Abstract
Cys2His2 zinc-fingers (C2H2 ZFs) mediate a wide variety of protein–DNA and protein–protein interactions. DNA-binding C2H2 ZFs can be shuffled to yield artificial proteins with different DNA-binding specificities. Here we demonstrate that shuffling of C2H2 ZFs from transcription factor dimerization zinc-finger (DZF) domains can also yield two-finger DZFs with novel protein–protein interaction specificities. We show that these synthetic protein–protein interaction domains can be used to mediate activation of a single-copy reporter gene in bacterial cells and of an endogenous gene in human cells. In addition, the synthetic two-finger domains we constructed can also be linked together to create more extended, four-finger interfaces. Our results demonstrate that shuffling of C2H2 ZFs can yield artificial protein-interaction components that should be useful for applications in synthetic biology.
Keywords: Cys2His2 zinc-finger, domain shuffling, protein engineering, protein–protein interactions
Introduction
Construction of complex synthetic cellular networks will require access to large sets of macromolecular components such as DNA-binding proteins and protein interaction domains. Cys2His2 zinc-fingers (C2H2 ZFs) provide an important framework for the design of synthetic proteins with novel interaction specificities. C2H2 ZFs are compact molecular recognition domains found in 2–3% of all human genes (Lander et al, 2001; Tupler et al, 2001; Venter et al, 2001; Muller et al, 2002). These domains consist of a short β-sheet and an α-helix (whose overall fold is stabilized by coordination of a zinc atom by conserved cysteines and histidines) and are typically found in proteins as tandem arrays. C2H2 ZFs mediate specific recognition of different DNA (Wolfe et al, 2000), RNA (Lu et al, 2003), and protein sequences (Mackay and Crossley, 1998). The functional versatility and widespread prevalence of these domains demonstrate that evolution has extensively utilized the C2H2 ZF fold to mediate interactions with a variety of different macromolecules.
A number of studies have shown that naturally occurring and engineered DNA-binding C2H2 ZFs can be ‘mixed and matched' to create synthetic multifinger arrays possessing novel DNA-binding specificities (Klug, 1999; Pabo et al, 2001; Falke and Juliano, 2003; Jamieson et al, 2003; Lee et al, 2003; Blancafort et al, 2004; Jantz et al, 2004). These synthetic DNA-binding domains (DBDs) can be fused to transcriptional regulatory domains to create artificial transcription factors capable of altering expression of specific endogenous genes in cell types ranging from yeast to human as well as in whole organisms (Klug, 1999; Pabo et al, 2001; Falke and Juliano, 2003; Jamieson et al, 2003; Lee et al, 2003; Blancafort et al, 2004; Jantz et al, 2004). In addition, recent work has demonstrated that artificial C2H2 ZFs can be used to construct synthetic two-dimensional gene networks that produce different patterns of gene expression in a cell-free environment (Isalan et al, 2005). Engineered C2H2 ZFs will undoubtedly play a central role in synthetic biology efforts and therefore improving our capabilities to engineer the interaction specificities of these domains is an important goal for future studies.
However, in contrast to our current detailed understanding of DNA-binding C2H2 ZFs, relatively little is understood at a physical–chemical level about protein-interacting C2H2 ZFs (Mackay and Crossley, 1998; McCarty et al, 2003; Westman et al, 2004). Our limited understanding of protein-interacting DZFs also contrasts with more detailed knowledge about dimerization and multimerization by leucine zippers, a widely distributed family of motifs found in eukaryotic transcription factors (Newman and Keating, 2003). Leucine zippers can interact as parallel or antiparallel coiled-coils, can mediate homo- and/or hetero-typic interactions, and can interact as dimers, trimers, or higher order oligomers (Vinson et al, 2002). These detailed insights have come from studies of naturally occurring motifs (O'Shea et al, 1989; O'Shea et al, 1991) but also from selection and design efforts that have successfully engineered artificial leucine zippers with novel interaction specificities, geometries, and oligomerization states (Hu et al, 1990; Harbury et al, 1993; Hu et al, 1993; O'Shea et al, 1993; Vinson et al, 1993; Harbury et al, 1994; Zeng et al, 1997; Moll et al, 2001).
We wished to determine whether protein-interacting C2H2 ZFs might be shuffled to engineer a repertoire of finger arrays with novel protein–protein interaction specificities. We chose to focus our initial efforts on C2H2 ZFs from dimerization zinc-finger (DZF) domains found in various transcription factors including members of the mammalian Ikaros family of transcription factors, the Drosophila melanogaster Hunchback protein, and the human TRPS-1 protein (Hahm et al, 1994; Sun et al, 1996; Morgan et al, 1997; Hahm et al, 1998; Perdomo et al, 2000; McCarty et al, 2003; Westman et al, 2003). DZFs consist of two C2H2 ZFs joined by a short linker (Figure 1) and are sufficient for mediating homo- and heterotypic interactions among these various transcription factors. Previous studies have shown that both C2H2 ZFs in a DZF are required for efficient interaction, suggesting that both fingers contribute binding energy (Sun et al, 1996; McCarty et al, 2003; Westman et al, 2004). We were particularly interested in focusing on DZFs because a recent report described a functional hybrid DZF constructed from portions of the human Ikaros and Drosophila Hunchback DZFs, a result which strongly suggested that shuffling of DZF-derived C2H2 ZF domains might yield synthetic DZFs with novel protein–protein interaction specificities (McCarty et al, 2003).
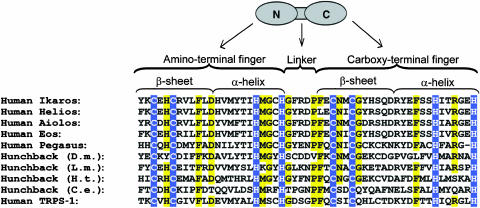
Figure 1.
DZF domains from various transcription factors. The top of the figure presents a schematic of a DZF domain with amino- (N) and carboxy- (C) terminal fingers represented as ovals and the interfinger linker as a connecting bar. The bottom portion of the figure shows an amino-acid sequence alignment of DZFs from human Ikaros, human Helios, human Aiolos, human Eos, human Pegasus, human TRPS-1, Drosophila melanogaster (D.m.) Hunchback, Locusta migratoria (L.m.) Hunchback, Helobdella triserialis (H.t.) Hunchback, and Caenorhabditis elegans (C.e.) Hunchback. Conserved cysteines and histidines are highlighted in blue. Positions showing 80% or greater conservation among the 10 DZF domains shown are highlighted in yellow.
An additional motivation for our studies was to gain greater insight into the interaction geometry of DZF-mediated interactions. The single synthetic hybrid Ikaros–Hunchback DZF described above exhibited a novel homotypic interaction specificity, which suggested that this DZF interacted in a ‘parallel' interaction mode with each of its C2H2 ZFs interacting with its counterpart in the opposing monomer (McCarty et al, 2003). We reasoned that creation of more synthetic DZFs by domain shuffling would provide an additional test of the idea that DZFs interact in a parallel manner.
In this report, we demonstrate that shuffling protein-interacting C2H2 ZFs from DZF domains can yield finger arrays with novel protein–protein interaction specificities and that these DZFs can be used to build synthetic transcription factors capable of altering gene expression in human and bacterial cells. To do this, we created libraries of two-finger units by shuffling C2H2 ZFs from DZFs found in transcription factors ranging from D. melanogaster to humans and then identified pairs of interacting two-finger domains using a bacterial two-hybrid (B2H) system (Dove et al, 1997; Dove and Hochschild, 1998; Joung et al, 2000). We show that the synthetic two-finger DZFs we identified can be used to construct a series of artificial transcriptional activators in Escherichia coli or to construct artificial transcriptional activators of the endogenous VEGF-A gene in human cells (by-passing normal physiologic mechanisms for regulating this gene). In addition, we show that our synthetic two-finger domains can be linked together to create more extended four-finger protein–protein interaction interfaces. Surprisingly, analysis of the interaction specificities of our synthetic domains suggests that DZFs can also interact in an antiparallel mode in addition to the parallel mode described previously (McCarty et al, 2003). Our findings have implications for understanding naturally occurring C2H2 ZF-mediated protein–protein interactions and expand the number and kind of protein–protein interaction ‘parts' potentially available to synthetic biologists for building or modifying cellular networks.
Results
Detecting and identifying DZF–DZF interactions using a B2H system
We reasoned that we could create libraries of synthetic DZFs with potentially altered interaction specificities by shuffling C2H2 ZFs from various wild-type DZFs. We also hypothesized that interacting pairs of synthetic DZFs from these shuffled libraries could be identified using a B2H selection system designed to detect protein–protein interactions (Joung, 2001; Dove and Hochschild, 2004). Therefore, we first sought to determine whether the B2H system could be used to detect interactions between DZF domains.
The B2H system is based on the observation that in an appropriately engineered E. coli strain, the interaction of two arbitrary protein domains (X and Y) can trigger transcriptional activation of a linked reporter gene(s) (e.g., the lacZ gene encoding β-galactosidase or a selectable marker gene; Dove et al, 1997; Dove and Hochschild, 1998; Figure 2A). This activation occurs when a hybrid protein consisting of a DBD (e.g., the DBD of the Zif268 transcription factor) fused to protein domain X binds upstream of a weak test promoter and recruits RNA polymerase (RNAP) complexes that have incorporated a second hybrid protein consisting of protein domain Y fused to the RNAP α-subunit (Figure 2A). In this configuration, the interaction of protein domains X and Y leads to an increase in reporter gene expression.
