Abstract
We have systematically made a set of precisely defined, single-gene deletions of all nonessential genes in Escherichia coli K-12. Open-reading frame coding regions were replaced with a kanamycin cassette flanked by FLP recognition target sites by using a one-step method for inactivation of chromosomal genes and primers designed to create in-frame deletions upon excision of the resistance cassette. Of 4288 genes targeted, mutants were obtained for 3985. To alleviate problems encountered in high-throughput studies, two independent mutants were saved for every deleted gene. These mutants—the ‘Keio collection'—provide a new resource not only for systematic analyses of unknown gene functions and gene regulatory networks but also for genome-wide testing of mutational effects in a common strain background, E. coli K-12 BW25113. We were unable to disrupt 303 genes, including 37 of unknown function, which are candidates for essential genes. Distribution is being handled via GenoBase (http://ecoli.aist-nara.ac.jp/).
Keywords: bacterial functional genomics, E. coli/gene, essential gene, knockout mutants, resources, systems biology
Introduction
The increased availability of genome sequences has provided the basis for comprehensive understanding of organisms at the molecular level. Besides sequence data, a large number of experimental and computational resources are required for genome-scale analyses. Escherichia coli K-12 has been one of the best-characterized organisms in molecular biology. Yet, many key resources for functional genomics and systems biology studies of E. coli are still lacking.
Whole genome sequences are now available for two closely related K-12 strains, MG1655 (Blattner et al, 1997) and W3110 (Hayashi et al, 2006). Whole-genome comparative sequencing and reconciliation of differences by re-sequencing selected regions from both strains have recently provided the most accurate genome of any organism (accompanying manuscript; Hayashi et al, 2006). Of 267 regions that were initially found to have short insertion or deletion (indel) and nucleotide (nt) disparities, only eight sites were found to be true differences. The vast majority (243) were due to errors in the original 4.5-Mb E. coli K-12 MG1655 genome (an error rate of less than 1 per 13 000 nt 8 years later); 16 were due to errors in the 2.6 Mb of the W3110 genome reported from 1992 to 1997. Sequence corrections resulted in major changes in the translation of 111 MG1655 open-reading frames (ORFs), mostly due to frame shifting (85), but also due to gene fissions (2), gene fusions (23), and inversion (1; Hayashi et al, 2006).
The availability of highly accurate E. coli K-12 genomes (Hayashi et al, 2006) provided an impetus for the cooperative re-annotation of both MG1655 and W3110 (Riley et al, 2006). Sequence corrections also changed many gene boundaries, which led to dropping 31 previously annotated genes and adding 66 new ones. The composite K-12 genome has 4453 genes, encoding 4296 ORFs (including 74 pseudogenes), 156 RNAs, and one annotated feature (oriC). Major differences between the MG1655 and W3110 genomes are the 12 additional sites of an insertion sequence (IS) in W3110, and one additional IS site and the defective CPZ-55 phage (seven prophage genes) only in MG1655. Consequently, MG1655 and W3110 have two and 17 extra copies of IS genes, respectively, and MG1655 has 11 and W3110 has 21 unique genes (including seven additional pseudogenes). Thus, on the basis of the 2005 annotation snapshot, MG1655 has a total of 4464 genes and W3110 has 4474 (Hayashi et al, 2006). In addition to updating annotations of gene functions, start sites were changed for 682 MG1655 ORFs (Riley et al, 2006). An additional 76 ORFs that have been predicted in W3110 have been targeted, for a total of 4550 genes encoding 4390 ORFs (Hayashi et al, 2006), although these have not been recognized as ORFs in the recent K-12 annotation workshops.
An E. coli K-12 functional genomics project was initiated in Japan to (1) create new experimental resources, (2) establish new analysis methods, (3) develop new computational approaches, (4) improve databases, and (5) analyze gene function through experimentation by using these resources, methods, approaches, and databases (Mori et al, 2000). Newly created experimental resources now include: (a) two E. coli K-12 ORFeome plasmid banks of nearly all predicted ORFs, the ASKA clone sets (Kitagawa et al, 2005), (b) a large collection of transposon-generated gene-disruption mutants (Mori et al, 2000), and (c) mutants individually deleted of all nonessential E. coli K-12 genes (this study). Newly established analysis methods have included DNA microarrays (Oshima et al, 2002b), proteome analysis tools (Katayama et al, 2002), and tagged genes for detecting protein–protein interactions (Arifuzzaman et al, 2006). Newly developed computational approaches have included tools for gene clustering and codon usage diversity (Kanaya et al, 2001). An improved E. coli K-12 GenoBase (version 5.0; http://ecoli.aist-nara.ac.jp/) supports data analysis based on these new resources, analysis methods, and computational approaches. These resources and methods have been helpful for assignment of new cellular roles to many genes of unknown or poorly described function (e.g. Oshima et al, 2002a).
In a Saccharomyces cerevisiae functional genomics project, a nearly complete set of single-gene deletions covering 96% of yeast annotated ORFs was constructed by using a PCR gene replacement method (Giaever et al, 2002). The yeast mutants were isolated by direct transformation with PCR products encoding kanamycin resistance and containing 45-nt flanking homologous sequences for adjacent chromosomal regions. Genome-scale disruption of Bacillus subtilis genes (Kobayashi et al, 2003) was carried out by inactivating each gene with a gene-specific plasmid clone. Comprehensive transposon mutagenesis of Pseudomonas aeruginosa was carried out by generating a large set (30 100) of sequence-defined mutants (Jacobs et al, 2003).
Two groups began projects to construct comprehensive transposon mutant libraries of E. coli K-12. In Japan, chromosomal segments in a phage λ library (Kohara et al, 1987) were subjected to transposon mutagenesis, after which the mutations were recombined onto the chromosome by homologous recombination (Mori et al, 2000; T Miki, personal communication). The other group subjected PCR products encoding ORFs to in vitro Tn5 transposition (Goryshin et al, 2000), and then recombined the mutations onto the chromosome by λ Red-mediated recombination (Datsenko and Wanner, 2000), which led to the creation of insertion alleles for 1976 ORFs (Kang et al, 2004).
Although transposon mutagenesis has yielded large unique collections of valuable mutants, the methodologies for building a comprehensive library are laborious. First, it is necessary to define the insertion sites by PCR or DNA sequencing. Second, rearrangements or genetic duplications can result when recombining mutations onto the chromosome, compounding results, and requiring additional testing. Third, complications resulting from transposon mutagenesis, such as incomplete disruption of the targeted gene and polarity effects on downstream genes, are unavoidable.
While the project for building a transposon library was underway in Japan, a highly efficient method for direct inactivation of chromosomal genes in E. coli K-12 was reported (Datsenko and Wanner, 2000). This breakthrough provided a simple and efficient method for gene deletion analogous to the one that has been used in yeast (Baudin et al, 1993), except by use of cells carrying an easily curable, low-copy-number plasmid expressing the λ Red recombinase. Advantages are being able to target genes for complete deletion, to design deletions arbitrarily and precisely, and to easily eliminate the antibiotic resistance marker subsequently. Here, we used the λ Red system for the systematic construction of a set E. coli K-12 mutants with precisely defined single-gene deletions, called the Keio collection, which upon release of the resistance marker will leave behind an in-frame deletion. For convenience of gene transfer, the Keio collection retains the resistance marker.
