Abstract
Cells respond to fatty acid exposure by metabolic reorganization and proliferation of peroxisomes. Described here is the development and application of a genome-wide screen to identify nonessential yeast genes necessary for efficient metabolism of myristic and oleic acids. Comparison of the resultant fitness data set with an integrated data set of genes transcriptionally responsive to fatty acids revealed very little overlap between the data sets. Furthermore, the fitness data set enriched for genes involved in peroxisome biogenesis and other processes related to cell morphology, whereas the expression data set enriched for genes related to metabolism. These data suggest that in response to fatty acid exposure, transcriptional control is biased towards metabolic reorganization, and structural changes tend to be controlled post-transcriptionally. They also suggest that fatty acid responsive metabolic networks are more robust than those related to cell structure. Statistical analyses of these and other global data sets suggest that the utilization of distinct control mechanisms for the execution of morphological versus metabolic responses is widespread.
Introduction
Understanding how discrete subcellular processes are coordinately regulated in response to stimuli and the relationship of this regulation to cell fitness are significant systems-level problems. Insight can come from large-scale data sets as their comprehensiveness, and the relative nature of the measurements made, that is, one component relative to another or one time point relative to another, make it appropriate to analyse the individual data together as ‘data sets', which through statistical analysis can reveal new insight into the relationships between processes.
Further insight can come from integrating large-scale data sets. The identification of multiple relationships among groups of genes, such as identifying both a correlation of transcription profiles and physical interactions among the encoded proteins, strengthens the confidence of molecular associations. However, there is mounting evidence that some data types are more complementary in nature. For example, genes required for cell fitness can be distinct from those that are transcriptionally responsive under the same condition (Birrell et al, 2002; Giaever et al, 2002). For this reason, it is important to comparatively analyse data sets that encompass different levels of biological control. Such analyses can both direct integration strategies and reveal insight into how different control mechanisms are utilized by the cell.
The response of yeast cells to growth in the presence of fatty acids includes upregulation of genes involved in fatty acid metabolism and an increase in the size and number of peroxisomes, subcellular organelles that house enzymes involved in fatty acid β-oxidation. Because the response includes not only metabolic reorganization, but also a distinguishable change in cell structure, this response is well suited to studying the coordinate regulation of different cellular processes.
Results and discussion
Identification of genes necessary for fatty acid metabolism
We developed and applied a screen to the Saccharomyces cerevisiae haploid, viable gene-deletion set (Resgen/Invitrogen, Carlsbad, CA) to comprehensively identify genes necessary for efficient fatty acid metabolism. The screen is based on an assay that measures the ability of strains to form a clear zone in the surrounding turbid agar medium containing oleic acid, a monounsaturated fatty acid (cis-C18:1(9)) (Gurvitz et al, 1997). However, because of the low opacity of the medium, clear zone formation was difficult to visualize and document on oleate-containing plates (Figure 1A). To complement this analysis, myristic acid, a C14 saturated fatty acid, was substituted for oleic acid. Like oleic acid, myristic acid requires functional peroxisomal β-oxidation for its metabolism, and the growth rate of S. cerevisiae on myristic acid was similar to that on oleic acid (data not shown). However, the contrast of the clear zones was dramatically improved on myristic acid medium (Figure 1B and C), and this data set is likely to have fewer false negatives than the oleate data set.
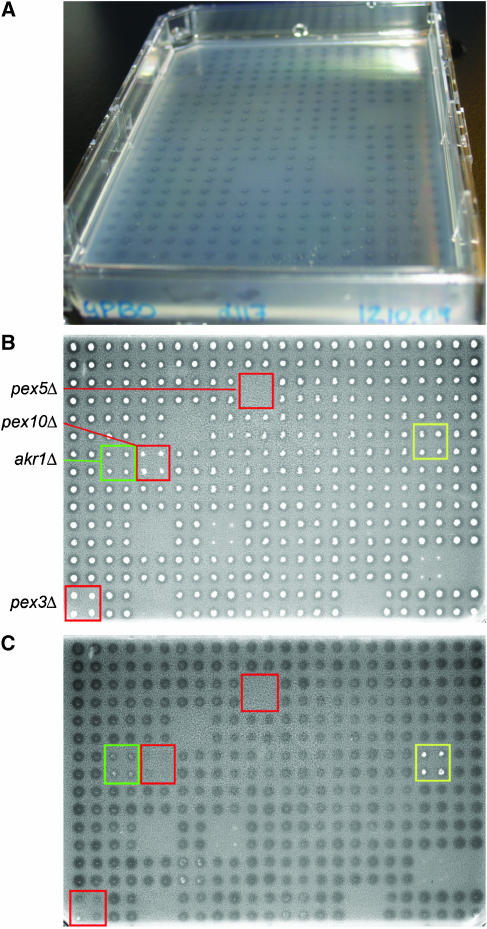
Figure 1.
Clear zone assays. (A) Digital photograph of clear zones on an oleate plate after cell material was removed from the agar surface by rinsing with running water. (B, C) Greyscale images of myristate plate number 128 scanned on a flatbed scanner with transmitted light before and after removal of cell material, respectively. The locations of previously characterized and novel mutants of fatty acid metabolism are boxed in red and green, respectively. A strain resistant to cell removal is boxed in yellow.