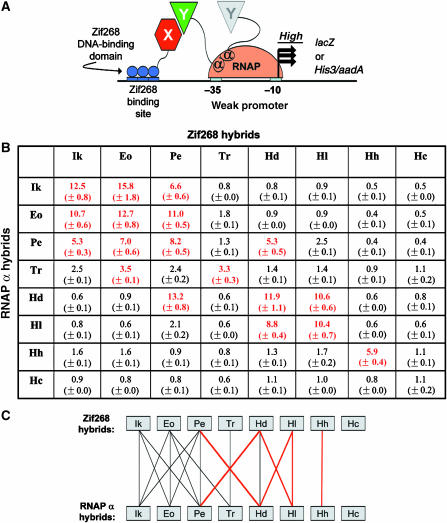
Figure 2.
Dimerization specificities of wild-type DZFs determined using a B2H system. (A) Schematic of B2H system used to study DZF interactions. Interaction of arbitrary protein domains X and Y fused to the Zif268 DBD and an E. coli RNAP α-subunit fragment, respectively, mediates recruitment of RNAP to a weak promoter bearing a Zif268 binding site. RNAP recruitment leads to increased transcription of an associated reporter gene (lacZ) or co-cistronic selectable marker genes (His3 and aadA). In the experiments of this report, domains X and Y are DZFs. Note that two α-subunits are present in an RNAP complex. (B) Pairwise combinations of eight wild-type DZF domains tested in the B2H system. Combinations of plasmids encoding wild-type DZFs were cotransformed into a B2H reporter strain harboring the lacZ gene and reporter gene expression was measured by β-galactosidase assay. Each combination was tested in triplicate with mean fold-activation values and standard errors of the means shown. Values highlighted in bold red text indicate positive interactions defined as >2.5-fold activation of LacZ expression. This cutoff was chosen based on the observation that the highest fold-activation observed in this assay for a noninteracting DZF pair (Ikaros and TRPS-1; McCarty et al, 2003) is 2.5. Ik=human Ikaros; Eo=human Eos; Pe=human Pegasus; Tr=human TRPS-1; Hd=Drosophila melanogaster Hunchback; Hl=Locusta migratoria Hunchback; Hh=Helobdella triserialis Hunchback; and Hc=Caenorhabditis elegans Hunchback. (C) Summary of DZF interaction specificity profiles determined using the B2H system. Lines indicate interactions detected among 64 pairwise combinations of eight wild-type DZFs (experiments of Figure 2B). Black lines indicate interactions that have also been detected by other methods and red lines indicate novel interactions that have not been described previously.
To test whether the B2H system could be used to detect interactions between DZFs, we transformed combinations of plasmids encoding various DZF-Zif268 DBD fusions and DZF-RNAP α-subunit fusions into a ‘B2H reporter strain' (in which the reporter gene is the E. coli lacZ gene encoding β-galactosidase) and then performed β-galactosidase assays. Using this approach, we tested all 64 possible pairwise combinations of eight DZFs from the human Ikaros, human Eos, human Pegasus, human TRPS-1, D. melanogaster Hunchback, and Hunchback homologs from grasshopper, leech, and worms (identified using database searches; Figure 1). Our results (Figure 2B) confirm DZF interactions previously detected by biochemical, immunoprecipitation, or yeast two-hybrid experiments (black lines, Figure 2C). Interactions were detected in both orientations with the exception of the TRPS-1 DZF and Eos DZF interaction, which was only detected in one orientation. Interestingly, our experiments also reveal new homo- and heterotypic DZF domain interactions that have not been previously tested and/or described (red lines, Figure 2C). (We do not believe that these interactions represent false-positives of the B2H method because they activate transcription of the reporter gene even if their locations in the Zif268 and RNAP α-subunit hybrid proteins are reversed and because noninteracting DZFs (e.g., from Ikaros and Drosophila Hunchback) fail to activate transcription of the reporter gene.) We conclude that the B2H system can be used as a rapid and reliable method to identify and test DZF–DZF interactions.
Construction of ‘shuffled' C2H2 ZF libraries
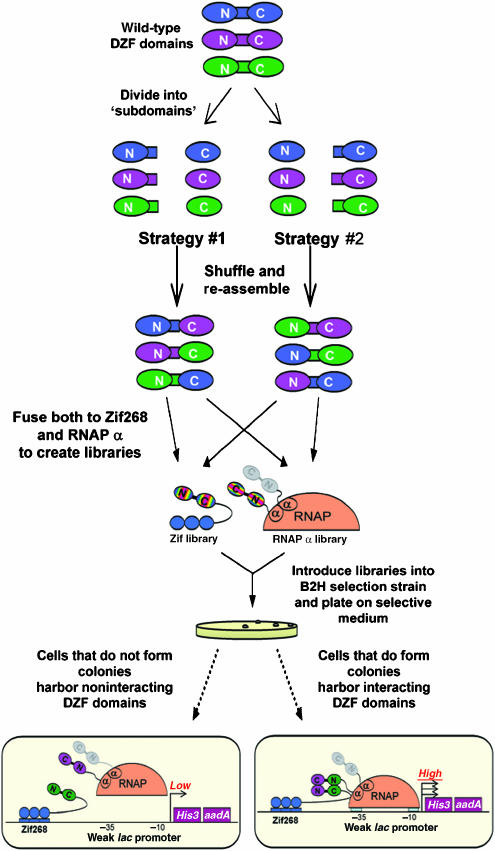
We used two different shuffling approaches to construct plasmid DNA-based libraries encoding synthetic DZFs consisting of shuffled combinations of C2H2 ZFs (Figure 3; also see Materials and methods). In one approach, the interfinger linker remained associated with the amino-terminal finger and in the other the linker remained associated with the carboxy-terminal finger. Our library construction strategies deliberately precluded re-formation of DNA molecules encoding the original wild-type DZFs. We created three different ‘sets' of shuffled DZF libraries, each derived from a different combinatorial subset of eight wild-type DZFs (details of theoretical and actual library sizes are provided in Materials and methods and Supplementary Table 2). As illustrated in Figure 3 (and described in Materials and methods), each shuffled ‘set' consists of four libraries: two with the synthetic DZFs expressed as fusions to the Zif268 DBD and two expressed as fusions to the RNAP α-subunit.
Figure 3.
Schematic overview of shuffled DZF library construction and B2H selections to identify interacting synthetic DZFs. DZFs are represented as in Figure 1. Note that although two α-subunits are present in an RNAP complex, the DZF–DZF interaction illustrated can occur presumably because either the DZF domains on the RNAP α-fusion protein fail to dimerize (perhaps owing to a geometrical constraint) or because the RNAP complexes that are recruited to the promoter harbor one DZF domain α hybrid subunit and one wild-type α-subunit. Additional details are provided in the Results and Materials and methods section.
Interacting synthetic DZF pairs identified using the B2H system
To identify interacting synthetic DZF pairs, we introduced pairwise combinations of our various shuffled DZF-encoding plasmid libraries into a ‘B2H selection strain' harboring the selectable, cocistronic His3 and aadA genes as reporters (Joung et al, 2000; Hurt et al, 2003). B2H selection strain cells expressing DZF-Zif268 and DZF-RNAP α-subunit hybrid proteins that interact with each other should activate transcription of the His3 and aadA genes, thereby permitting survival and colony formation on appropriate selective media (see details in Materials and methods). Sufficient numbers of transformed B2H selection strain cells were plated to ensure at least 1000-fold oversampling of the theoretical number of DZF combinations for each selection performed (see Materials and methods and Supplementary Table 2 for additional details). DZF-encoding plasmids were isolated from surviving colonies and sequenced to determine the identities of C2H2 ZFs and linkers present in the synthetic selected DZFs. We believe that for all selections performed, we identified nearly all possible interacting pairs as we frequently obtained multiple isolates containing the same pair of DZFs (Table I).
Table 1.