Results and discussion
Keio collection mutants
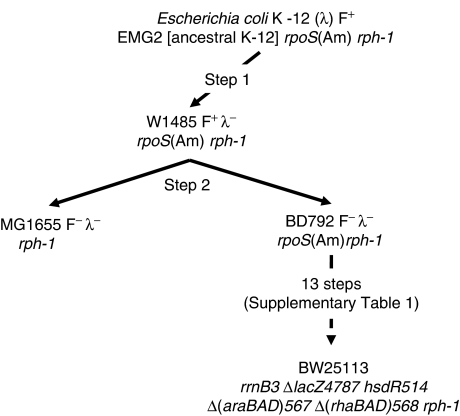
The Keio collection is comprised of 3985 deletions in duplicate (7970 total) of E. coli K-12 strain BW25113 (Datsenko and Wanner, 2000), a strain with a well-defined pedigree that has not been subjected to mutagens (Figure 1; Supplementary Table 1). Mutants were directly selected as kanamycin-resistant (KmR) colonies after electroporation of BW25113 carrying the λ Red expression plasmid pKD46 (Datsenko and Wanner, 2000). To alleviate problems that can arise in high-throughput experiments, resulting from handling errors, crosscontamination, and accumulation of secondary mutations, two independent mutants were saved for each deletion.
Figure 1.
Derivation of E. coli K-12 BW25113. Strain BD792, like MG1655, is a two-step descendent of ancestral E. coli K-12, EMG2, originally called WG1 (Bachmann, 1996; Hayashi et al, 2006; late BJ Bachmann, personal communication). Like its predecessor W1485F+ (Hayashi et al, 2006), BD792 has the rpoS396(Am) allele (codon 33, TAG (Am); unpublished data). Strain BW25113 was derived from BD792 in a series of steps involving generalized transduction and allele replacements, which included introducing the pseudoreversion rpoS (Q33) allele from MG1655 into a predecessor of BW25113 (Supplementary Table 1). The derivation of W3110 is shown in Figure 1 of accompanying manuscript (Hayashi et al, 2006).
Design of in-frame, single-gene deletion mutants
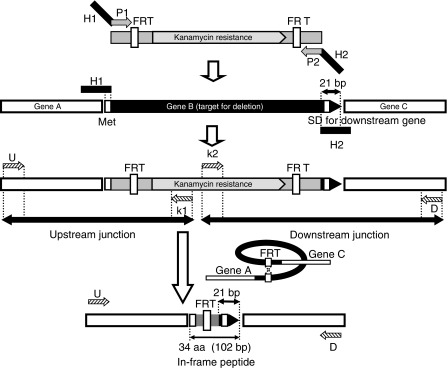
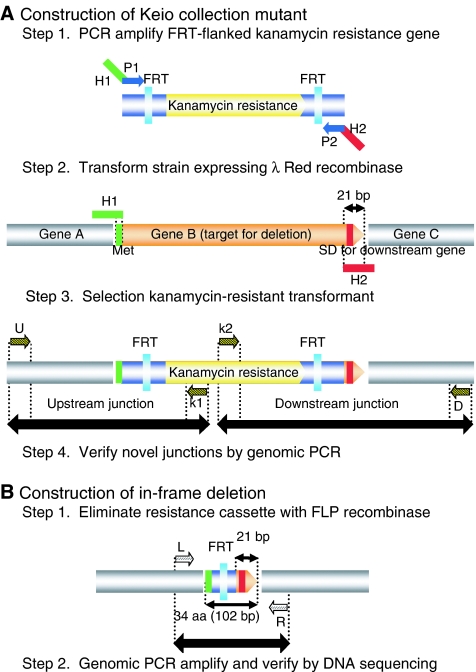
Chromosomal genes were targeted for mutagenesis with PCR products containing a resistance cassette flanked by FLP recognition target (FRT) sites and 50-bp homologies to adjacent chromosomal sequences (Figure 2). To reduce polar effects on downstream gene expression, primers were designed so that excision of the resistance cassette with the FLP recombinase would create an in-frame deletion of the respective chromosomal gene (Figure 3). Primer sequences were based on the highly accurate E. coli K-12 genome (Hayashi et al, 2006), in which the majority of the corrections to coding regions and start codon re-assignments had been made in accordance with the November 2003 E. coli K-12 annotation workshop (Riley et al, 2006).
Figure 2.
Primer design and construction of single-gene deletion mutants. Gene knockout primers have 20-nt 3′ ends for priming upstream (P1) and downstream (P2) of the FRT sites flanking the kanamycin resistance gene in pKD13 and 50-nt 5′ ends homologous to upstream (H1) and downstream (H2) chromosomal sequences for targeting the gene deletion (Supplementary Table 2). H1 includes the gene B (target) initiation codon. H2 includes codons for the six C-terminal residues, the stop codon, and 29-nt downstream. The same primer design with respect to gene B was used to target deletions regardless of whether gene B lies in an operon with genes A and C, as shown, or in different chromosomal arrangements. Novel junctions created between the resistance cassette and neighboring upstream (gene A) and downstream (gene C) sequences were verified by PCR with kanamycin (k1 or k2) and locus-specific (U or D) primers. Structures created after excision of the resistance gene are verified by PCR with neighboring gene-specific primers and by direct DNA sequencing of the region encompassing the H1-P1-FRT-P2-H2 scar to verify correct ones, as described elsewhere (Datsenko and Wanner, 2000). SD, Shine–Dalgarno ribosome binding sequence.
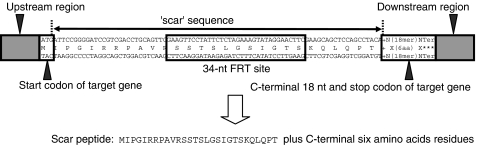
Figure 3.
Structure of in-frame deletions. FLP-mediated excision of the FRT-flanked resistance gene is predicted to create a translatable scar sequence in-frame with the gene B target initiation codon and its C-terminal 18-nt coding region. Translation from the authentic gene B SD and start codon is expected to produce a 34-residue scar peptide with an N-terminal Met, 27 scar-specific residues, and six C-terminal, gene B-specific residues.
The targeting PCR products were designed to create in-frame deletions of the 2nd through the 7th codon from the C-terminus, leaving the ORF start codon and translational signal for a downstream gene intact (Figure 2). However, according to its latest genome annotation, E. coli K-12 has 742 overlapping genes, ranging in length from 1 to 260 nt, with the longest being for ytfP and yzfA. Although the majority are short (1–8 nt), 191 genes overlap by at least 9 nt. Thus, our standard design for construction of in-frame deletions can in some cases simultaneously affect the coding of two overlapping ORFs, which can be especially important when evaluating gene essentiality.