The deletion set was thus assayed for clear zone formation in the presence of oleate or myristate. Strains were also assayed for growth on acetate-containing plates, as peroxisomal fatty acid β-oxidation demands a functional mitochondrial electron transport chain for energy production, and thus this condition serves as a control for the ability of cells to metabolize nonfermentable carbon sources. Growth and clear zone data for the entire deletion set are reported in Supplementary Table 1. A sample myristate plate is shown before and after removal of cell material (Figure 1B and C, respectively). Three deletion strains previously known to have defects in peroxisome biogenesis (pex5Δ, pex10Δ and pex3Δ) have defects in clear zone formation and are boxed in red. It should be noted that pex5Δ cells were unable to grow on acetate medium. akr1Δ, a deletion strain not previously implicated in fatty acid metabolism, but with a defect in clear zone formation, is boxed in green.
A total of 212 strains had fatty acid-specific fitness defects (i.e. wild-type growth on acetate and defective clear zones on oleate or myristate), 31 of which have previously been implicated in fatty acid metabolism or peroxisome biology (Table I). Out of the 212 strains, 203 were identified by the myristate screen, whereas 103 were identified by the oleate screen. This was not surprising considering the poor visualization of clear zones and the conservative nature of the analysis using oleic acid as the carbon source. Despite their differences in size, 91% of the oleate data set was included in the myristate data set, suggesting that the requirements for metabolizing myristate and oleate are similar.
Table 1.
List of genes of the fitness data set
| Gene identified by both screens | Gene identified by one screen | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Ole | Myr | a | Gene | Ole | Myr | a | Gene | Ole | Myr | a | Gene | Ole | Myr | a | Gene | Ole | Myr | a |
| ACS1 | 2 | 2 | PEX14 | 1 | 1 | ADE2 | 3 | 2 | MLS1 | 3 | 1 | YIP4 | 3 | 2 | |||||
| ADO1 | 1 | 2 | PEX15 | 1 | 1 | ADY3 | 3 | 2 | a | MRS3 | 3 | 4 | YMR31 | 3 | 2 | ||||
| ADR1 | 1 | 2 | PEX17 | 1 | 1 | ALF1 | 2 | 3 | NGG1 | 3 | 1 | YPT11 | 3 | 2 | |||||
| AKR1 | 1 | 1 | a | PEX18 | 2 | 2 | ARF1 | 3 | 2 | a | NOP6 | 3 | 2 | a | ZWF1 | 3 | 2 | ||
| ARP1 | 2 | 2 | PEX19 | 1 | 2 | ARN1 | 3 | 2 | NPL6 | 3 | 2 | YBL012C | 3 | 2 | |||||
| ASR1 | 2 | 2 | PEX3 | 1 | 1 | ARP5 | 3 | 2 | PAC10 | 3 | 2 | a | YDR049W | 3 | 1 | a | |||
| BCK1 | 1 | 2 | a | PEX4 | 1 | 1 | ATG12 | 2 | 3 | PAP2 | 3 | 2 | a | YDR290W | 3 | 2 | |||
| BOR1 | 2 | 2 | PEX6 | 2 | 2 | AVL9 | 3 | 2 | PCL1 | 3 | 2 | YDR442W | 3 | 2 | |||||
| BPT1 | 2 | 2 | PEX7 | 1 | 2 | a | BDF1 | 3 | 2 | PEX22 | 3 | 1 | YDR532C | 3 | 2 | ||||
| CBS1 | 1 | 1 | PEX8 | 1 | 1 | BLM3 | 3 | 1 | a | PIM1 | 3 | 2 | a | YHL044W | 3 | 2 | a | ||
| CDC10 | 1 | 1 | PIP2 | 1 | 1 | BUB3 | 3 | 2 | POP2 | 3 | 2 | a | YIL029C | 3 | 4 | ||||
| CKB2 | 2 | 1 | PMP3 | 2 | 2 | a | CAP1 | 3 | 2 | a | PPR1 | 3 | 2 | YIL165C | 3 | 4 | |||
| CLA4 | 2 | 2 | POT1 | 1 | 2 | CBC2 | 3 | 2 | a | PRE9 | 3 | 1 | a | YJL149W | 3 | 2 | |||