Interacting pairs of synthetic shuffled DZF domains identified by genetic selection
| Zif268 hybrids | RNAP α hybrids | Frequency |
|---|---|---|
| Library set A | ||
| Ik-Eo-Eo | Eo-Ik-Ik | 2 |
| Ik-Eo-Eo | Eo-Eo-Ik | 1 |
| Ik-Eo-Eo | Ik-Eo-Eo | 4 |
| Ik-Eo-Eo | Ik-Ik-Eo | 1 |
| Ik-Ik-Eo | Eo-Ik-Ik | 2 |
| Ik-Ik-Eo | Eo-Eo-Ik | 3 |
| Ik-Ik-Eo | Ik-Ik-Eo | 1 |
| Ik-Ik-Eo | Ik-Eo-Eo | 1 |
| Ik-Ik-Eo | Pe-Eo-Eo | 2 |
| Ik-Ik-Eo | Pe-Ik-Ik | 1 |
| Ik-Eo-Eo | Pe-Eo-Eo | 1 |
| Ik-Eo-Eo | Eo-Pe-Pe | 1 |
| Pe-Eo-Eo | Ik-Ik-Eo | 3 |
| Pe-Pe-Hd | Hd-Hd-Pe | 4 |
| Pe-Hd-Hd | Hl-Eo-Eo | 1 |
| Ik-Ik-Hd | Pe-Pe-Eo | 1 |
| Pe-Pe-Eo | Eo-Eo-Hd | 2 |
| Ik-Ik-Hd | Pe-Pe-Eo | 1 |
| Library set B | ||
| Hl-Hd-Hd | Hl-Hd-Hd | 2 |
| Hd-Hl-Hl | Hd-Hl-Hl | 1 |
| Pe-Hd-Hd | Hl-Hd-Hd | 2 |
| Pe-Pe-Hd | Hl-Hl-Hd | 2 |
| Pe-Pe-Hd | Hd-Pe-Pe | 4 |
| Pe-Hd-Hd | Hd-Hd-Hl | 1 |
| Pe-Pe-Hd | Hd-Hd-Pe | 6 |
| Library set C | ||
| Tr-Eo-Eo | Eo-Eo-Hd | 3 |
| Identities of the amino-terminal C2H2 ZF, the interfinger linker, and the carboxy-terminal C2H2 ZF (shown from left to right) for each of the synthetic DZFs identified from the three different selection ‘sets' A, B, and C are shown. Abbreviations describing the parental source of fingers and linkers are as defined in the legend to Figure 2. Location of the synthetic DZF on the Zif268 or RNAP α-subunit and the number of times each pair of DZFs were isolated are shown. Four DZF pairs chosen for additional analysis are in bold. | ||
As described in Materials and methods, our initial shuffled library sets were constructed using DZFs from both the Ikaros and Hunchback families (Library sets A and B). As shown in Table I, many of the DZFs we obtained from these initial selections were composed of C2H2 ZFs from wild-type DZFs known to interact with each other (e.g., the Ik-Eo-Eo and Eo-Ik-Ik DZF pair at the top of Table I (see legend for abbreviations)). However, in three of the synthetic DZF pairs we identified (bold letters, Table I), at least one DZF in the pair was composed of C2H2 ZFs from wild-type DZFs that do not interact with each other (as defined in the experiment of Figure 2B above). Reasoning that synthetic DZFs from this latter category would be more likely to possess novel specificities, we constructed an additional shuffled library (Library C) derived from DZFs that, with one exception (TRPS-1 and Eos, which interact weakly in the B2H), do not interact with each other. Our selections with Library set C yielded only a single interacting DZF pair (Table I). The four pairs of DZFs in bold in Table I were chosen for further characterization. (Note that these four pairs consist of a total of six unique DZFs because the Pe-Pe-Eo and Eo-Eo-Hd DZFs were each identified twice with different partners.)
Selected synthetic DZFs possess novel interaction specificities
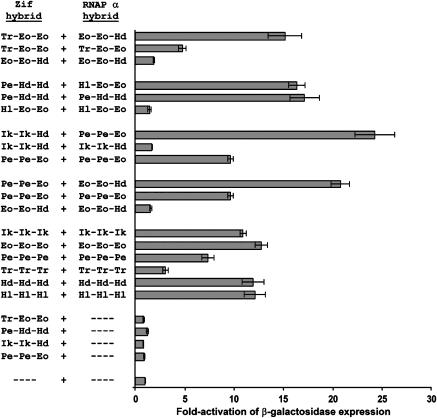
We examined the abilities of the synthetic DZFs we selected to mediate protein–protein interactions using ‘B2H reporter strains' harboring lacZ as the reporter gene. For each of the four DZF pairs examined, we assessed the ability of each DZF in the pair to interact with its selected partner (heterotypic interaction) and with itself (homotypic interaction). The results of these assays (together with control experiments, which demonstrate that expression of only the Zif268-DZF fusion protein alone fails to activate lacZ expression) are shown in Figure 4. In these experiments, a lack of reporter gene activity can be interpreted as the absence of a productive DZF–DZF interaction because all DZF fusion proteins used in this experiment are known to be stably expressed and active in the B2H system as judged by their abilities to interact with at least one other DZF fusion protein (Figure 4 and data not shown). Interestingly, our results suggest that three of the four DZF pairs we tested appear to mediate preferentially heterotypic interactions. The preferentially heterotypic interaction specificities of these three selected DZF pairs differs from all wild-type DZFs tested to date, which possess either exclusively homotypic or a combination of homo- and heterotypic interaction specificities. The fourth pair consists of one DZF (Pe-Hd-Hd) that mediates homotypic and heterotypic interactions with equal efficiency and another DZF (Hl-Eo-Eo) that mediates only heterotypic interaction. The ability of the Pe-Hd-Hd to self-interact is not entirely surprising as its parental DZFs (Pegasus and Drosophila Hunchback) interact with each other (Figure 2B).
Figure 4.
Homo- and heterotypic interaction specificities of synthetic DZFs assessed in the B2H system. DZFs from four pairs were tested for hetero- and homotypic interaction using the B2H system. Controls testing the self-interaction of wild-type DZF domains (from which C2H2 ZFs present in the synthetic DZFs were derived) were also performed. ‘- - - -' indicates a plasmid encoding either the Zif268 DBD or the RNAP α-subunit with no DZF fused. Each experiment was performed at least three times. Bars shown represent mean fold-activations of transcription in the B2H system and error bars represent standard errors of the mean. Identities of the synthetic DZFs tested are abbreviated as in Table I.
For certain applications in synthetic biology, one may wish to simultaneously express multiple pairs of DZFs in a single cell. For these applications, minimal crossreactivity between DZFs in different pairs will be critical. Ideally, each synthetic DZF should preferentially interact with its intended partner and not with any other synthetic DZFs expressed in the cell. Therefore, we tested for potential crossinteractions among the six different synthetic DZFs we selected using the B2H system. The results of these experiments (shown in Supplementary Table 1) demonstrate that five of the six synthetic DZFs show excellent specificity—they interact most strongly with the partner DZF(s) they were selected together with. One synthetic DZF (Pe-Hd-Hd) interacts most strongly both with the partner DZF it was selected with (Hl-Eo-Eo) and with itself, a finding consistent with the results of Figure 4.
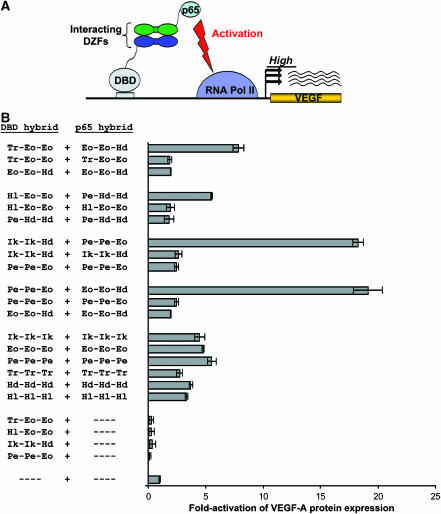
Synthetic DZFs can mediate protein–protein interactions in the nucleus of a mammalian cell
To obtain additional evidence that our synthetic DZFs mediate specific protein–protein interactions and to test whether they could be used to create synthetic gene regulatory networks, we constructed artificial transcription factors using our DZF pairs and tested their activities in the nucleus of human cells. To do this, we adapted a previously described ‘activator reconstitution' assay. In this assay (Pollock et al, 2002), an artificial, bi-partite transcriptional activator of the endogenous human VEGF-A gene can be created by constructing two interacting hybrid proteins (Figure 5A): a synthetic DBD that binds to a region of open chromatin in the human VEGF-A gene (originally termed ‘VZ+434b') (Liu et al, 2001) and a NF-κB p65 transcriptional activation domain (termed ‘p65'). Thus, to test the interaction of any given DZF pair, we fused individual DZFs to each of the two ‘halves' of the bi-partite artificial activator and then tested the ability of these DZFs to mediate transcriptional activation of the endogenous VEGF-A gene. To do this, we transiently transfected human 293 cells, which express low levels of VEGF-A, with plasmids that express various DZF–DBD and DZF–p65 hybrid proteins (each harboring a nuclear localization signal tag) and then measured levels of VEGF-A expression by ELISA.
Figure 5.
Synthetic DZFs can mediate assembly of an artificial bi-partite transcription factor and activation of the endogenous VEGF-A gene in human cells. (A) Schematic overview of a mammalian cell-based ‘activator reconstitution' assay for testing DZF interactions. Productive interaction of DZFs (green and dark blue ovals) fused to an NF-κB p65 (p65) transcriptional activation domain and to a synthetic DBD that binds the human VEGF-A gene is expected to mediate reconstitution of a synthetic activator protein capable of stimulating VEGF-A expression. (B) Homo- and heterotypic interaction of synthetic DZFs assessed using the activator reconstitution assay. Controls testing the homotypic interaction of wild-type DZF domains (from which the synthetic DZFs were derived) are shown. ‘- - - -' indicates a plasmid encoding either the DBD or p65 domains with no DZF fused. DZFs tested are represented as in Table I. Fold-stimulation of VEGF-A protein expression was calculated using ELISA measurements of secreted VEGF-A that were performed at least three times. Bars shown represent mean fold-activations of VEGF-A expression and error bars indicate standard errors of the mean.
As expected, control experiments demonstrated that interacting wild-type DZFs can mediate assembly of the bi-partite activator, thereby stimulating VEGF-A expression (Figure 5B). This increase is dependent on the presence of both DZF hybrid proteins (i.e., neither hybrid alone can activate VEGF-A expression; data not shown). In addition, no significant increase in VEGF-A expression is seen when noninteracting DZFs were present in the hybrid proteins (data not shown).
Having established that DZF interactions can function to mediate specific assembly of an artificial bi-partite activator, we next assessed whether our four synthetic DZF pairs could stimulate VEGF-A in the activator reconstitution assay. As shown in Figure 5B, our results demonstrate that all four synthetic DZF pairs can mediate efficient stimulation of VEGF-A expression in this assay. Control experiments show that the expression of only the DZF–DBD hybrid protein alone does not activate VEGF-A expression (Figure 5B). All four pairs of our synthetic DZFs interact in a preferentially heterotypic manner as judged by this assay. This result contrasts with experiments performed using the B2H reporter system (compare Figures 4 and 5B) in which only three of the four DZF pairs exhibit preferentially heterotypic interaction specificities. We do not know precisely why the mammalian ‘activator reconstitution' assay does not detect self-interaction of the Pe-Hd-Hd DZF but speculate that it may related to the observation that homodimeric interactions (e.g., by wild-type DZFs) appear to mediate less efficient VEGF-A activation as compared with heterodimeric interactions (e.g., by our synthetic DZFs) (Figure 5B; also see Discussion below).