For example, folC encodes bifunctional folylpolyglutamate and dihydrofolate synthases and has an 11-nt overlap with the downstream dedD, encoding a conserved protein of unknown function. In agreement with an earlier study (Pyne and Bognar, 1992), we found folC to be essential (on the basis of the criteria below). Preliminary results suggested that dedD was also essential. However, due to the folC-dedD gene overlap, it was conceivable that the lethality of a dedD deletion was due to alteration of the folC C-terminus. To address these kinds of issues, a small number of primers were redesigned to avoid altering two genes simultaneously, by taking into account gene overlaps. Indeed, dedD was successfully deleted with a PCR product that was synthesized with an N-terminal primer that was redesigned to prevent altering the folC coding region. Primer extensions are given in Supplementary Table 2.
Construction and verification of deletion mutants
Our standard protocol usually yielded 10–1000 KmR colonies when cells were incubated aerobically at 37°C on Luria broth (LB) agar containing 30 μg/ml kanamycin. The most critical step was preparation of highly electrocompetent cells (>109 transformants per 1 μg of plasmid DNA under standard conditions). Mutants were isolated in batches, in which each batch included a PCR product for disruption of ydhQ as a positive control as well as a no PCR product negative control. The latter usually gave only 10–100 tiny colonies. From every gene deletion experiment, four or eight KmR colonies were chosen and checked for ones with the correct structure by PCR using a combination of locus- and kanamycin-specific primers (Figure 2), as described elsewhere (Datsenko and Wanner, 2000). Mutants were scored as correct if two or more colonies had the expected structure based on PCR tests for both junction fragments.
Keio collection deletions
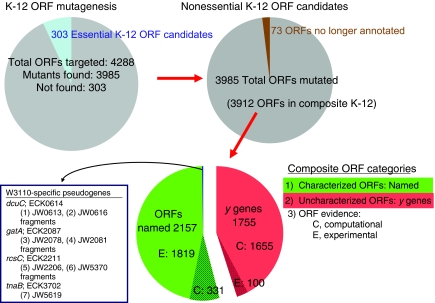
Of 4288 genes targeted, deletions were obtained for 3985 ORFs (Supplementary Table 3). Based on finding mutants with the predicted structure, these 3985 genes are (probably) nonessential, while the 303 genes (including 37 genes of unknown function), for which no mutants were found, are candidates for essential genes (Figure 4; Table I). Our ORF deletions include 3912 genes annotated in both E. coli K-12 MG1655 and W3110 and 73 previously annotated genes (Supplementary Table 4). The 3912 composite K-12 ORF deletions include 2157 characterized genes and 1755 genes of uncharacterized or unknown function. ORFs not targeted include 79 IS genes, four genes for small toxic polypeptides (ldrA, ldrB, ldrC, and ldrD), and seven genes already disrupted in BW25113 (araBAD, lacZ, and rhaBAD; Datsenko and Wanner, 2000). No in-frame deletion was targeted to 12 ORFs whose coding region was changed at the March 2005 annotation workshop (Riley et al, 2006) after completion of the Keio collection (Supplementary Table 5). RNA genes were also not targeted.
Figure 4.
Mutagenesis of E. coli K-12 ORFs. See text.
Table 1.
Mutant summary
| Class | ORFs |
|---|---|
| Total | 4390 |
| Targeteda | 4288 |
| Nonessential | 3985 |
| Essential | 303 |
| Not targetedb | 102 |
| ORF=open-reading frame. | |
aAll targeted ORFs are in given Supplementary Table 2.
bORFs not targeted are given in the text.
Evaluation of gene essentiality
Several causes can contribute to finding too many or too few nonessential genes. One way to evaluate gene essentiality is to examine our knockout efficiency (Table II), that is, the percent of the KmR colonies with the correct structure. For nearly 50% of the targeted ORFs, all KmR colonies had the expected structure for the correct deletion; for 93% of the ORFs, at least 50% were correct; and, with one exception, for all Keio mutants, at least 25% were correct. The exceptional case, secM, has a translational arrest sequence within its C-terminus that is required for expression of the downstream secA, encoding an essential preprotein translocase SecA subunit (Murakami et al, 2004; Nakatogawa et al, 2005). Thus, it is reasonable to suggest that the sole secM mutant arose because it acquired a suppressor allowing secA expression. Essential gene candidates are given in Supplementary Table 6.
Table 2.
| Percent correctb | ORFs | Essentiality scorec | ||
|---|---|---|---|---|
| <−1 | −1 to +1 | ⩾+1 | ||
| 100 | 1946 | 1916 | 30 | 0 |
| 87.5 | 729 | 719 | 10 | 0 |
| 75 | 499 | 487 | 12 | 0 |
| 62.5 | 316 | 307 | 9 | 0 |
| 50 | 219 | 211 | 8 | 0 |
| 37.5 | 116 | 112 | 4 | 0 |
| 25 | 160 | 149 | 11 | 0 |
| 12.5 | 1 | 0 | 1 | 0 |
| 0 | 302 | 0 | 88 | 214 |
| Total | 4288 | 3901 | 173 | 214 |
| ORF=open-reading frame. | ||||
aData are in given Supplementary Table 3.
bPercent of the four or eight KmR colonies shown to have the correct structure by PCR as described in text is given.
cThe number of ORFs with different essentiality scores is given. Scores less than −1 or greater than +1 mean that the gene is nonessential (<−1) or essential (>+1) with no inconsistency with previous studies. Scores between −1 and +1 mean some inconsistency exists.
The ability to select directly for knockout mutants may have led to other mutants with suppressors. For example, the same mutagenesis strategy has been used elsewhere to create a deletion of mreB (Kruse et al, 2003), an essential gene, in which case, the mutant was later shown to carry a suppressor (Kruse et al, 2005). Yet, we repeatedly failed to recover a ΔmreB mutant, even when using the identical primers and host. We also confirmed the absence of mreB coding sequences in their ΔmreB mutant, thus ruling out the possibility of a duplicate mreB sequence (data not shown). Clearly, secM and mreB are examples of ‘quasi-essential' genes, for suppressors allow viability of mutants with the respective deletions. By definition, deletions of truly essential genes cannot be mutationally suppressed.
In addition to suppressors, a functional redundancy or duplication can hide gene essentiality. It is difficult to assess functional redundancy without further experimentation. However, gene duplications can explain why we recovered mutants with deletions of some genes, like ileS and glyS, encoding isoleucyl-tRNA and glycine (β-subunit) tRNA synthetases, which are essential. In these cases, the mutants carry intact copies of the respective deleted gene elsewhere (R D'Ari and K Nakahihashi, personal communication), presumably resulting from gene duplications. Nevertheless, because the vast majority of mutants were recovered at a high frequency (Table II), neither suppressors nor duplications seem to be major concerns. Genetic duplications resulting from gene amplification have been well documented in bacteria; however, the frequency is low; under ordinary conditions, about one in 400 genes is on average duplicated in a culture (Anderson and Roth, 1977). If we assume similar values, then no more than about 10 of our mutants is likely to have a gene duplication. Even though about 1.5% of the yeast mutants were eliminated due to duplications (Giaever et al, 2002), most studies on gene essentiality fail to consider this issue.