| DID2 | 1 | 1 | POX1 | 1 | 1 | CBF1 | 3 | 2 | PTK2 | 3 | 2 | YJL160C | 3 | 2 | a | ||||
| DJP1 | 2 | 2 | PPE1 | 2 | 1 | CCH1 | 3 | 2 | PXA1 | 2 | 3 | YJL175W | 3 | 2 | |||||
| DYN2 | 1 | 1 | PUF6 | 2 | 2 | CCZ1 | 3 | 2 | PXA2 | 2 | 3 | YKR035C | 3 | 2 | |||||
| ELP3 | 2 | 1 | PUT3 | 2 | 2 | CIN8 | 3 | 2 | RCY1 | 3 | 2 | YLL020C | 2 | 3 | |||||
| FAB1 | 1 | 2 | RAM1 | 2 | 2 | CLG1 | 3 | 2 | RPL13B | 3 | 2 | YLR065C | 3 | 2 | |||||
| FAT1 | 2 | 2 | RMD1 | 2 | 2 | CSM1 | 3 | 2 | RPL2B | 3 | 2 | YLR358C | 3 | 2 | |||||
| FEN1 | 1 | 2 | RPL31A | 1 | 2 | DMA1 | 3 | 4 | RSC1 | 3 | 1 | a | YLR412W | 3 | 2 | a | |||
| FOX2 | 1 | 1 | RPL34B | 2 | 2 | DUN1 | 3 | 2 | a | RTS1 | 3 | 2 | YNL045W | 3 | 2 | ||||
| FYV6 | 1 | 2 | RVS161 | 2 | 1 | a | EAF7 | 5 | 2 | RTT106 | 3 | 2 | YNL305C | 3 | 2 | ||||
| HNT3 | 2 | 2 | SAC7 | 1 | 2 | EAP1 | 3 | 2 | SAK1 | 3 | 2 | YOR246C | 1 | 3 | |||||
| KEX1 | 2 | 2 | SAP190 | 2 | 2 | ECI1 | 1 | 3 | SEC22 | 3 | 1 | VPL107W | 3 | 2 | |||||
| KNS1 | 2 | 1 | SGO1 | 1 | 1 | ELF1 | 3 | 1 | a | SEL1 | 3 | 1 | |||||||
| LEA1 | 2 | 2 | SIN3 | 1 | 2 | ELM1 | 3 | 2 | SHE4 | 3 | 2 | ||||||||
| LPE10 | 1 | 1 | SLA2 | 1 | 1 | a | ERG3 | 3 | 2 | SOH1 | 3 | 2 | |||||||
| LSC2 | 1 | 1 | SLT2 | 2 | 2 | a | ERG6 | 2 | 3 | SOK1 | 3 | 2 | |||||||
| LYS4 | 1 | 2 | SLX8 | 2 | 2 | FIG4 | 3 | 2 | STO1 | 3 | 2 | a | |||||||
| MDH2 | 1 | 2 | SMI1 | 2 | 2 | FMP13 | 3 | 2 | TIF3 | 3 | 1 | ||||||||
| MDH3 | 1 | 2 | SNF7 | 1 | 1 | FMT1 | 3 | 2 | TNA1 | 3 | 2 | a | |||||||
| MDM20 | 2 | 2 | SPR6 | 2 | 2 | GUP1 | 3 | 1 | TOF2 | 3 | 2 | ||||||||
| MRS2 | 1 | 1 | SRB2 | 1 | 2 | HDA2 | 3 | 2 | TPM1 | 3 | 2 | ||||||||
| MUB1 | 2 | 2 | TOF1 | 2 | 2 | HIR2 | 5 | 2 | TRM9 | 3 | 1 | ||||||||
| NAT3 | 2 | 2 | a | UBI4 | 2 | 2 | HMF1 | 2 | 3 | UBP6 | 3 | 2 | a | ||||||
| NCA3 | 2 | 1 | UBR2 | 1 | 1 | HNT1 | 3 | 2 | UGA3 | 3 | 2 | ||||||||
| NCS6 | 1 | 2 | a | VPS41 | 1 | 1 | HOT13 | 3 | 2 | a | UPF3 | 3 | 2 | ||||||
| NIP100 | 2 | 1 | YPT7 | 2 | 2 | HSP31 | 3 | 2 | a | URA4 | 3 | 2 | |||||||
| NPL4 | 2 | 2 | YER134W | 2 | 1 | a | HTZ1 | 3 | 2 | URM1 | 3 | 2 | a | ||||||
| OAF1 | 1 | 1 | YCL005W | 2 | 1 | ICL1 | 3 | 1 | VAM10 | 3 | 2 | ||||||||
| OPT1 | 1 | 2 | YDR149C | 2 | 2 | IXR1 | 3 | 2 | VAM7 | 3 | 2 | ||||||||
| OSH2 | 2 | 2 | YEL007W | 1 | 2 | KRE6 | 3 | 2 | a | VID21 | 3 | 2 | a | ||||||
| PEX1 | 1 | 1 | YGL152C | 1 | 1 | KTI12 | 3 | 2 | VPS64 | 3 | 2 | ||||||||
| PEX10 | 1 | 1 | YIL102C | 2 | 2 | LDB7 | 3 | 2 | a | VPS65 | 3 | 2 | a | ||||||
| PEX11 | 1 | 2 | YJL211C | 1 | 1 | LSM6 | 3 | 2 | a | VPS68 | 3 | 2 | |||||||
| PEX12 | 1 | 1 | YKL187C | 1 | 2 | LSM7 | 3 | 2 | a | VPS69 | 3 | 2 | a | ||||||
| PEX13 | 1 | 1 | YNL285W | 1 | 2 | MET14 | 3 | 2 | WHI4 | 3 | 2 | a | |||||||
Genes previously implicated in fatty acid metabolism and/or peroxisome biology are underlined. For oleate (Ole) and myristate (Myr) assays, numbers reflect clear zone sizes (4, bigger than wild-type; 3, wild-type; 2, moderately small; 1, very small or not detectable). Those annotated with ‘a' adhered to the agar after removal of cell material.
