES (1955) Transduction of linked genetic characters of the host by bacteriophage P1. Virology 1: 190–206 [DOI] [PubMed] [Google Scholar]
- Mori H (2004) From the sequence to cell modeling: comprehensive functional genomics in Escherichia coli . J Biochem Mol Biol 37: 83–92 [DOI] [PubMed] [Google Scholar]
- Naas T, Blot M, Fitch WM, Arber W (1994) Insertion sequence-related genetic variation in resting Escherichia coli K-12. Genetics 136: 721–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima T, Aiba H, Baba T, Fujita K, Hayashi K, Honjo A, Ikemoto K, Inada T, Itoh T, Kajihara M, Kanai K, Kashimoto K, Kimura S, Kitagawa M, Makino K, Masuda S, Miki T, Mizobuchi K, Mori H, Motomura K, Nakamura Y, Nashimoto H, Nishio Y, Saito N, Horiuchi T (1996) A 718-kb DNA sequence of the Escherichia coli K-12 genome corresponding to the 12.7–28.0 min region on the linkage map. DNA Res 3: 137–155 [DOI] [PubMed] [Google Scholar]
- Plunkett G III, Burland V, Daniels DL, Blattner FR (1993) Analysis of the Escherichia coli genome. III. DNA sequence of the region from 87.2 to 89.2 min. Nucleic Acids Res 21: 3391–3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley M, Abe T, Arnaud MB, Berlyn MB, Blattner FR, Chaudhuri RR, Glasner JD, Mori H, Horiuchi T, Keseler IM, Kosuge T, Perna NT, Plunkett G III, Rudd KE, Serres MH, Thomas GH, Thomson NR, Wishart DS, Wanner BL (2006) Escherichia coli K-12: a cooperatively developed annotation snapshot—2005. Nucleic Acids Res 34: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rod ML, Alam KY, Cunningham PR, Clark DP (1988) Accumulation of trehalose by Escherichia coli K-12 at high osmotic pressure depends on the presence of amber suppressors. J Bacteriol 170: 3601–3610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofia HJ, Burland V, Daniels DL, Plunkett G III, Blattner FR (1994) Analysis of the Escherichia coli genome. V. DNA sequence of the region from 76.0 to 81.5 min. Nucleic Acids Res 22: 2576–2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatum EL (1959) A case history in biological research. Science 129: 1711–1715 [DOI] [PubMed] [Google Scholar]
- Wanner BL, Finney A, Hucka M (2005) Modeling the E. coli cell: the need for computing, cooperation, and consortia. Top Curr Genet 13: 163–189 [Google Scholar]
- Yamamoto Y, Aiba H, Baba T, Hayashi K, Inada T, Isono K, Itoh T, Kimura S, Kitagawa M, Makino K, Miki T, Mitsuhashi N, Mizobuchi K, Mori H, Nakade S, Nakamura Y, Nashimoto H, Oshima T, Oyama S, Saito N, Sampei G, Satoh Y, Sivasundaram S, Tagami H, Takahashi H, Takeda J, Takemoto K, Uehara K, Wada C, Yamagata S, Horiuchi T (1997) Construction of a contiguous 874 kb sequence of the Esherichia coli K-12 genome corresponding to 50.0–68.8 min region on the linkage map and analysis of its sequence features. DNA Res 4: 91–113 [DOI] [PubMed] [Google Scholar]
- Yura T, Mori H, Nagai H, Nagata T, Ishihama A, Fujita N, Isono K, Mizobuchi K, Nakata A (1992) Systematic sequencing of the Escherichia coli genome: analysis of the 0–2.4 min region. Nucleic Acids Res 20: 3305–3308 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1
Supplementary Figure 1
Supplementary Table 2
Supplementary Information