Special cases
A few discrepancies exist between our results and those of earlier studies. For example, we were able to delete hlpA, encoding a periplasmic chaperone for outer membrane proteins, which had been reported to be essential (Dicker and Seetharam, 1992). This can be explained by the location of hlpA immediately upstream of lpxD, encoding an essential UDP-3-O-(3-hydroxymyristoyl)-glucosamine N-acyltransferase. A polar effect of the hlpA disruption on lpxD expression was likely responsible for the earlier evidence of gene essentiality. Mutants described here are initially nonpolar because downstream genes can be expressed from the resistance gene promoter (Figure 2), and from the upstream native promoter upon elimination of the resistance cassette.
A number of factors can cause a nonessential gene to appear to be essential. The absence of diaminopimelic acid from standard laboratory media is surely why no mutants requiring this supplement (dapA, B, or E) were recovered. Likewise, our inability to recover particular mutants in central metabolic pathways, for example, gapA, is due to our use of media on which mutants lacking (nonessential) glycolysis genes fail to grow (Fraenkel, 1996), which can be due to the accumulation of toxic intermediates.
Occasional technical problems can also interfere with the isolation of deletions. In a few instances, PCR products failed to target a gene due to the presence of IS elements at sites that were previously unrecognized. Such deletions were successfully made when the primer(s) was redesigned to take the IS element into account. Primer quality is also important. In rare cases, we failed to isolate a deletion for no apparent explanation, yet we were able to do so with a new batch of primers.
Toxin–antitoxin (TA) systems
Deletion of a single gene can lead to aberrant behavior in certain gene contexts. Well-studied examples are the prokaryotic TA stress response loci (Gerdes et al, 2005). For example, RelE and MazF are toxins that cleave mRNA in response to a nutritional stress. Under nonstress conditions, a specific antitoxin (RelB or MazE) prevents cleavage, allowing normal growth. E. coli K-12 encodes six such TA systems, three belonging to the RelE (toxin)/RelB (antitoxin) family, RelE/RelB, YafQ/DinJ, and YoeB/YefM; two belonging to the MazF/MazE family, ChpA(MazF)/ChpR(MazE) and ChpB/ChpS; and one belonging to the TA-like system, HipA/HipB. Our failure to find deletions of yefM, chpR, or chpS is likely because they encode TA system antitoxins.
Categories of essential genes
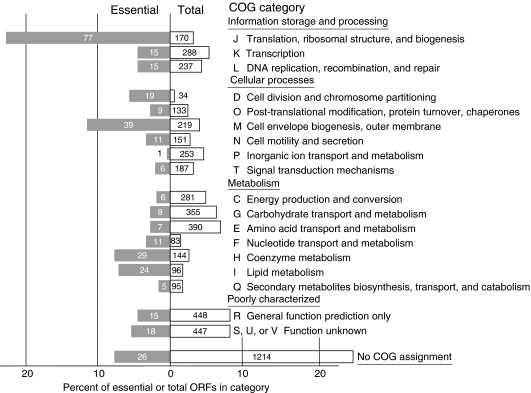
ORFs can be classified into clusters of orthologous groups (COGs) belonging to different functional categories (Figure 5; Supplementary Table 7). It is natural for multidomain proteins to be comprised of more than one COG. Some COGs also belong to more than one functional class. Consequently, the 4390 ORFs in E. coli K-12 strain W3110 correspond to 4011 COGs (and 1214 with no COG assignment), while the 303 essential ORF candidates correspond to 315 COGs (and 26 with no COG assignment). The fraction of essential genes varies widely with the COG classification. The greatest fractions are for COGs with roles in translation, ribosomal structure, and biogenesis. The vast majority of essential genes belong to COGs with roles in cell division, lipid metabolism, translation, transcription, and cell envelope biogenesis. For example, our results showed that rpoE and rpoH, encoding RNA polymerase heat-shock sigma factors E and H, respectively, are essential, in agreement with earlier studies (Zhou et al, 1988; Hiratsu et al, 1995). Our data also showed that we were able to disrupt genes for five ribosomal proteins (S6, S20, L1, L11, and L33), which had been previously shown to be nonessential (Dabbs, 1991). Discrepancies for 11 others may have resulted from use of different growth conditions or strain.
Figure 5.
COG classification of K-12 genes. See Supplementary Table 7.
Comparison with other E. coli gene essentiality studies
Genetic footprinting (Gerdes et al, 2003; Tong et al, 2004) revealed 620 genes to be essential for robust aerobic growth of E. coli K-12. Yet, only 67% (205 genes) overlap with the predicted essential genes in this study. Striking differences can be attributed to the use of different mutagenesis strategies (transposon insertion versus deletion), different growth conditions (broth versus agar), or the approach for discriminating essential versus nonessential genes. Because genetic footprinting measures cell populations, a mutation causing slow growth can lead to under-representation of the mutant and hence false classification of many genes as essential. In contrast, we sought deletion mutants as survivors without regard to growth rate. Supplementary Table 6 has a comparison of our results with those from genetic footprinting (Gerdes et al, 2003), the PEC database (Hashimoto et al, 2005), and transposon mutagenesis (Kang et al, 2004), in which an ‘essentiality score' is computed for all 303 essential gene candidates from our study.
We also examined the conservation of the K-12 essential genes in genomes of other organisms in the Microbial Genome Database (http://mbgd.genome.ad.jp/; Uchiyama, 2003). Comparison with three other E. coli genomes revealed that more than 90% (282) of the essential genes are universally present. About one-half (147) are conserved among 20 different Enterobacteriaceae genomes. One-third (85) are conserved among 74 Proteobacteria and less than 15% (42) are conserved among 171 bacteria (Supplementary Tables 6 and 8).
Comparison with gene essentiality in other free-living bacteria
B. subtilis has a 4.2-Mb genome and 271 essential genes (Kobayashi et al, 2003). About one-half (150) of the orthologous genes are also essential in E. coli. Another 67 genes that are essential in E. coli are not essential in B. subtilis, while 86 E. coli essential genes have no B. subtilis ortholog. Details are given in Supplementary Table 6.