Intersecting the data sets provides a qualitative measurement of significance, as the confidence is higher for genes identified by both screens rather than by only one. In a few cases, however, the genes are likely to have bona fide membership exclusively in one group. For example, ECI1, a gene necessary for the metabolism of unsaturated but not saturated fatty acids, and PXA1 and PXA2, genes required for long- but not medium-chain fatty acid metabolism (reviewed in Hiltunen et al, 2003), are exclusive to the oleate data set, as expected.
In the analysis of clear zones, we noted that for some strains, cell material remained on the agar surface after rinsing with water (e.g. yellow box in Figure 1B and C). Distribution analysis of strains adhering to myristate plates (Table I and Supplementary Table 1) revealed that adherent strains tend to be those with myristate-specific metabolism defects (>13 × the number expected by chance; binomial distribution probability P-value=1 × 10−35). Agar adhesion is a characteristic of yeast cells that undergo differentiation to a filamentous form consisting of chains of polarized, elongated cells (Gimeno et al, 1992). Although the laboratory strain used here does not undergo the complete differentiation to this form, these data might reflect functional links that have been identified between the dimorphic transition, and peroxisomes and fatty acid metabolism (Prinz et al, 2004; reviewed in Titorenko and Rachubinski, 2004).
Genes of the fitness data set are distinct from those that transcriptionally respond to fatty acids
A comprehensive data set of 202 genes that are transcriptionally responsive to fatty acids was generated using two complementary time-course microarray data sets from the literature measuring the early (up to ∼95 min; Koerkamp et al, 2002) and long-term (up to 26 h; Smith et al, 2002) transcriptional response to oleate (see Materials and methods). In order to compare these genes to those of the fitness data set (representing only nonessential genes), the expression data set was reduced to include only the 172 nonessential genes.
Remarkably, of the 212 genes of the fitness data set, only 14 genes (MDH3, POX1, PEX18, POT1, PXA2, FOX2, ECI1, MLS1, MDH2, ICL1, ACS1, HSP31, YKL187c and PEX11) were also present in the expression data set. These results are consistent with studies demonstrating that only ∼10% of genes predicted to be peroxisome related by transcriptome profiling are necessary for growth on oleic acid (Smith et al, 2002). The results also support evidence in the literature suggesting that genes required for optimal growth in a new environment, including DNA damage (Birrell et al, 2002), high salt, sorbitol, pH 8 and galactose (Giaever et al, 2002), are distinct from those that are transcriptionally responsive to that environment. To understand the functional relevance of this phenomenon, we comparatively analysed the expression and fitness data sets.
Expression and fitness data sets represent different peroxisome-related networks
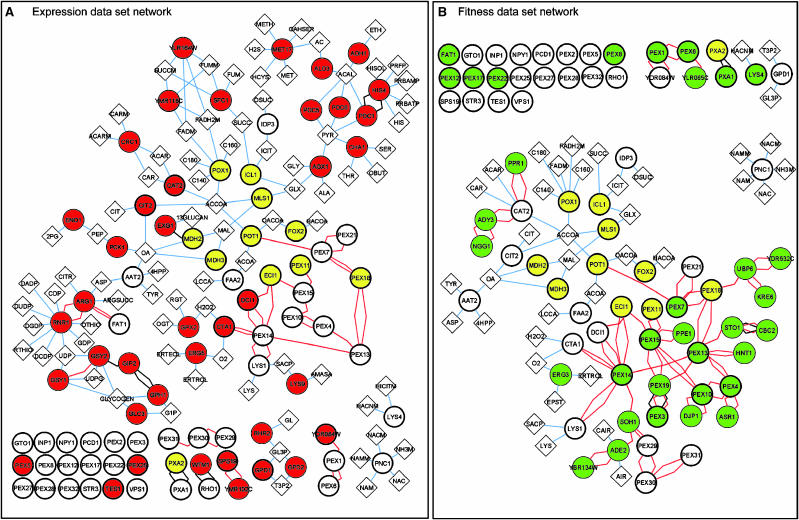
Eleven of the 14 genes common to both data sets encode peroxisomal proteins; this includes Mdh2p (Huh et al, 2003) and Icl1p, the latter of which has been shown to be peroxisomal only in other organisms (Titorenko et al, 1996; Igamberdiev and Lea, 2002). We therefore first analysed peroxisomal proteins of each data set. Cytoscape software (version 2.1; Shannon et al, 2003) (www.cytoscape.org) and large-scale physical interaction data (see Materials and methods) were used to generate and analyse a peroxisome-related physical interaction network for each data set (Figure 2). The networks show known peroxisomal proteins (bold circles) and data set proteins that interact with them (coloured circles). To increase connectivity, metabolites (diamonds) that are either substrates or products of proteins in the network are also included.
Figure 2.