For applications in synthetic biology, it is important to consider the possibility that certain endogenous proteins (e.g., other DZF-containing proteins) might also mediate competing interactions with our synthetic DZF domains. If this were the case, it is possible that the observed interaction specificities of our synthetic DZF fusions might be dependent on their overexpression relative to these other hypothetical competing proteins. (In our VEGF-A activator reconstitution assays, our synthetic DZF–DBD and DZF–p65 fusion proteins are likely to be overexpressed because their expression is driven by a strong CMV promoter and by an optimized Kozak translation initiation sequence.) To rule out this possibility, we performed experiments showing that (1) the interactions of synthetic DZFs are not critically dependent on overexpression and (2) the specificities of DZFs are maintained even when they and their potential interaction partners are both overexpressed. The results of the experiments are presented in the Supplementary information section and in Supplementary Figures 2 and 3.
Synthetic DZFs can mediate protein–protein interactions in the cytoplasm of mammalian cells
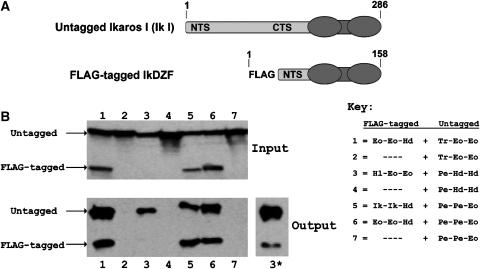
Having established that our engineered DZFs can interact in the nucleus, we next sought to determine whether they could also interact in the cytoplasm of a human cell. To do this, we used a previously developed co-immunoprecipitation method to assess DZF interactions (McCarty et al, 2003). As illustrated in Figure 6A and described in Materials and methods, a pair of DZFs can be tested for interaction by coexpressing DZFs in HEK293 cells as two fusion proteins of different molecular weights: the smaller size DZF fusion harbors a FLAG tag epitope, whereas the larger size DZF fusion does not. (Note that in contrast to the activator reconstitution assay, none of the DZF fusion proteins used in this experiment harbors a nuclear localization signal.) To test for interaction, immunoprecipitation of cytoplasmic cell lysates is performed using an anti-FLAG antibody and then the precipitated DZF fusion proteins are visualized using Western blotting performed with an antibody that recognizes an epitope present in both fusions. As shown in Figure 6B, we found that each of our synthetic DZF pairs mediate interaction as judged by this assay. Control experiments in which only the untagged larger size DZF fusion is expressed demonstrate that this protein is only ‘pulled down' when an interacting FLAG-tagged fusion is also expressed (bottom panel, Figure 6B, lanes 2, 4, and 7). We conclude that our co-immunoprecipitation results together with our ‘activator reconstitution' results show that these DZFs can function in two different compartments of a human cell.
Figure 6.
Synthetic DZFs mediate interactions in a mammalian cell-based co-immunoprecipitation assay. (A) Schematic illustrating tagged and untagged DZF fusion proteins used in the co-immunoprecipitation assay. NTS and CTS represent amino- and carboxy-terminal portions of the Ikaros isoform I protein, respectively (see Materials and methods for details). DZFs are represented as a pair of ovals and are present at the carboxy-terminal end of both fusion proteins. (B) Synthetic DZF interactions analyzed using the co-immunoprecipitation assay. Cell extracts from Flp-In TRex 293 cells (Invitrogen) cotransfected with plasmids encoding combinations of tagged and untagged DZFs or only untagged DZFs as controls (defined in the key; abbreviations as in Table I) were immunoprecipitated using an anti-FLAG antibody (see Materials and methods for details). Fusion proteins in these extracts were visualized before (‘Input') and after (‘Output') immunoprecipitation by Western blotting with an antibody that recognizes the Ikaros NTS sequence present in both types of fusion proteins. For the ‘output' of combination #3, a longer exposure (labeled as 3* and shown to the right of the output blot) was required to visualize the untagged DZF, presumably because the FLAG-tagged Hl-Eo-Eo DZF fusion is expressed at a low level (accounting for its absence in the Input blot where a smaller fraction of the total cellular extract is loaded relative to the Output blot).
Interaction geometry of synthetic DZFs
Analysis of the homo- and heterotypic interaction specificities of our synthetic DZFs suggests a potential antiparallel interaction mode for these domains. Previous studies suggested that DZFs may dimerize in a parallel mode (i.e., that amino-terminal C2H2 ZFs in each monomer interact with each other and carboxy-terminal C2H2 ZFs in each monomer interact with each other) (McCarty et al, 2003). This conclusion was based on the observation that a synthetic Ikaros–Hunchback hybrid DZF could efficiently homodimerize and on the assumption that the protein fragments in this synthetic hybrid retain the interaction specificities of the DZFs from which they were derived (McCarty et al, 2003). However, applying a similar analysis to the results of our bacterial cell- and mammalian cell-based interaction assays (Figures 4 and 5B) suggests that the interaction mode of the four synthetic DZF pairs we analyzed is more consistent with an antiparallel interaction mode. For example, the Ik-Ik-Hd and Pe-Pe-Eo DZFs interact with each other, but each interact significantly less efficiently or not at all with themselves. Assuming that the individual C2H2 ZFs in these shuffled domains retain the interaction specificities of their parental DZFs (summarized in Figure 2C above), the observed specificities can only be explained by an antiparallel (and not a parallel) interaction mode.
Engineering of more extended C2H2 ZF-mediated protein–protein interfaces
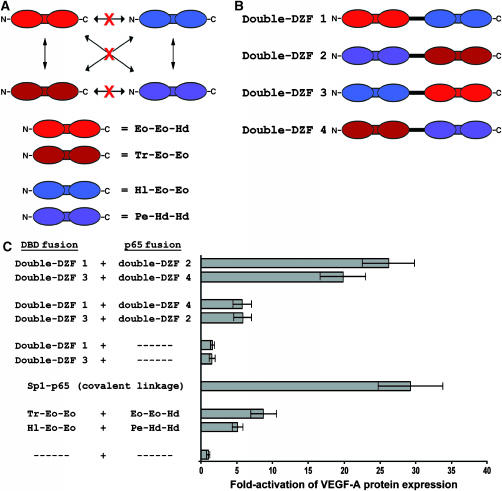
Previous studies have shown that DNA-binding C2H2 ZFs can be linked together into tandem arrays capable of recognizing extended DNA sequences. For example, units composed of two C2H2 ZFs have been joined together by linkers to create four- and six-finger proteins capable of binding 12 and 18 bp DNA sequences, respectively (Moore et al, 2001; Tan et al, 2003; Urnov et al, 2005). We were therefore interested in testing the hypothesis that, by analogy, extended protein–protein interfaces might be constructed by linking together two synthetic DZFs to create ‘double-DZFs' composed of four C2H2 ZFs. To do this, we used two pairs of DZFs from our selections. These particular pairs were chosen because both B2H reporter system and mammalian cell ‘activator reconstitution' experiments demonstrated that each DZF in these pairs interacts only with the partner it was selected with and does not crossinteract with either of the DZFs in the other pair (depicted in Figure 7A; data in Supplementary Table 1 and Figure 1). We chose these particular DZFs to minimize the occurrence of unwanted, complicating inter- and intramolecular DZF–DZF interactions. As illustrated in Figure 7B, by varying the linear order of these DZFs, we constructed four double-DZFs: double-DZF1, double-DZF2, double-DZF3, and double-DZF4.
Figure 7.
Synthetic double-DZFs interact in mammalian cells. (A) Interaction specificities of synthetic DZFs used to construct double-DZFs. Interaction specificities shown (arrows) were determined using the B2H system and the mammalian cell-based ‘activator reconstitution' assay as reporters (Supplementary Table 1 and Figure 1). Red X's indicate no significant interaction was detectable in either system. (B) Schematic of synthetic double-DZFs. Inter-DZF linkers are shown as black lines and are described in Materials and methods. (C) Interaction of double-DZF pairs assessed using the mammalian cell-based activator reconstitution assay. Controls testing the interaction of single DZF pairs and of an intact synthetic VEGF-A activator (consisting of the DBD that binds the VEGF-A gene covalently fused to the NF-κB p65 activation domain) are also shown. Fold-stimulation of VEGF-A protein expression was calculated using ELISA measurements of secreted VEGF-A that were performed at least three times. Bars shown represent mean fold-activations of VEGF-A expression and error bars indicate standard errors of the mean.