Profiling contributions of individual genes during growth on rich and minimal media
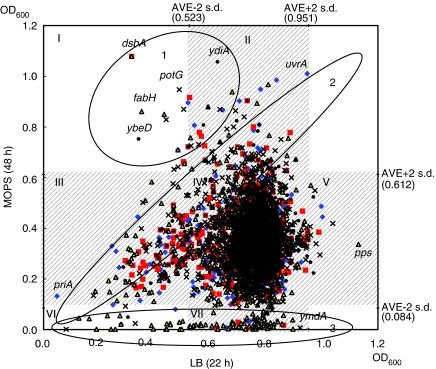
All mutants were profiled for growth yield in both rich (LB) and minimal glucose MOPS media (Figure 6). Growth data in Figure 6 are summarized according to COG category in Table III. Complete information is in Supplementary Table 3. Many factors can contribute to how efficiently cells convert nutrients into biomass. The vast majority showed no differences from wild type. Mutants in circled area 1 gave higher yield in minimal than rich; those in area 2 gave similar yields in both media; and those in area 3 gave higher yields in rich than minimal. No correlation with mutant class was seen for those in areas 1 or 2. As expected, the majority in area 3 has defects in biosynthesis, for example, for amino acids, purines, pyrimidines, and vitamins. Curiously, a subset of these auxotrophs showed modest growth after 48 h, suggesting that suppressors arose. The trivial explanation of crosscontamination is unlikely because similar results were obtained in replica cultures. A few mutants with deletions of genes of unknown function also grew well in rich but not in minimal, which may provide a handle on determination of their function. Some grew after 24 h but showed no growth after 48 h, suggesting lysis, for example, ddlB (D-alanine:D-alanine ligase), csgC (predicted curli production protein), rsxC (predicted 4Fe–4S ferredoxin-type protein), and others. Many grew poorly on both rich and minimal media, for example, priA (primosome factor), atp (ATP synthase components), and cyaA (adenylate cyclase). Nevertheless, the majority showed no striking growth defect.
Figure 6.
Profiling gene contribution for growth. Mutants of all 3985 genes in the Keio collection were grown 22 h in LB and 24 and 48 h in 0.4% glucose MOPS 2 mM Pi medium (Wanner, 1994). Maximum cell density values are plotted. Circled areas 1, 2, and 3 are discussed in text. Grayed areas show 2 × s.d. Groups labeled I–VII differ by more than 2 × s.d. Summary data are given in Table III. Additional time-course data are given in Supplementary Figure 1. Data are in given Supplementary Table 3. Major COG categories: blue diamond, information storage and processing; red square, cellular processes; yellow triangle, metabolism; black filled circle, poorly characterized; and black cross, no COG assignment.
Table 3.
Summary of growth data for Keio collection according to COG category
| COG category | Groupa | |||||||
|---|---|---|---|---|---|---|---|---|
| Medium | I | II | III | IV | V | VI | VII | |
| LB | <0.52 | 0.52–0.95 | <0.52 | 0.52–0.95 | 0.95< | <0.52 | 0.52–0.95 | |
| MOPS | 0.61< | 0.61< | 0.08–0.61 | 0.08–0.61 | 0.08–0.61 | <0.08 | <0.08 | |
| Information storage and processing | J | 0 | 1 | 8 | 84 | 0 | 0 | 0 |
| K | 1 | 12 | 8 | 248 | 1 | 1 | 2 | |
| L | 1 | 6 | 9 | 127 | 1 | 1 | 1 | |
| D | 0 | 0 | 0 | 15 | 0 | 0 | 0 | |
| O | 2 | 2 | 11 | 110 | 0 | 0 | 0 | |
| Cellular processes | M | 0 | 3 | 12 | 163 | 0 | 0 | 1 |
| N | 2 | 5 | 33 | 100 | 0 | 0 | 0 | |
| P | 0 | 11 | 4 | 234 | 1 | 0 | 2 | |
| T | 0 | 5 | 9 | 165 | 0 | 0 | 0 | |
| C | 1 | 12 | 26 | 229 | 0 | 2 | 4 | |
| G | 2 | 15 | 13 | 311 | 1 | 0 | 0 | |
| E | 0 | 12 | 7 | 305 | 1 | 5 | 53 | |
| Metabolism | F | 0 | 1 | 3 | 47 | 0 | 12 | 9 |
| H | 1 | 2 | 5 | 88 | 0 | 2 | 17 | |
| I | 1 | 2 | 0 | 69 | 0 | 0 | 0 | |
| Q | 0 | 1 | 0 | 88 | 0 | 0 | 1 | |
| R | 0 | 15 | 11 | 403 | 0 | 1 | 2 | |
| Poorly characterized | S | 1 | 7 | 8 | 259 | 3 | 0 | 0 |
| U | 1 | 4 | 15 | 88 | 0 | 0 | 0 | |
| V | 0 | 2 | 0 | 41 | 0 | 0 | 0 | |
| No COG | 5 | 34 | 36 | 1091 | 6 | 2 | 7 | |
| COG=clusters of orthologous groups; LB=Luria broth; MOPS=0.4% glucose MOPS 2 mM Pi medium (Wanner, 1994). | ||||||||
aGroups are shown in Figure 6. Number of mutants in each COG category are given.
Use and distribution of the Keio collection
Several complete sets of the Keio collection as well as thousands of individual mutants have already been distributed worldwide. Distribution is being handled via GenoBase (http://ecoli.aist-nara.ac.jp/) together with supporting data and other key resources, including the ASKA (A complete Set of E. coli K-12 ORF Archive) clone sets (Kitagawa et al, 2005). Several studies have already reported use of these mutants. For example, single-gene deletion mutants of the Keio collection were utilized for the study of uncharacterized gene function (Melnick et al, 2004) and the analysis of metabolism (Jiao et al, 2003; Hua et al, 2003, 2004; Yang et al, 2003; Zhao et al, 2004a, 2004b). The use of subsets of Keio collection mutants has substantiated the value of systematical approaches for the understanding of cellular systems (Tenorio et al, 2003; Ito et al, 2005; Perrenoud and Sauer, 2005).
Summary
We have undertaken a large-scale project for systematic construction of a set of precisely defined single-gene, knockout mutants of all nonessential genes in E. coli K-12. These mutants were designed to create in-frame (nonpolar) deletions upon elimination of the resistance cassette. Our analysis of these mutants has provided new key information on E. coli biology. First, the vast majority of the genes that were independently disrupted at least twice are probably nonessential, at least under the conditions of selection. Second, those genes that we repeatedly failed to disrupt are candidates for essential E. coli genes. Lastly, by comparing the effects of these mutations in the same E. coli K-12 genetic background, we profiled the contribution of these genes to growth on synthetic minimal and rich medium.
The Keio collection should provide not only a basic resource for systematic functional genomics but also experimental data source for systems biology approaches. The mutants can serve as fundamental tools for a number of reverse genetics approaches, permitting analysis of the consequences of the complete loss of gene function, in contrast to forward genetics approaches in which mutant phenotypes are associated with a corresponding gene(s). By providing this resource to the research community, the authors hope to contribute to worldwide efforts directed towards a comprehensive understanding of the E. coli K-12 model cell. Because many E. coli gene products are well conserved in nature, the Keio collection is likely to be useful not only for studying E. coli and other bacteria but also for examining properties of genes from a wide range of living organisms.
Materials and methods
Bacteria and plasmids
E. coli BW25141 (rrnB3 DElacZ4787 DEphoBR580 hsdR514 DE(araBAD)567 DE(rhaBAD)568 galU95 DEendA9::FRT DEuidA3::pir(wt) recA1 rph-1) was used for maintenance of the template plasmid pKD13 (GenBank™ Accession number AY048744). pKD46 (GenBank™ Accession number AY048746; Datsenko and Wanner, 2000) was made by PCR amplification of the Red recombinase genes from phage λ and cloning into pKD16, a derivative of INT-ts (Haldimann and Wanner, 2001) carrying araC and araBp from pBAD18 (Guzman et al, 1995).