Physical interactions among peroxisomal proteins and proteins of the expression and fitness data sets are shown in (A) and (B), respectively. Red and green circles represent proteins exclusive to the expression and fitness data sets, respectively, and yellow circles represent proteins common to both data sets. Edges representing two-hybrid and affinity precipitation protein–protein interaction data are red and black, respectively. Interactions between proteins and metabolites (diamonds) are shown as blue edges.
Both networks show the 11 peroxisomal proteins found in both data sets (yellow circles), most of which are involved in key processes integral to fatty acid utilization, including fatty acid transport (Pxa2p), peroxisomal β-oxidation (Pot1p, Fox2p, Pox1p, Eci1p and Pex11p) and the glyoxylate cycle (Mls1p, Mdh3p and Icl1p). However, proteins unique to the fitness data set (green circles in Figure 2B) are primarily involved in peroxisome biogenesis (including 14 peroxins encoded by PEX genes, and the chaperone Djp1p), whereas those proteins unique to the expression data set (red circles in Figure 2A) are primarily involved in metabolism (including Cta1p, Dci1p, Cit2p, Cat2p, Gpd1p and Tes1p) and include only two peroxins. This trend is also reflected by the interaction types for each network; 78% (31 of 40) of the proteins unique to the expression data set interact with metabolites, as compared to only 9% (three of 33) of the proteins unique to the fitness data set. Overall, visualization of the networks in this manner reveals that for peroxisome-related proteins, the expression data set primarily represents metabolic processes, whereas the fitness data set mainly represents biogenesis.
The fitness and expression data sets represent dissimilar gene ontologies
To test whether the complementary nature of the functional classes represented by each data set extends beyond peroxisomes, the data sets were compared with respect to their gene ontology (GO) annotation frequencies (see Materials and methods). This analysis showed that not only were the genes of each data set different, but the functional categories they represented were also different.
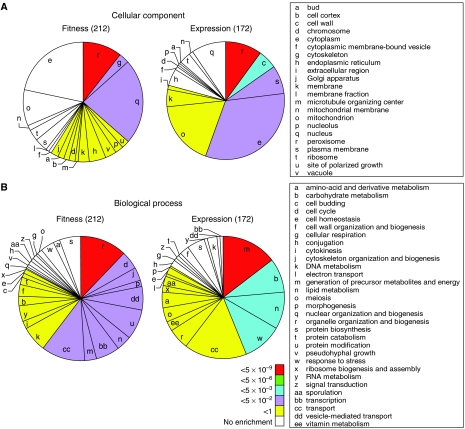
GO slim terms represented in each data set are depicted in pie charts (Figure 3) (see also Supplementary Table 3). Cellular component terms for each data set are presented in Figure 3A. Although ‘peroxisome' (r) was significantly enriched in both data sets as expected, the data sets were otherwise very different. Cytoskeleton (g) and nucleus (q) were most significantly enriched in the fitness data set, compared with cell wall (c), plasma membrane (s) and cytoplasm (e) for the fitness data set. Biological process term enrichments were also very different (Figure 3B). Significantly over-represented terms in the expression data set are related to metabolism (m, b and n) and stress response (w), whereas the enriched terms of the fitness data set relate to the organization and biogenesis of cell structures (r, j, p and dd) and cell regulatory events (bb and u). Molecular function data are not shown, but for this category the expression data set had a significant enrichment of ‘oxidoreductase', whereas the fitness data set was significantly enriched for the terms ‘protein binding' and ‘protein kinase activity'.
Figure 3.
Pie graphs comparing GO slim terms of genes in the fitness and expression data sets. Terms for cellular component (A) and biological process (B) that are represented in the data sets are depicted as slices, the sizes of which are proportional to the term's frequency in the data set. Pies start at 0° with significantly over-represented terms (in a clockwise order of decreasing significance) and end with under-represented terms. Colours represent binomial distribution probability P-values for over-represented terms (see key in panel B), which reflect the statistical significance of enrichments.
This analysis supports and extends the conclusion that the fitness and expression data sets represent different gene classes. The fitness data set enriched for terms related to the organization and dynamics of subcellular structures, including not only peroxisomes, but also (actin) cytoskeleton and vesicle-mediated transport. These terms appear to reflect the dynamic nature of peroxisomes, which may involve their budding from the endoplasmic reticulum and other fission and fusion events (reviewed in Titorenko and Rachubinski, 2001; Tabak et al, 2003). Indeed, the actin cytoskeleton has recently been identified as having a role in peroxisome fission, movement and inheritance (Hoepfner et al, 2001; Marelli et al, 2004; Fagarasanu et al, 2005). The fitness data set was also enriched for regulatory proteins including ubiquitin-related proteins (Ubi4p, Urm1p, Ubr2p and Ubp6p), which might relate to the recently identified roles of ubiquitin in protein import into peroxisomes (Purdue and Lazarow, 2001; Kiel et al, 2005; Kragt et al, 2005).