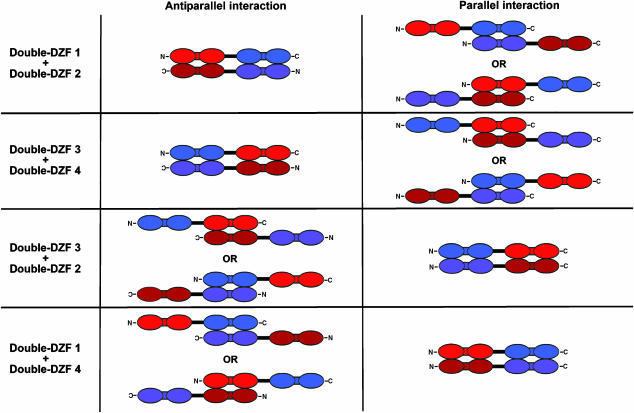
To test whether double-DZFs could mediate specific interactions, we used our mammalian cell-based activator reconstitution assay. As shown in Figure 7C, double-DZF1/double-DZF2 and double-DZF3/double-DZF4 pairs each mediate robust activation of VEGF-A expression greater than that observed with the single DZF pairs from which they were constructed. This high level of activation is similar to that observed when the two parts of the artificial VEGF-A activator are covalently linked together into a single molecule (Figure 7C; Liu et al, 2001). By contrast, different pairings of the same double-DZFs (double-DZF2/double-DZF3 and double-DZF1/double-DZF4) stimulate VEGF-A expression less efficiently, to a level similar to that observed with the single DZF pairs used to construct the double-DZFs (Figure 7C). We note that these lower levels of VEGF-A activation are not likely to be owing to poor expression or stability of the double-DZFs as these same proteins mediate higher levels of activation when tested in different combinations. A simple interpretation of these results is that double-DZF-1/double-DZF-2 and double-DZF-3/double-DZF-4 strongly interact using both pairs of DZFs, whereas double-DZF-2/double-DZF-3 and double-DZF-1/double-DZF-4 interact less strongly because they use only one of the two DZF pairs. Consistent with this possibility, if we assume that the DZFs within the double-DZF interact in an antiparallel manner, we would expect the double-DZF1/double-DZF2 and double-DZF3/double-DZF4 pairs to interact more strongly because they would be able to use both DZFs (Figure 8, left panel). Using the same assumption, we would also expect the double-DZF2/double-DZF3 and double-DZF1/double-DZF4 pairs to interact less strongly because they would be able to use only one DZF at a time (Figure 8, left panel). By contrast, if the DZFs in the double-DZFs interact in a parallel manner, these predictions of relative affinity would be reversed (Figure 8, right panel). Thus, the pattern of relative interaction strengths observed with double-DZFs in the activator reconstitution assay provides additional compelling evidence for the antiparallel interaction of the synthetic DZFs we selected in this study.
Figure 8.
Antiparallel and parallel interaction models for synthetic double-DZFs. Interaction models illustrated assume that the individual DZFs interact only with the partner DZF with which they were selected (as in Figure 7A). Identities of double-DZFs and the single DZFs from which they were constructed are color-coded as in Figure 7. Note that in certain cases more than one potential interaction mode is predicted.
Discussion
Shuffling of DZF-derived C2H2 ZFs yields novel protein–protein interactions
Before this study, it was not known whether protein-interacting C2H2 ZFs, like their DNA-binding counterparts, could be mixed and matched to create synthetic proteins with novel binding specificities. Intrigued by a previous report that described a functional chimeric DZF (McCarty et al, 2003) and by the sequence variation and functional diversity of wild-type DZFs, we systematically investigated whether C2H2 ZFs and DZFs could be ‘mixed and matched' to create domains with novel interaction specificities. Our results show that shuffling of DZF-derived C2H2 ZFs can yield synthetic DZFs with new specificities that can in turn be linked together to create more extended interaction interfaces. (We presume that the DZFs we have created interact as dimers, although we cannot rule out that they may interact as higher-order oligomers because recent studies suggest that the Eos DZF may self-interact as a multimer of as many as 10 molecules (Westman et al, 2003).) The ability to create domain-shuffled DZFs is consistent with recent biophysical studies showing that C2H2 ZFs in the Eos DZF fold independently of each other (Westman et al, 2004).
Although our results clearly demonstrate that certain DZF-derived C2H2 ZFs can be shuffled, we note that only a small number (26 pairs; Table I) of the potential combinations of DZFs produced by our shuffling strategies (5344 combinations; Supplementary Table 2) actually were identified by our selections. We believe that our experiments identified nearly all interacting pairs of synthetic DZFs in our libraries because we vastly oversampled the theoretical number of combinations in our selections and because our sequencing results revealed that we identified many of the interacting pairs multiple times. The fact that less than 0.5% (26 selected DZF pairs/5344 total potential pairs) of the potential interacting DZF pairs were positive for interaction emphasizes the importance of the B2H selection method for the success of our experiments. This low frequency also explains why initial attempts by our group to create synthetic DZFs without the use of the B2H selection method were unsuccessful (A Giesecke and JK Joung, unpublished data). Although it is possible that at least some of the synthetic DZFs encoded in our libraries are poorly expressed, unfolded, unstable, or toxic, we hypothesize that the small number of interacting DZF pairs identified may be a consequence of the fact that DZF-derived C2H2 ZFs do not always behave in a fully modular manner. DNA-binding C2H2 ZFs exhibit context-dependent effects (Wolfe et al, 1999, 2001; Hurt et al, 2003) and it is therefore not surprising that protein-interacting fingers might also be subject to such effects.
Important future studies will include obtaining detailed thermodynamic and kinetic information regarding DZF interactions. Determination of these values has proven to be challenging owing to technical barriers (e.g., insolubility, aggregation) that have thwarted efforts to purify intact, untagged DZFs (A Giesecke, R Fang, and JK Joung, unpublished data). We have encountered these issues with both wild-type and synthetic DZF domains and other laboratories have also reported encountering technical difficulties in isolating intact DZFs for biochemical and structural studies (McCarty et al, 2003; Westman et al, 2003, 2004). However, despite this limitation, we can estimate an approximate upper boundary for dissociation constants of DZF–DZF interactions based on the observation that transcriptional activation in the B2H has been observed with dissociation constants as high as ∼1 μM (Dove et al, 1997). Although previous studies have suggested that the magnitude of transcriptional activation observed in the B2H system correlates with the strength of the interaction (Dove et al, 1997), we hesitate to apply such a correlation to DZF–DZF interactions owing to potentially confounding effects of self-interaction by each of the DZF hybrid proteins.
An alternative ‘antiparallel' interaction mode for DZFs
Our selection experiments revealed a potential antiparallel interaction mode for DZF domains. Previous studies suggested that DZFs might dimerize in a parallel mode with the amino-terminal C2H2 ZFs in each monomer interacting with each other and the carboxy-terminal C2H2 ZFs in each monomer interacting with each other (McCarty et al, 2003). Two observations suggest that our synthetic DZFs interact in an alternative antiparallel geometry. First, as noted above, the identities of C2H2 ZFs and the homo- and heterotypic specificities of our synthetic DZFs (determined in bacterial cell- and mammalian cell-based interaction assays; Figures 4 and 5B) strongly suggests that these DZFs interact in an antiparallel manner. Second, the interaction specificities of the four-finger double-DZFs we constructed are most consistent with a model in which the component DZFs interact in an anti-parallel manner (Figures 7 and 8).
We note that our findings do not preclude the existence of a previously proposed parallel DZF–DZF interaction mode (McCarty et al, 2003). In fact, inspection of the sequences of our selected DZF pairs identifies at least one synthetic DZF pair whose C2H2 ZF composition suggests a parallel interaction mode (Pe-Hd-Hd and Hd-Hd-Hl; Table I). The finding of both parallel and antiparallel interaction geometries for DZFs is reminiscent of the behavior of leucine zippers, which can mediate homo- and heterotypic interactions among eukaryotic transcription factors and can interact as parallel or antiparallel coiled-coils (Light et al, 1996; Walshaw and Woolfson, 2001; Vinson et al, 2002). The existence of two potential interaction modes for synthetic DZFs raises the important question of which orientation(s) is utilized by naturally occurring DZFs. Future biophysical and structural studies (once purified DZF domains can be obtained) should provide important insights into the physical–chemical parameters that dictate the geometric orientation of DZF–DZF interactions.
Applications of artificial protein-interacting C2H2 ZF domains to synthetic biology
We have demonstrated that our synthetic interacting DZF pairs can potentially be useful for applications in synthetic biology. First, we have shown that our synthetic DZFs can be used to construct synthetic, bi-partite transcriptional activators capable of stimulating expression of the endogenous VEGF-A gene in human cells even in the absence of normal regulatory signals such as hypoxia. This represents an example of synthetic control as we have by-passed the normal regulatory mechanisms of the VEGF-A gene. Furthermore, because this activation depends upon the presence of two DZF-containing proteins, it provides a mechanism for making VEGF-A expression codependent on two inputs (e.g., as in an ‘AND' circuit). Our results are particularly relevant because an important goal of synthetic biology is the creation of artificial proteins and networks that interface with, and influence the behavior of, endogenous gene expression patterns. Second, we have also shown that synthetic DZF domains (together with DNA-binding C2H2 ZFs) can be used to activate expression of a specific gene in a bacterial cell. Thus, artificial DNA-binding C2H2 ZFs and DZFs provide the synthetic biologist with potential parts for creating multiple transcriptional activators, a capability useful for constructing synthetic circuits in bacteria.
Because our synthetic DZF domains mediate preferentially heterotypic interactions, they provide important additional functionality compared with naturally occurring wild-type DZFs, which mediate preferentially homotypic or mixed homo- and heterotypic interactions (Figure 2C). Heterotypic interactions may be particularly useful for applications requiring asymmetric complex assembly. For example, our experiments suggest that most of our synthetic preferentially heterotypic DZFs are more efficient at mediating assembly of a bi-partite transcription factor in our mammalian cell-based ‘activator reconstitution' assay than naturally occurring homotypic DZFs (Figure 5B). One possible reason for this difference may be the formation of undesired homodimers of DBD fusions or activation domain fusions mediated by the wild-type homotypic DZFs that compete with the formation of the desired DBD/activation domain heterodimer.