Media, chemicals, and other reagents
Cells were routinely grown in LB medium containing 1% Bacto Tryptone (Difco), 0.5% yeast extract (Difco), and 0.5% NaCl with or without antibiotics at 50 μg/ml for ampicillin (Wako, Osaka, Japan) and 30 μg/ml for kanamycin (Wako, Osaka, Japan). Glucose, L-arabinose, and other chemicals were from Wako (Osaka, Japan). DpnI was from New England Biolabs (MA, USA); Taq polymerase, TaKaRa Ex Taq, and agarose, SeaKem GTG Agarose from Takara Shuzo Inc. E-Gel 96 systems were from Invitrogen. MOPS medium was prepared as described elsewhere (Wanner, 1994).
PCR primers
With a few exceptions, N-terminal deletion primers had a 50-nt 5′ extension including the gene initiation codon (H1) and the 20-nt sequence 5′-ATTCCGGGGATCCGTCGACC-3′ (P1), and C-terminal deletion primers consisted of 21 nt for the C-terminal region, the termination codon and 29-nt downstream (H2), and the 20-nt sequence 5′-TGTAGGCTGGAGCTGCTTCG-3′ (P2; Figure 2). All extensions are in given Supplementary Table 2.
Generation of PCR fragments
PCR reactions were carried out in 96-well microplates in 50 μl reactions containing 2.5 U of TaKaRa Ex Taq polymerase, 1 pg pKD13 DNA, 1.0 μM of each primer, and 200 μM dNTPs. Reactions were run for 30 cycles: 94°C for 30 s, 59°C for 30 s, 72°C for 2 min, plus an additional 2 min at 72°C. PCR products were digested with DpnI, ethanol precipitated, resuspended in 6 μl H2O, and analyzed by 1% agarose gel electrophoresis using 0.5 × Tris-acetate buffer or the E-Gel 96 system.
Electroporation and mutant selection
E. coli K-12 BW25113 carrying the Red helper plasmid pKD46 was grown in 100 ml SOB medium with ampicillin and 1 mM L-arabinose at 30°C to an OD600 of 0.3, and electroporation-competent cells were prepared as described elsewhere (Sambrook et al, 1998). A measure of 50 μl of competent cells was mixed with 400 ng of the PCR fragment in an ice-cold 0.2 cm cuvette (Bio-Rad Inc.). Cells were electroporated at 2.5 kV with 25 mF and 200 Ω, immediately followed by the addition of 1 ml of SOC medium (2% Bacto Tryptone (Difco), 0.5% yeast extract (Difco), 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 10 mM MgSO4, 20 mM glucose) with 1 mM L-arabinose. After incubation for 2 h at 37°C, one-tenth portion was spread onto agar plate to select KmR transformants at 37°C.
PCR verification of deletions
Two PCR reactions were carried out to test for correct chromosomal structures. Eight independent colonies were transferred into 150-μl LB medium with kanamycin in 96-well microplates and incubated overnight at 37°C without shaking. A measure of 1 μl of each culture was separately examined in 20-μl PCR reactions following 2-min ‘hot start' at 95°C. PCR verification with kanamycin-specific primers k1 and k2 and locus-specific primers U and D (Figure 2) was carried out as described previously (Datsenko and Wanner, 2000). PCR products were analyzed by 1% agarose gel electrophoresis as above.
Storage of mutants
Mutants were stored at −80°C in 96-well microplates containing 150-μl LB medium with kanamycin and 15% glycerol.
Growth tests
Mutants were tested for growth in 200-μl LB medium with kanamycin as a rich medium in 96-well microplates inoculated directly with 96 inoculation pins (Genetix Limited, UK) and incubated for 22 h at 37°C without shaking. Absorbance at 600 nm was measured after mixing for 5 s in a 96-well plate reader (Molecular Dynamics). Mutants were transferred with 96-inoculation pins (Genetix Limited, UK) from LB into 200-μl 0.4% glucose MOPS medium with 2 mM Pi (Wanner, 1994) and kanamycin as minimal medium and incubated for 24 and 48 h at 37°C without shaking. Absorbances were similarly measured.
Supplementary Material
Supplementary Figure 1
Legend to Supplementary Figure and Tables
Supplementary Table 1
Supplementary Table 2
Supplementary Table 3
Supplementary Table 4
Supplementary Table 5
Supplementary Table 6
Supplementary Table 7
Supplementary Table 8
Figure 7.
Electron micrograph of E. coli K-12 by Melvin L Demphilis and Julius Adler. Republished with permission (Holden, 2002).
Figure 8.
PCR gene replacement strategy. (A) Gene targeting fragment encoding kanamycin resistance with short homology extensions (H1 and H2) is generated by PCR by using priming sites P1 and P2 (Step 1). Gene targeting fragment is introduced into E. coli K-12 BW25113 expressing the Red recombinase from pKD46 (Step 2). Kanamycin-resistant transformants are selected (Step 3). Transformants are verified by PCR (Step 4). (B) Elimination of the resistance cassette by use of the FLP recombinase plasmid pCP20 is expected to leave behind a 102-nt ‘scar' encoding a 34-residue peptide (Step 1). The scar region is amplified and sequenced to be sure no mutations, especially 1-nt deletions (Datsenko and Wanner, 2000), were introduced (Step 2).
Acknowledgments
This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, a grant from CREST, JST (Japan Science and Technology), and in part from NEDO (New Energy and Industrial Technology Development Organization) and from Tsuruoka City and Yamagata Prefecture governments. BLW is supported by NIH GM62662. We thank Miki Naba, Daisuke Kido, Narith Chy, Toru Kodama, Koji Komatsu, and Prof. Kazuyuki Shimizu from the Kyushu Institute of Technology for help in measuring growth of the glycolysis gene deletion mutants.
References
- Anderson RP, Roth JR (1977) Tandem genetic duplications in phage and bacteria. Annu Rev Microbiol 31: 473–505 [DOI] [PubMed] [Google Scholar]
- Arifuzzaman M, Maeda M, Itoh A, Nishikata K, Takita C, Saito R, Ara T, Nakahigashi K, Hirai A, Tsuzuki K, Nakamura S, Altaf-Ul-Amin M, Oshima T, Baba T, Yamamoto N, Kawamura T, Ioka-Nakamichi T, Kitagawa M, Tomita M, Kanaya S, Wada C, Mori H (2006) Protein interaction networks of Escherichia coli K-12 (submitted) [DOI] [PMC free article] [PubMed]
- Bachmann BJ (1996) Derivations and genotypes of some mutant derivatives of Escherichia coli K-12. In Escherichia coli and Salmonella typhimurium Cellular and Molecular Biology, Neidhardt FC, Curtiss III R, Ingraham JL, Lin ECC, Low Jr KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (eds), 2 edn, pp 2460–2488. Washington, DC: ASM Press [Google Scholar]
- Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C (1993) A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae . Nucleic Acids Res 21: 3329–3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y (1997) The complete genome sequence of Escherichia coli K-12. Science 277: 1453–1462 [DOI] [PubMed] [Google Scholar]
- Crick F (2002) Letter to Barry L. Wanner dated October 1, 2002.