In contrast to the fitness data set, the expression data set reflects a reallocation of resources to fit the new metabolic state of the cell (enrichment of precursors of metabolism and energy, and carbohydrate metabolism) and detoxification (enrichment of oxidoreductases and stress response). The component term enrichments are in agreement with these findings; for example, many of the genes annotated as plasma membrane are related to metabolite transport (including ATO3, PHO89, PMA2, HXT2, PTR2, FRE1 and PNS1) or stress response and detoxification (PDR5, HSP30, ATR1, TPO1, TPO4 and AQR1).
In general, transcriptional control is biased towards genes with metabolic roles
The above analysis suggests that for the cellular response to fatty acids, which involves both morphological and metabolic changes, transcriptional control is biased towards genes with metabolic roles. To test the global applicability of this bias, we estimated the representation of transcriptionally regulated genes in a comprehensive data set of genes whose products interact with metabolites (Prinz et al, 2004), and in the fatty acid-related expression and fitness data sets (see Materials and methods). For this analysis, transcriptionally regulated genes were defined as those occurring as DNA targets at least once in the global genome localization data set (P<0.001; Harbison et al, 2004). This data set contains protein–DNA interactions for virtually all of the yeast DNA-binding transcriptional regulators (203 activators and repressors) under a variety of conditions (rich media, and for 84 regulators, at least one of 12 other environmental conditions not including fatty acid exposure). Interestingly, the expression and metabolic data sets had 56 and 44% more transcriptionally regulated genes than expected by chance (binomial distribution probability P-values of 6.0 × 10−10 and 7.0 × 10−20, respectively); by contrast, the fitness data set did not show any enrichment, suggesting that transcriptional control is biased towards genes with metabolic roles.
Conclusion
Comparative analysis of fitness and expression data sets related to fatty acid exposure identified a relationship between the functional class of a gene and the mechanism of its regulation. The enrichment of proteins related to the dynamics and assembly of cell structures in the fitness and not the expression data set suggests that these processes tend to be regulated post-transcriptionally. Perhaps other regulatory mechanisms such as protein modification are favoured for structural responses because they make feasible the regulation of complex assemblies rather than of individual components, which reduces the biosynthetic cost and increases the speed of the response. The bias of transcriptional regulation towards metabolically related genes suggests that coordination rather than speed is important for metabolic reorganization. Coordinate changes to multiple metabolic pathways might be critical for reducing toxicity and for balancing levels of small molecules and intermediates involved in multiple pathways.
This study also revealed a relationship between the functional class of a gene and its requirement for cell fitness as genes necessary for fatty acid utilization tended to be related to cell structure, but not metabolism. These data do not necessarily suggest that transcriptionally controlled metabolic reorganization is not important for cell fitness; indeed, genes annotated with the biological process ‘transcription' were enriched in the fitness data set. It does suggest, however, that at least with respect to fatty acid metabolism, metabolic networks are more robust and resistant to perturbation, than structural complexes involved in the response.
Although the focus here was on the cellular response to fatty acid exposure, these trends appear to be more widespread. Further global analysis, data integration and assessment of cellular responses to differing environmental cues will help to understand the relationships in more detail.
Materials and methods
Myristate screen and generation of the ‘fitness' data set
The entire matα haploid viable yeast deletion set from S. cerevisiae strain BY4742 (Resgen/Invitrogen) was assayed for the formation of clear zones in turbid medium containing myristate or oleate, and for growth on medium containing acetate. The entire deletion set was pinned in quadruplicate on YEPD agar (1% yeast extract, 2% peptone, 2% glucose, 2% agar) in omnitrays using a 96-floating-pin replicator and colony copier (V and P Scientific, San Diego, CA) resulting in a total of 384 colonies on each plate. Colonies were replicated in triplicate onto acetate, oleate or myristate agar omnitrays. Plates were incubated for 3–4 days at 30°C. Omnitrays contained 40 ml of YPBA agar (0.67% yeast nitrogen base, 0.1% yeast extract, 0.5% potassium phosphate buffer, pH 6.0, 2% agar, 2% acetate), 20 ml of YPBO agar (0.3% yeast extract, 0.5% potassium phosphate buffer, pH 6.0, 0.5% peptone, 0.2% Tween 40, 2% agar, 0.1% oleic acid) or 20 ml of YPBM agar (0.67% yeast nitrogen base, 0.1% yeast extract, 0.5% potassium phosphate buffer, pH 6.0, 2% agar, 0.5% Tween 40, 0.125% myristic acid). For turbid fatty acid media, detergent and fatty acid were mixed together, warmed to 60°C and added to media after autoclaving.
Growth and clear zone formation around cell patches were scored by visual inspection. Growth was scored as 3, 2 or 1 for patches with wild type, moderate or little/no growth, respectively. Clear zone sizes around cell patches were scored as 4 for larger than wild type, 3 for wild type, 2 for less than wild type and 1 for small or not detectable. To facilitate the analysis of clear zones, plates were scored before and after cell material was removed from the agar surface by rinsing the plate under running water. A 600 dpi greyscale image of each myristate plate was generated by placing the plate face up on a flatbed scanner and scanning with transmitted light. Images were saved as jpeg files. Oleate and myristate data were merged. The 212 genes for which the corresponding deletion strain had wild-type growth on acetate and a defective clear zone on oleate and/or myristate make up the ‘fitness' data set.