An important goal for future studies will be to obtain more synthetic C2H2 ZF domains with novel specificities using the finger shuffling and selection approaches described in this report. Additional sources of C2H2 ZFs for future shuffled libraries include other naturally occurring DZFs or protein-interacting domains. Alternatively, by analogy with engineering work carried out with DNA-binding C2H2 ZFs and with leucine zipper interaction domains, it may be possible to randomize residues within DZF-derived fingers that have been shown to be important for mediating protein–protein interactions (McCarty et al, 2003; Westman et al, 2004) to create C2H2 ZFs with completely novel specificities. Additional mutagenesis and high-resolution structures of DZFs will help further refine the choice of residues for randomization. Shuffling of these various naturally occurring and re-engineered C2H2 ZFs combined with selections in the B2H could yield finger arrays with novel interaction specificities different from the domains already obtained in this report. We note that our studies indicate that the B2H system functions as an effective selection method for identifying synthetic DZFs that function well in a mammalian cell context.
Another interesting issue for future experiments will be to examine to what degree, if any, our synthetic DZFs interact with endogenous proteins in different cellular contexts. Our data show that synthetic DZFs function in the cytoplasm of bacterial cells and in the nucleus and cytoplasm of human cells. These results demonstrate that any potential interactions with the hundreds, if not thousands, of endogenous proteins expressed in these two cellular contexts (including C2H2 ZF and DZF proteins) do not interfere with the abilities of our synthetic DZF pairs to interact, even when the expression level of our domains is lowered by seven-fold or more. Given that little is known about protein–protein interactions mediated by C2H2 ZFs and given the large numbers of C2H2 ZFs encoded in eukaryotic cells (>3000 domains in the human genome alone), a challenge for future studies will be to develop methods for identifying all possible protein interaction partners of our synthetic DZFs.
The demonstration that DZFs can be joined together into more extended arrays (to create double-DZFs) also expands the utility of our synthetic protein-interacting domains. Our results suggest that interfaces of variable affinities can be produced by varying the number of fingers present at the interface. In addition, we speculate that it may be possible to engineer synthetic, multi-DZF ‘scaffolds' or ‘adaptors' upon which various DZF-linked proteins might be assembled. Successful creation of such scaffolds could permit the creation of ‘circuit components' in which various cellular pathway inputs (e.g., kinases, transcription factors) might be integrated into a single output (Park et al, 2003). The capability to engineer synthetic C2H2 ZFs with novel protein–protein interaction specificities, particularly when coupled with existing technologies for constructing designer C2H2 ZF DNA-binding proteins, should expand the range of potential ‘parts' available to synthetic biologists for constructing artificial cellular networks.
Materials and methods
B2H plasmids and strains
DNA fragments encoding DZF domains were either amplified from a human cDNA library (Panomics) using polymerase chain reaction or assembled using synthetic overlapping oligonucleotides. Each DZF-encoding DNA fragment was cloned into plasmid pACYC-α (for expression as a carboxy-terminal fusion with the amino-terminal domain and intersubunit linker of the E. coli RNAP α subunit) and plasmid pBR-UV5-Zif268 (for expression as an amino-terminal fusion with the Zif268 DBD). Plasmid pACYC-α encodes the amino-terminal domain and linker (residues 1–248) of the E. coli RNA polymerase α subunit expressed from an isopropyl β-D-thiogalactosidase (IPTG)-inducible lpp/lacUV5 tandem promoter. Plasmid pBR-UV5-Zif268 encodes the DNA-binding C2H2 ZF domain of the murine Zif268 protein (residues 327–421) expressed from the IPTG-inducible lacUV5 promoter (Joung et al, 2000).
Strains used for B2H experiments were derived from strain KJ1C (Joung et al, 2000) and harbor a recombinant F′ (constructed as previously described (Hurt et al, 2003)) bearing the Zif268 binding site centered at position −65 relative to the transcription start point of a modified lac promoter directing expression of either the lacZ gene (in the ‘B2H reporter strain') or the cocistronic His3 and aadA genes (in the ‘B2H selection strain'). The F′ reporter is strictly maintained at single copy in E. coli strains.
β-Galactosidase reporter assays
β-Galactosidase assays were performed in triplicate as described previously (Thibodeau et al, 2004). Briefly, this method involves lysing logarithmic phase bacterial cultures grown in LB (containing 50 μg/ml carbenicillin, 30 μg/ml chloramphenicol, 30 μg/ml kanamycin, 10 μM ZnCl2 or ZnSO4, and 50 μM IPTG) using a commercially available lysis reagent (BugBuster, Novagen) and then assaying these extracts for β-galactosidase activity by monitoring the ability of this enzyme to process a substrate (ONPG) that yields a yellow color upon cleavage.
Construction of shuffled DZF domain libraries
For each library we constructed, two strategies were used to create shuffled combinations of DZF-derived C2H2 ZFs: one in which the interfinger linker remained associated with the amino-terminal C2H2 ZF and another in which the linker remained associated with the carboxy-terminal C2H2 ZF (Figure 3). These two different types of combinatorial pools were constructed by performing directional ligation of DNA fragments in a way that specifically precluded the formation of DNAs encoding wild-type DZFs. (Additional details of library construction are available upon request.) Each of these two different shuffled pools was then ligated into the compatible pACYC-α and pBR-UV5-Zif268 plasmids so that these domains could be expressed as fusions to the E. coli RNAP α-subunit and the Zif268 DBD, respectively (Figure 3). This approach yields a ‘set' of four different plasmid libraries that can be used for selections in the B2H system (Figure 3): two libraries expressed as E. coli RNAP α-subunit hybrid proteins and two libraries expressed as Zif268 DBD hybrid proteins.
Different ‘sets' of libraries were constructed by shuffling combinations of C2H2 ZFs derived from various subsets of eight wild-type DZFs: set A consisted of shuffled combinations of the human Ikaros, human Eos, human Pegasus, D. melanogaster Hunchback, Locusta migratoria Hunchback, and Helobdella triserialis Hunchback DZFs; set B consisted of shuffled combinations of the human Pegasus, D. melanogaster Hunchback, and L. migratoria Hunchback DZFs; and set C consisted of shuffled combinations of human Eos, human TRPS-1, D. melanogaster Hunchback, H. triserialis Hunchback, and Caenorhabditis elegans Hunchback. Statistics describing the theoretical and actual number of candidates in each of the four libraries of each ‘set' are provided in the first two columns of Supplementary Table 2. Note that the actual number of candidates in each library we constructed exceeded the theoretical number of potential combinations by at least 500-fold, ensuring that nearly all possible shuffled DZFs should be present in each of our libraries.
B2H selections
Three independent ‘groups' of selection experiments were performed, each using one of the three library ‘sets' (A, B, and C). For each selection ‘group,' four different selections testing all possible pairwise combinations of the four libraries in a ‘set' were tested for potential interactions. The theoretical numbers of combinations for each selection in each set are shown in column 3 of Supplementary Table 2. Thus, the total number of combinations interrogated by all three ‘groups' of selections is 5344=(900 DZF combinations × 4 selections)+(36 DZF combinations × 4 selections)+(400 DZF combinations × 4 selections). Pairs of libraries were introduced serially into ‘B2H selection strain' cells with the RNAP α-fusion library introduced first by electroporation followed by the Zif268 fusion library introduced by phagemid infection as described previously (Joung et al, 2000). Note that for each selection performed, the number of transformants plated (typically ∼106–107) exceeded the theoretical number of potential combinations by at least 1000-fold (see column 4 of Supplementary Table 2), thereby ensuring that essentially all possible DZF–DZF combinations were tested for interaction. Increased His3 expression in ΔhisB KJ1C cells permits growth on histidine-deficient medium and increased aadA gene expression permits growth on medium containing streptomycin (Joung et al, 2000). Thus, transformed cells were plated on histidine-deficient NM medium (Hurt et al, 2003) containing 50 μM IPTG, 25mM 3-aminotriazole (a competitive inhibitor of HIS3), and 40 μg/ml streptomycin. Following 2 days of growth at 37°C, four to 12 surviving colonies were picked from each selection for linkage analysis.
Linkage analysis was performed by isolating plasmid DNA from candidates that survived the selection, purifying each of the two plasmids independently, and then introducing pairs of purified plasmids into ‘B2H reporter strains' and performing β-galactosidase assays. Plasmid pairs that result in elevated level of β-galactosidase expression were then sequenced to determine the identities of the C2H2 ZFs and linkers.
Mammalian cell-based ‘activator reconstitution' assay
We constructed mammalian expression plasmids that encode hybrid proteins consisting of one or two DZF domains joined to the carboxy-terminus of the human NF-κB p65 subunit (residues 283–551) by a short Gly-Ser linker. A similar series of plasmids were constructed to express fusion proteins consisting of one or two DZF domains joined by a short linker of sequence GPGS to the carboxy-terminus of an engineered C2H2 ZF DBD that binds to the human VEGF-A gene (the VZ+434b domain from Liu et al (2001)). In all of these plasmids, expression of the fusion protein is driven by a modified CMV promoter that can be repressed by tetracycline repressor (from plasmid pcDNA5; Invitrogen) and all fusions harbor a nuclear localization signal from the simian virus 40 (SV40) large T antigen at their amino-terminus. The DZFs in double-DZF1, double-DZF2, and double-DZF4 were joined by a linker of sequence GEKP, whereas the DZFs in double-DZF3 were joined by a linker of sequence EFPKPSTPPGSSGGAP.