- Crick FHC (1973) Project K: ‘The complete solution of E. coli'. Perspectives in Biology and Medicine (17): 67–70; Baltimore: Johns Hopkins University Press. [Google Scholar]
- Dabbs ER (1991) Mutants lacking individual ribosomal proteins as a tool to investigate ribosomal properties. Biochimie 73: 639–645 [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicker IB, Seetharam S (1992) What is known about the structure and function of the Escherichia coli protein FirA. Mol Microbiol 6: 817–823 [DOI] [PubMed] [Google Scholar]
- Fraenkel DG (1996) Glycolysis. In Escherichia coli and Salmonella Cellular and Molecular Biology, Neidhardt FC, Curtiss III R, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (eds), 2 edn, pp 189–198. Washington, DC: ASM Press [Google Scholar]
- Gerdes K, Christensen SK, Lobner-Olesen A (2005) Prokaryotic toxin–antitoxin stress response loci. Nat Rev Microbiol 3: 371–382 [DOI] [PubMed] [Google Scholar]
- Gerdes SY, Scholle MD, Campbell JW, Balazsi G, Ravasz E, Daugherty MD, Somera AL, Kyrpides NC, Anderson I, Gelfand MS, Bhattacharya A, Kapatral V, D'Souza M, Baev MV, Grechkin Y, Mseeh F, Fonstein MY, Overbeek R, Barabasi AL, Oltvai ZN, Osterman AL (2003) Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J Bacteriol 185: 5673–5684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, Arkin AP, Astromoff A, EL Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A, Entian KD, Flaherty P, Foury F, Garfinkel DJ, Gerstein M, Gotte D, Guldener U, Hegemann JH, Hempel S, Herman Z, Jaramillo DF, Kelly DE, Kelly SL, Kotter P, LaBonte D, Lamb DC, Lan N, Liang H, Liao H, Liu L, Luo C, Lussier M, Mao R, Menard P, Ooi SL, Revuelta JL, Roberts CJ, Rose M, Ross-Macdonald P, Scherens B, Schimmack G, Shafer B, Shoemaker DD, Sookhai-Mahadeo S, Storms RK, Strathern JN, Valle G, Voet M, Volckaert G, Wang CY, Ward TR, Wilhelmy J, Winzeler EA, Yang Y, Yen G, Youngman E, Yu K, Bussey H, Boeke JD, Snyder M, Philippsen P, Davis RW, Johnston M (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391 [DOI] [PubMed] [Google Scholar]
- Goryshin IY, Jendrisak J, Hoffman LM, Meis R, Reznikoff WS (2000) Insertional transposon mutagenesis by electroporation of released Tn5 transposition complexes. Nat Biotechnol 18: 97–100 [DOI] [PubMed] [Google Scholar]
- Guzman L-M, Belin D, Carson MJ, Beckwith J (1995) Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177: 4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldimann A, Wanner BL (2001) Conditional-replication, integration, excision, and retrieval plasmid–host systems for gene structure–function studies in bacteria. J Bacteriol 183: 6384–6393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Ichimura T, Mizoguchi H, Tanaka K, Fujimitsu K, Keyamura K, Ote T, Yamakawa T, Yamazaki Y, Mori H, Katayama T, Kato J (2005) Cell size and nucleoid organization of engineered Escherichia coli cells with a reduced genome. Mol Microbiol 55: 137–149 [DOI] [PubMed] [Google Scholar]
- Hayashi K, Morooka N, Yamamoto Y, Fujita K, Isono K, Choi S, Ohtsubo E, Baba T, Wanner BL, Mori H, Horiuchi T (2006) Highly accurate genome sequences of the Escherichia coli K-12 strains MG1655 and W3110. Mol Syst Biol 2006.0007. doi:10.1038/msb4100049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu K, Amemura M, Nashimoto H, Shinagawa H, Makino K (1995) The rpoE gene of Escherichia coli, which encodes σE, is essential for bacterial growth at high temperature. J Bacteriol 177: 2918–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden C (2002) Cell biology: alliance launched to model E. coli. Science 297: 1459–1460 [DOI] [PubMed] [Google Scholar]
- Hua Q, Yang C, Baba T, Mori H, Shimizu K (2003) Responses of the central metabolism in Escherichia coli to phosphoglucose isomerase and glucose-6-phosphate dehydrogenase knockouts. J Bacteriol 185: 7053–7067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Q, Yang C, Oshima T, Mori H, Shimizu K (2004) Analysis of gene expression in Escherichia coli in response to changes of growth-limiting nutrient in chemostat cultures. Appl Environ Microbiol 70: 2354–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Baba T, Mori H, Mori H (2005) Functional analysis of 1440 Escherichia coli genes using the combination of knock-out library and phenotype microarrays. Metab Eng 7: 318–327 [DOI] [PubMed] [Google Scholar]
- Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R, Chun-Rong L, Guenthner D, Bovee D, Olson MV, Manoil C (2003) Comprehensive transposon mutant library of Pseudomonas aeruginosa . Proc Natl Acad Sci USA 100: 14339–14344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Z, Baba T, Mori H, Shimizu K (2003) Analysis of metabolic and physiological responses to gnd knockout in Escherichia coli by using C-13 tracer experiment and enzyme activity measurement. FEMS Microbiol Lett 220: 295–301 [DOI] [PubMed] [Google Scholar]
- Kanaya S, Kinouchi M, Abe T, Kudo Y, Yamada Y, Nishi T, Mori H, Ikemura T (2001) Analysis of codon usage diversity of bacterial genes with a self-organizing map (SOM): characterization of horizontally transferred genes with emphasis on the E. coli O157 genome. Gene 276: 89–99 [DOI] [PubMed] [Google Scholar]
- Kang Y, Durfee T, Glasner JD, Qiu Y, Frisch D, Winterberg KM, Blattner FR (2004) Systematic mutagenesis of the Escherichia coli genome. J Bacteriol 186: 4921–4930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama A, Tsujii A, Wada A, Nishino T, Ishihama A (2002) Systematic search for zinc-binding proteins in Escherichia coli . Eur J Biochem 269: 2403–2413 [DOI] [PubMed] [Google Scholar]