Generation of the ‘expression data set'
A comprehensive data set of genes transcriptionally responsive to fatty acids was generated by analysing two complementary time-course data sets in the literature (Koerkamp et al, 2002; Smith et al, 2002). For the Smith and Koerkamp data sets, the data analysed were for effectively all the genes in the genome and for the published differentially expressed genes, respectively. For each time-course data set, gene expression profiles comprised of the gene expression ratios (oleate versus reference carbon source) for each time point were created. For each profile, as a statistical measure, we used the integration of the expression ratios over the time course. The significance of differential expression across the time course was calculated by a hypothesis test using an empirical probability density function determined from kernel density estimation (Bowman and Azzalini, 1997). For each gene, the P-values from the two data sets were then combined using Stouffer's Z (Hedges and Olkin, 1985). The combined P-value represents the significance of differential expression in response to fatty acids in both experiments. P-values for all genes are listed in Supplementary Table 2. We selected the 202 genes that had combined P-values <0.025 as genes that have significant differential expression in response to fatty acids. From these, we selected the 172 genes that were among the 4770 nonessential genes analysed in the clear zone assay. These genes make up the expression data set.
Physical interaction data sets
Protein–protein interactions were downloaded from the SGD website on 05/26/05 in Cytoscape format using the Batch Download tool. Metabolites that are substrates or products of proteins in the network were obtained from Prinz et al (2004), which is a modified version of interactions compiled previously (Forster et al, 2003).
Gene Ontology slim term enrichment
GO slim terms for each gene in the yeast genome were downloaded from the Saccharomyces Genome Database (SGD) website (www.yeastgenome.org) on 05/26/05. For each term, the observed frequencies in each data set (expression or fitness) were compared with those expected by chance (the frequency of annotation for the 4770 nonessential genes). For enriched terms, the probability that the observed distribution would be found by chance was determined by calculating the binomial distribution probability using Microsoft Excel and the probability mass function. This algorithm has been used by others to estimate the significance of term enrichments with similar population and sample sizes (Begley et al, 2004).
Analysis of general transcriptional regulation
All yeast genes in SGD were annotated with their respective regulators identified by a previously published large-scale genome-wide location analysis (P-values <0.001; Harbison et al, 2004). Genes annotated with at least one regulator were termed ‘regulated', whereas genes with no annotations were termed ‘non-regulated'. The observed frequency of regulated genes in each data set (expression or fitness) was compared with that expected by chance (the frequency of annotation to the 4770 nonessential genes). For over-represented terms, the significance of enrichment was calculated as outlined for GO slim terms (above). The analysis of the transcriptional regulation of metabolic genes (those that are reported to interact with metabolites; Prinz et al, 2004) was the same except that the expected frequencies were calculated using the frequency of annotation to all yeast genes rather than the nonessential genes.
Supplementary Material
Supplementary Table 1
Supplementary Table 2
Supplementary Table 3
Acknowledgments
We thank Ramsey Saleem and Richard Rogers for their contributions. This work was supported by a grant from the National Institutes of Health to JDA (GM067228) and by a grant from the Canadian Institutes of Health Research (53326) to RAR and JDA. RAR is Canada Research Chair in Cell Biology and an International Research Scholar of the Howard Hughes Medical Institute. JJS is the recipient of a Postdoctoral Fellowship from the CIHR.
References
- Begley TJ, Rosenbach AS, Ideker T, Samson LD (2004) Hot spots for modulating toxicity identified by genomic phenotyping and localization mapping. Mol Cell 16: 117–125 [DOI] [PubMed] [Google Scholar]
- Birrell GW, Brown JA, Wu HI, Giaever G, Chu AM, Davis RW, Brown JM (2002) Transcriptional response of Saccharomyces cerevisiae to DNA-damaging agents does not identify the genes that protect against these agents. Proc Natl Acad Sci USA 99: 8778–8783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman AW, Azzalini A (1997) Applied Smoothing Techniques for Data Analysis. Oxford University Press: Oxford [Google Scholar]