Flp-In TRex 293 cells (Invitrogen) expressing tetracycline repressor were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum in a 5% CO2 incubator at 37°C. To perform transient transfections, cells were plated in 24-well plates (Corning) at a density of 150 000 cells/well 24 h before transfection. Plasmids encoding DZF–DBD and DZF–p65 fusions were transfected into cells using 1 μl of Lipofectamine (Invitrogen) and 0.5 μg total plasmid DNA per well. At ∼16 h following transfection, medium was removed and replaced with fresh medium containing doxycycline (1 μg/μl) to induce the expression of the fusion proteins. At 24 h after induction, the culture medium was harvested and secreted VEGF-A levels in the culture medium quantified using ELISA (R&D Systems) performed according to the manufacturer's protocol, except that values were normalized to the number of viable cells (determined using WST-1 reagent; Roche).
Western blot analysis
FLAG-tagged DZF hybrid proteins were created by inserting an amino-terminal FLAG tag-coding sequence into the DZF–DBD and DZF–p65 fusion plasmids. The DZF–DBD and DZF–p65 constructs were cotransfected into HEK293 cells (seeded in 24-well plates) using Lipofectamine with 0.25 μg of the DZF–DBD plasmid and either 0.25 or 0.06 μg of the DZF–p65 fusion plasmid (0.19 μg of pcDNA5 plasmid DNA was used to keep the total DNA amount of DNA added constant at 0.5 μg). The medium was removed and replaced with fresh medium containing doxycycline (1 μg/μl)∼16 h after transfection. At 24 h after induction, the culture medium and the cells were harvested. Secreted VEGF-A levels in the culture medium were quantified using ELISA (R&D Systems).
For Western blot analysis of the DZF hybrid protein expression level, cells were lysed using Laemmli sample loading buffer. The amounts of lysate loaded on each SDS–polyacrylamide gel electrophoresis (PAGE) gel were normalized using the number of viable cells in each transfection (determined using WST-1 reagent; Roche). Western blot analysis was performed using the anti-FLAG M2 monoclonal antibody (Sigma). Anti-mouse IgG (Amersham Biosciences) served as a secondary antibody and ECL (Amersham Biosciences) was used for visualization. Band intensities were quantified using a Biorad Fluor-S MultiImager and Quantity One software.
Mammalian cell-based co-immunoprecipitation assay
DZFs were expressed as one of two different fusion proteins, one untagged and one tagged with a FLAG epitope. These proteins (illustrated schematically in Figure 6A) contain various portions of the murine Ikaros protein as described previously (McCarty et al, 2003). None of the DZF fusion proteins made for these experiments harbored a nuclear localization signal. Sequences encoding these proteins were cloned into plasmid pcDNA3 (Invitrogen) such that their expression is directed by the constitutive CMV promoter.
Co-immunoprecipitation assays were performed essentially as described previously (Cupit et al, 2003; McCarty et al, 2003). Confluent Flp-In TRex 293 cells were transfected with pairs of plasmid DNAs (10 μg total) using the calcium phosphate method. Cells were harvested 48 h after transfection and resuspended in cold buffer A (10 mM HEPES (pH 7.8), 1.5 mM MgCl2, 10 mM KCl, 20 μM ZnCl2, 1 mM DTT, 1 mM benzamidine, one tablet of protease inhibitor per 5 ml). Cells were homogenized and cytoplasmic extracts were clarified before use by microcentrifugation. 200 μl of cold RIPA buffer (150 mM NaCl, 50 mM Tris (pH 7.5), 0.1% SDS, 1% NP40, 0.025% sodium deoxylate, 1 mM DTT) was added to 10 μl lysate. RIPA buffer-equilibrated Anti-FLAG M2 Affinity Gel (Sigma) was then added and the sample was rotated for 1.5 h and 4°C. The beads were collected by centrifugation, washed three times with 1 ml of cold RIPA buffer, mixed with 30 μl 2 × SDS sample buffer, boiled, and centrifuged. The resulting supernatants were resolved by SDS–PAGE. Western blot analysis was performed using the Ikaros M-20 antibody (Santa Cruz Biotechnology) directed against the amino-terminal 53 amino acids of Ikaros. Monoclonal anti-goat IgG (Sigma) was used as secondary antibody and ECL (Amersham) was used for visualization.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2A
Supplementary Figure 2B
Supplementary Figure 3A
Supplementary Figure 3B
Supplementary Table 1
Supplementary Table 2
Supplemental Figure and Table Legends
Acknowledgments
We thank Andrew Hirsh for many helpful discussions and comments on the manuscript and Stacey Thibodeau for help with figures. AVG, RF, and JKJ were supported by grants from the NIH (K08 DK002883, R01 GM069906, and R01 GM072621) and by the MGH Department of Pathology. JKJ dedicates this paper to the memory of Robert L Burghoff, PhD, an outstanding scientist, patient teacher, and kind friend.
References
- Blancafort P , Segal DJ , Barbas CF III (2004) Designing transcription factor architectures for drug discovery. Mol Pharmacol 66: 1361–1371 [DOI] [PubMed] [Google Scholar]
- Cupit PM , Hansen JD , McCarty AS , White G , Chioda M , Spada F , Smale ST , Cunningham C (2003) Ikaros family members from the Agnathan Myxine glutinosa and the Urochordate Oikopleura dioica: emergence of an essential transcription factor for adaptive immunity. J Immunol 171: 6006–6013 [DOI] [PubMed] [Google Scholar]
- Dove SL , Hochschild A (1998) Conversion of the omega subunit of Escherichia coli RNA polymerase into a transcriptional activator or an activation target. Genes Dev 12: 745–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove SL , Hochschild A (2004) A bacterial two-hybrid system based on transcription activation. Methods Mol Biol 261: 231–246 [DOI] [PubMed] [Google Scholar]
- Dove SL , Joung JK , Hochschild A (1997) Activation of prokaryotic transcription through arbitrary protein-protein contacts. Nature 386: 627–630 [DOI] [PubMed] [Google Scholar]
- Falke D , Juliano RL (2003) Selective gene regulation with designed transcription factors: implications for therapy. Curr Opin Mol Ther 5: 161–166 [PubMed] [Google Scholar]
- Hahm K , Cobb BS , McCarty AS , Brown KE , Klug CA , Lee R , Akashi K , Weissman IL , Fisher AG , Smale ST (1998) Helios, a T cell-restricted Ikaros family member that quantitatively associates with Ikaros at centromeric heterochromatin. Genes Dev 12: 782–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahm K , Ernst P , Lo K , Kim GS , Turck C , Smale ST (1994) The lymphoid transcription factor LyF-1 is encoded by specific, alternatively spliced mRNAs derived from the Ikaros gene. Mol Cell Biol 14: 7111–7123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbury PB , Kim PS , Alber T (1994) Crystal structure of an isoleucine-zipper trimer. Nature 371: 80–83 [DOI] [PubMed] [Google Scholar]
- Harbury PB , Zhang T , Kim PS , Alber T (1993) A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science 262: 1401–1407 [DOI] [PubMed] [Google Scholar]
- Hu JC , Newell NE , Tidor B , Sauer RT (1993) Probing the roles of residues at the e and g positions of the GCN4 leucine zipper by combinatorial mutagenesis. Protein Sci 2: 1072–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JC , O'Shea EK , Kim PS , Sauer RT (1990) Sequence requirements for coiled-coils: analysis with lambda repressor-GCN4 leucine zipper fusions. Science 250: 1400–1403 [DOI] [PubMed] [Google Scholar]
- Hurt JA , Thibodeau SA , Hirsh AS , Pabo CO , Joung JK (2003) Highly specific zinc finger proteins obtained by directed domain shuffling and cell-based selection. Proc Natl Acad Sci USA 100: 12271–12276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isalan M , Lemerle C , Serrano L (2005) Engineering gene networks to emulate Drosophila embryonic pattern formation. PLoS Biol 3: e64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson AC , Miller JC , Pabo CO (2003) Drug discovery with engineered zinc-finger proteins. Nat Rev Drug Disc 2: 361–368 [DOI] [PubMed] [Google Scholar]
- Jantz D , Amann BT , Gatto GJ Jr , Berg JM (2004) The design of functional DNA-binding proteins based on zinc finger domains. Chem Rev 104: 789–799 [DOI] [PubMed] [Google Scholar]
- Joung JK (2001) Identifying and modifying protein–DNA and protein–protein interactions using a bacterial two-hybrid selection system. J Cell Biochem Suppl, (Suppl 37): 53–57 [DOI] [PubMed] [Google Scholar]
- Joung JK , Ramm EI , Pabo CO (2000) A bacterial two-hybrid selection system for studying protein–DNA and protein–protein interactions. Proc Natl Acad Sci USA 97: 7382–7387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug A (1999) Zinc finger peptides for the regulation of gene expression. J Mol Biol 293: 215–218 [DOI] [PubMed] [Google Scholar]