- Kitagawa M, Ara T, Nakamichi T, Inamoto E, Toyonaga H, Mori H (2005) Complete set of ORF clones of Escherichia coli: unique resources for biological research. DNA Res (doi:10.1093/dnares/dsi012) [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, Asai K, Ashikaga S, Aymerich S, Bessieres P, Boland F, Brignell SC, Bron S, Bunai K, Chapuis J, Christiansen LC, Danchin A, Debarbouille M, Dervyn E, Deuerling E, Devine K, Devine SK, Dreesen O, Errington J, Fillinger S, Foster SJ, Fujita Y, Galizzi A, Gardan R, Eschevins C, Fukushima T, Haga K, Harwood CR, Hecker M, Hosoya D, Hullo MF, Kakeshita H, Karamata D, Kasahara Y, Kawamura F, Koga K, Koski P, Kuwana R, Imamura D, Ishimaru M, Ishikawa S, Ishio I, Le CD, Masson A, Mauel C, Meima R, Mellado RP, Moir A, Moriya S, Nagakawa E, Nanamiya H, Nakai S, Nygaard P, Ogura M, Ohanan T, O'Reilly M, O'Rourke M, Pragai Z, Pooley HM, Rapoport G, Rawlins JP, Rivas LA, Rivolta C, Sadaie A, Sadaie Y, Sarvas M, Sato T, Saxild HH, Scanlan E, Schumann W, Seegers JF, Sekiguchi J, Sekowska A, Seror SJ, Simon M, Stragier P, Studer R, Takamatsu H, Tanaka T, Takeuchi M, Thomaides HB, Vagner V, van Dijl JM, Watabe K, Wipat A, Yamamoto H, Yamamoto M, Yamamoto Y, Yamane K, Yata K, Yoshida K, Yoshikawa H, Zuber U, Ogasawara N (2003) Essential Bacillus subtilis genes. Proc Natl Acad Sci USA 100: 4678–4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohara Y, Akiyama K, Isono K (1987) The physical map of the whole E. coli chromosome: Application of a new strategy for rapid analysis and sorting of a large genomic library. Cell 50: 495–508 [DOI] [PubMed] [Google Scholar]
- Kruse T, Bork-Jensen J, Gerdes K (2005) The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol Microbiol 55: 78–89 [DOI] [PubMed] [Google Scholar]
- Kruse T, Moller-Jensen J, Lobner-Olesen A, Gerdes K (2003) Dysfunctional MreB inhibits chromosome segregation in Escherichia coli . EMBO J 22: 5283–5292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnick J, Lis E, Park JH, Kinsland C, Mori H, Baba T, Perkins J, Schyns G, Vassieva O, Osterman A, Begley TP (2004) Identification of the two missing bacterial genes involved in thiamine salvage: thiamine pyrophosphokinase and thiamine kinase. J Bacteriol 186: 3660–3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori H, Isono K, Horiuchi T, Miki T (2000) Functional genomics of Escherichia coli in Japan. Res Microbiol 151: 121–128 [DOI] [PubMed] [Google Scholar]
- Murakami A, Nakatogawa H, Ito K (2004) Translation arrest of SecM is essential for the basal and regulated expression of SecA. Proc Natl Acad Sci USA 101: 12330–12335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt FC (1996) The enteric bacterial cell and the age of bacteria. In Escherichia coli and Salmonella Cellular and Molecular Biology, Neidhardt FC, Curtiss III R, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M and Umbarger HE (eds), 2 edn, pp 1–4. Washington, DC: ASM Press [Google Scholar]
- Nakatogawa H, Murakami A, Mori H, Ito K (2005) SecM facilitates translocase function of SecA by localizing its biosynthesis. Genes Dev 19: 436–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima T, Aiba H, Masuda Y, Kanaya S, Sugiura M, Wanner BL, Mori H, Mizuno T (2002a) Transcriptome analysis of all two-component regulatory systems of Escherichia coli . Mol Microbiol 46: 281–291 [DOI] [PubMed] [Google Scholar]
- Oshima T, Wada C, Kawagoe Y, Ara T, Maeda M, Masuda Y, Hiraga S, Mori H (2002b) Genome-wide analysis of deoxyadenosine methyltransferase-mediated control of gene expression in Escherichia coli . Mol Microbiol 45: 673–695 [DOI] [PubMed] [Google Scholar]
- Perrenoud A, Sauer U (2005) Impact of global transcriptional regulation by ArcA, ArcB, Cra, Crp, Cya, Fnr, and Mlc on glucose catabolism in Escherichia coli. J Bacteriol 187: 3171–3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyne C, Bognar AL (1992) Replacement of the folC gene, encoding folylpolyglutamate synthetase–dihydrofolate synthetase in Escherichia coli, with genes mutagenized in vitro . J Bacteriol 174: 1750–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley M, Abe T, Arnaud MB, Berlyn MB, Blattner FR, Chaudhuri RR, Glasner JD, Mori H, Horiuchi T, Keseler IM, Kosuge T, Perna NT, Plunkett G III, Rudd KE, Serres MH, Thomas GH, Thomson NR, Wishart DS, Wanner BL (2006) Escherichia coli K-12: a cooperatively developed annotation snapshot—2005. Nucleic Acids Res 34: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1998) Molecular Cloning, A Laboratory Manual, 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory [Google Scholar]
- Tenorio E, Saeki T, Fujita K, Kitakawa M, Baba T, Mori H, Isono K, Horiuchi T, Wada C, Kanaya S, Kitagawa M, Ara T, Ohshima H, Miki T (2003) Systematic characterization of Escherichia coli genes/ORFs affecting biofilm formation. FEMS Microbiol Lett 225: 107–114 [DOI] [PubMed] [Google Scholar]
- Tong X, Campbell JW, Balazsi G, Kay KA, Wanner BL, Gerdes SY, Oltvai ZN (2004) Genome-scale identification of conditionally essential genes in E. coli by DNA microarrays. Biochem Biophys Res Commun 322: 347–354 [DOI] [PubMed] [Google Scholar]
- Uchiyama I (2003) MBGD: microbial genome database for comparative analysis. Nucleic Acids Res 31: 58–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner BL (1994) Gene expression in bacteria using TnphoA and TnphoA′ elements to make and switch phoA gene, lacZ (op), and lacZ (pr) fusions. In Methods in Molecular Genetics, Adolph KW (ed), Vol. 3, pp 291–310. Orlando: Academic Press [Google Scholar]
- Yang C, Hua Q, Baba T, Mori H, Shimizu K (2003) Analysis of Escherichia coli anaplerotic metabolism and its regulation mechanisms from the metabolic responses to altered dilution rates and phosphoenolpyruvate carboxykinase knockout. Biotechnol Bioeng 84: 129–144 [DOI] [PubMed] [Google Scholar]
- Zhao J, Baba T, Mori H, Shimizu K (2004a) Effect of zwf gene knockout on the metabolism of Escherichia coli grown on glucose or acetate. Metab Eng 6: 164–174 [DOI] [PubMed] [Google Scholar]
- Zhao J, Baba T, Mori H, Shimizu K (2004b) Global metabolic response of Escherichia coli to gnd or zwf gene-knockout, based on 13C-labeling experiments and the measurement of enzyme activities. Appl Microbiol Biotechnol 64: 91–98 [DOI] [PubMed] [Google Scholar]
- Zhou YN, Kusukawa N, Erickson JW, Gross CA, Yura T (1988) Isolation and characterization of Escherichia coli mutants that lack the heat shock sigma factor sigma 32. J Bacteriol 170: 3640–3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Legend to Supplementary Figure and Tables
Supplementary Table 1
Supplementary Table 2
Supplementary Table 3
Supplementary Table 4
Supplementary Table 5
Supplementary Table 6
Supplementary Table 7
Supplementary Table 8