- Fagarasanu M, Fagarasanu A, Tam YY, Aitchison JD, Rachubinski RA (2005) Inp1p is a peroxisomal membrane protein required for peroxisome inheritance in Saccharomyces cerevisiae. J Cell Biol 169: 765–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster J, Famili I, Fu P, Palsson BO, Nielsen J (2003) Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Res 13: 244–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, Arkin AP, Astromoff A, El-Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A, Entian KD, Flaherty P, Foury F, Garfinkel DJ, Gerstein M, Gotte D, Guldener U, Hegemann JH, Hempel S, Herman Z, Jaramillo DF, Kelly DE, Kelly SL, Kotter P, LaBonte D, Lamb DC, Lan N, Liang H, Liao H, Liu L, Luo C, Lussier M, Mao R, Menard P, Ooi SL, Revuelta JL, Roberts CJ, Rose M, Ross-Macdonald P, Scherens B, Schimmack G, Shafer B, Shoemaker DD, Sookhai-Mahadeo S, Storms RK, Strathern JN, Valle G, Voet M, Volckaert G, Wang CY, Ward TR, Wilhelmy J, Winzeler EA, Yang Y, Yen G, Youngman E, Yu K, Bussey H, Boeke JD, Snyder M, Philippsen P, Davis RW, Johnston M (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391 [DOI] [PubMed] [Google Scholar]
- Gimeno CJ, Ljungdahl PO, Styles CA, Fink GR (1992) Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68: 1077–1090 [DOI] [PubMed] [Google Scholar]
- Gurvitz A, Rottensteiner H, Kilpelainen SH, Hartig A, Hiltunen JK, Binder M, Dawes IW, Hamilton B (1997) The Saccharomyces cerevisiae peroxisomal 2,4-dienoyl-CoA reductase is encoded by the oleate-inducible gene SPS19. J Biol Chem 272: 22140–22147 [DOI] [PubMed] [Google Scholar]
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, Jennings EG, Zeitlinger J, Pokholok DK, Kellis M, Rolfe PA, Takusagawa KT, Lander ES, Gifford DK, Fraenkel E, Young RA (2004) Transcriptional regulatory code of a eukaryotic genome. Nature 431: 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges L, Olkin I (1985) Statistical Method for Meta-analysis. San Diego, CA: Academic Press [Google Scholar]
- Hiltunen JK, Mursula AM, Rottensteiner H, Wierenga RK, Kastaniotis AJ, Gurvitz A (2003) The biochemistry of peroxisomal beta-oxidation in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev 27: 35–64 [DOI] [PubMed] [Google Scholar]
- Hoepfner D, van den Berg M, Philippsen P, Tabak HF, Hettema EH (2001) A role for Vps1p, actin, and the Myo2p motor in peroxisome abundance and inheritance in Saccharomyces cerevisiae. J Cell Biol 155: 979–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK (2003) Global analysis of protein localization in budding yeast. Nature 425: 686–691 [DOI] [PubMed] [Google Scholar]
- Igamberdiev AU, Lea PJ (2002) The role of peroxisomes in the integration of metabolism and evolutionary diversity of photosynthetic organisms. Phytochemistry 60: 651–674 [DOI] [PubMed] [Google Scholar]
- Kiel JA, Emmrich K, Meyer HE, Kunau WH (2005) Ubiquitination of the peroxisomal targeting signal type 1 receptor, Pex5p, suggests the presence of a quality control mechanism during peroxisomal matrix protein import. J Biol Chem 280: 1921–1930 [DOI] [PubMed] [Google Scholar]
- Koerkamp MG, Rep M, Bussemaker HJ, Hardy GP, Mul A, Piekarska K, Szigyarto CA, De Mattos JM, Tabak HF (2002) Dissection of transient oxidative stress response in Saccharomyces cerevisiae by using DNA microarrays. Mol Biol Cell 13: 2783–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kragt A, Voorn-Brouwer T, van den Berg M, Distel B (2005) The Saccharomyces cerevisiae peroxisomal import receptor Pex5p is monoubiquitinated in wild type cells. J Biol Chem 280: 7867–7874 [DOI] [PubMed] [Google Scholar]
- Marelli M, Smith JJ, Jung S, Yi E, Nesvizhskii AI, Christmas RH, Saleem RA, Tam YY, Fagarasanu A, Goodlett DR, Aebersold R, Rachubinski RA, Aitchison JD (2004) Quantitative mass spectrometry reveals a role for the GTPase Rho1p in actin organization on the peroxisome membrane. J Cell Biol 167: 1099–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz S, Avila-Campillo I, Aldridge C, Srinivasan A, Dimitrov K, Siegel AF, Galitski T (2004) Control of yeast filamentous-form growth by modules in an integrated molecular network. Genome Res 14: 380–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdue PE, Lazarow PB (2001) Pex18p is constitutively degraded during peroxisome biogenesis. J Biol Chem 276: 47684–47689 [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Marelli M, Christmas RH, Vizeacoumar FJ, Dilworth DJ, Ideker T, Galitski T, Dimitrov K, Rachubinski RA, Aitchison JD (2002) Transcriptome profiling to identify genes involved in peroxisome assembly and function. J Cell Biol 158: 259–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabak HF, Murk JL, Braakman I, Geuze HJ (2003) Peroxisomes start their life in the endoplasmic reticulum. Traffic 4: 512–518 [DOI] [PubMed] [Google Scholar]
- Titorenko VI, Eitzen GA, Rachubinski RA (1996) Mutations in the PAY5 gene of the yeast Yarrowia lipolytica cause the accumulation of multiple subpopulations of peroxisomes. J Biol Chem 271: 20307–20314 [DOI] [PubMed] [Google Scholar]
- Titorenko VI, Rachubinski RA (2001) The life cycle of the peroxisome. Nat Rev Mol Cell Biol 2: 357–368 [DOI] [PubMed] [Google Scholar]
- Titorenko VI, Rachubinski RA (2004) The peroxisome: orchestrating important developmental decisions from inside the cell. J Cell Biol 164: 641–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1
Supplementary Table 2
Supplementary Table 3