- Lander ES , Linton LM , Birren B , Nusbaum C , Zody MC , Baldwin J , Devon K , Dewar K , Doyle M , FitzHugh W , Funke R , Gage D , Harris K , Heaford A , Howland J , Kann L , Lehoczky J , LeVine R , McEwan P , McKernan K , Meldrim J , Mesirov JP , Miranda C , Morris W , Naylor J , Raymond C , Rosetti M , Santos R , Sheridan A , Sougnez C , Stange-Thomann N , Stojanovic N , Subramanian A , Wyman D , Rogers J , Sulston J , Ainscough R , Beck S , Bentley D , Burton J , Clee C , Carter N , Coulson A , Deadman R , Deloukas P , Dunham A , Dunham I , Durbin R , French L , Grafham D , Gregory S , Hubbard T , Humphray S , Hunt A , Jones M , Lloyd C , McMurray A , Matthews L , Mercer S , Milne S , Mullikin JC , Mungall A , Plumb R , Ross M , Shownkeen R , Sims S , Waterston RH , Wilson RK , Hillier LW , McPherson JD , Marra MA , Mardis ER , Fulton LA , Chinwalla AT , Pepin KH , Gish WR , Chissoe SL , Wendl MC , Delehaunty KD , Miner TL , Delehaunty A , Kramer JB , Cook LL , Fulton RS , Johnson DL , Minx PJ , Clifton SW , Hawkins T , Branscomb E , Predki P , Richardson P , Wenning S , Slezak T , Doggett N , Cheng JF , Olsen A , Lucas S , Elkin C , Uberbacher E , Frazier M , Gibbs RA , Muzny DM , Scherer SE , Bouck JB , Sodergren EJ , Worley KC , Rives CM , Gorrell JH , Metzker ML , Naylor SL , Kucherlapati RS , Nelson DL , Weinstock GM , Sakaki Y , Fujiyama A , Hattori M , Yada T , Toyoda A , Itoh T , Kawagoe C , Watanabe H , Totoki Y , Taylor T , Weissenbach J , Heilig R , Saurin W , Artiguenave F , Brottier P , Bruls T , Pelletier E , Robert C , Wincker P , Smith DR , Doucette-Stamm L , Rubenfield M , Weinstock K , Lee HM , Dubois J , Rosenthal A , Platzer M , Nyakatura G , Taudien S , Rump A , Yang H , Yu J , Wang J , Huang G , Gu J , Hood L , Rowen L , Madan A , Qin S , Davis RW , Federspiel NA , Abola AP , Proctor MJ , Myers RM , Schmutz J , Dickson M , Grimwood J , Cox DR , Olson MV , Kaul R , Shimizu N , Kawasaki K , Minoshima S , Evans GA , Athanasiou M , Schultz R , Roe BA , Chen F , Pan H , Ramser J , Lehrach H , Reinhardt R , McCombie WR , de la Bastide M , Dedhia N , Blocker H , Hornischer K , Nordsiek G , Agarwala R , Aravind L , Bailey JA , Bateman A , Batzoglou S , Birney E , Bork P , Brown DG , Burge CB , Cerutti L , Chen HC , Church D , Clamp M , Copley RR , Doerks T , Eddy SR , Eichler EE , Furey TS , Galagan J , Gilbert JG , Harmon C , Hayashizaki Y , Haussler D , Hermjakob H , Hokamp K , Jang W , Johnson LS , Jones TA , Kasif S , Kaspryzk A , Kennedy S , Kent WJ , Kitts P , Koonin EV , Korf I , Kulp D , Lancet D , Lowe TM , McLysaght A , Mikkelsen T , Moran JV , Mulder N , Pollara VJ , Ponting CP , Schuler G , Schultz J , Slater G , Smit AF , Stupka E , Szustakowski J , Thierry-Mieg D , Thierry-Mieg J , Wagner L , Wallis J , Wheeler R , Williams A , Wolf YI , Wolfe KH , Yang SP , Yeh RF , Collins F , Guyer MS , Peterson J , Felsenfeld A , Wetterstrand KA , Patrinos A , Morgan MJ , Szustakowki J , de Jong P , Catanese JJ , Osoegawa K , Shizuya H , Choi S , Chen YJ (2001) Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Lee DK , Seol W , Kim JS (2003) Custom DNA-binding proteins and artificial transcription factors. Curr Top Med Chem 3: 645–657 [DOI] [PubMed] [Google Scholar]
- Light J , Maki R , Assa-Munt N (1996) Expression cloning of cDNA by phage display selection. Nucleic Acids Res 24: 4367–4368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PQ , Rebar EJ , Zhang L , Liu Q , Jamieson AC , Liang Y , Qi H , Li PX , Chen B , Mendel MC , Zhong X , Lee YL , Eisenberg SP , Spratt SK , Case CC , Wolffe AP (2001) Regulation of an endogenous locus using a panel of designed zinc finger proteins targeted to accessible chromatin regions. Activation of vascular endothelial growth factor A. J Biol Chem 276: 11323–11334 [DOI] [PubMed] [Google Scholar]
- Lu D , Searles MA , Klug A (2003) Crystal structure of a zinc-finger–RNA complex reveals two modes of molecular recognition. Nature 426: 96–100 [DOI] [PubMed] [Google Scholar]
- Mackay JP , Crossley M (1998) Zinc fingers are sticking together. Trends Biochem Sci 23: 1–4 [DOI] [PubMed] [Google Scholar]
- McCarty AS , Kleiger G , Eisenberg D , Smale ST (2003) Selective dimerization of a C2H2 zinc finger subfamily. Mol Cell 11: 459–470 [DOI] [PubMed] [Google Scholar]
- Moll JR , Ruvinov SB , Pastan I , Vinson C (2001) Designed heterodimerizing leucine zippers with a ranger of pIs and stabilities up to 10(–15) M. Protein Sci 10: 649–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M , Klug A , Choo Y (2001) Improved DNA binding specificity from polyzinc finger peptides by using strings of two-finger units. Proc Natl Acad Sci USA 98: 1437–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan B , Sun L , Avitahl N , Andrikopoulos K , Ikeda T , Gonzales E , Wu P , Neben S , Georgopoulos K (1997) Aiolos, a lymphoid restricted transcription factor that interacts with Ikaros to regulate lymphocyte differentiation. EMBO J 16: 2004–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller A , MacCallum RM , Sternberg MJ (2002) Structural characterization of the human proteome. Genome Res 12: 1625–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JR , Keating AE (2003) Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science 300: 2097–2101 [DOI] [PubMed] [Google Scholar]
- O'Shea EK , Klemm JD , Kim PS , Alber T (1991) X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science 254: 539–544 [DOI] [PubMed] [Google Scholar]
- O'Shea EK , Lumb KJ , Kim PS (1993) Peptide ‘Velcro': design of a heterodimeric coiled coil. Curr Biol 3: 658–667 [DOI] [PubMed] [Google Scholar]
- O'Shea EK , Rutkowski R , Stafford WF III , Kim PS (1989) Preferential heterodimer formation by isolated leucine zippers from fos and jun. Science 245: 646–648 [DOI] [PubMed] [Google Scholar]
- Pabo CO , Peisach E , Grant RA (2001) Design and selection of novel Cys2His2 zinc finger proteins. Annu Rev Biochem 70: 313–340 [DOI] [PubMed] [Google Scholar]
- Park SH , Zarrinpar A , Lim WA (2003) Rewiring MAP kinase pathways using alternative scaffold assembly mechanisms. Science 299: 1061–1064 [DOI] [PubMed] [Google Scholar]
- Perdomo J , Holmes M , Chong B , Crossley M (2000) Eos and Pegasus, two members of the Ikaros family of proteins with distinct DNA binding activities. J Biol Chem 275: 38347–38354 [DOI] [PubMed] [Google Scholar]
- Pollock R , Giel M , Linher K , Clackson T (2002) Regulation of endogenous gene expression with a small-molecule dimerizer. Nat Biotechnol 20: 729–733 [DOI] [PubMed] [Google Scholar]
- Sun L , Liu A , Georgopoulos K (1996) Zinc finger-mediated protein interactions modulate Ikaros activity, a molecular control of lymphocyte development. EMBO J 15: 5358–5369 [PMC free article] [PubMed] [Google Scholar]
- Tan S , Guschin D , Davalos A , Lee YL , Snowden AW , Jouvenot Y , Zhang HS , Howes K , McNamara AR , Lai A , Ullman C , Reynolds L , Moore M , Isalan M , Berg LP , Campos B , Qi H , Spratt SK , Case CC , Pabo CO , Campisi J , Gregory PD (2003) Zinc-finger protein-targeted gene regulation: genomewide single-gene specificity. Proc Natl Acad Sci USA 100: 11997–12002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibodeau SA , Fang R , Joung JK (2004) High-throughput beta-galactosidase assay for bacterial cell-based reporter systems. Biotechniques 36: 410–415 [DOI] [PubMed] [Google Scholar]
- Tupler R , Perini G , Green MR (2001) Expressing the human genome. Nature 409: 832–833 [DOI] [PubMed] [Google Scholar]
- Urnov FD , Miller JC , Lee YL , Beausejour CM , Rock JM , Augustus S , Jamieson AC , Porteus MH , Gregory PD , Holmes MC (2005) Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 435: 646–651 [DOI] [PubMed] [Google Scholar]
- Venter JC , Adams MD , Myers EW , Li PW , Mural RJ , Sutton GG , Smith HO , Yandell M , Evans CA , Holt RA , Gocayne JD , Amanatides P , Ballew RM , Huson DH , Wortman JR , Zhang Q , Kodira CD , Zheng XH , Chen L , Skupski M , Subramanian G , Thomas PD , Zhang J , Gabor Miklos GL , Nelson C , Broder S , Clark AG , Nadeau J , McKusick VA , Zinder N , Levine AJ , Roberts RJ , Simon M , Slayman C , Hunkapiller M , Bolanos R , Delcher A , Dew I , Fasulo D , Flanigan M , Florea L , Halpern A , Hannenhalli S , Kravitz S , Levy S , Mobarry C , Reinert K , Remington K , Abu-Threideh J , Beasley E , Biddick K , Bonazzi V , Brandon R , Cargill M , Chandramouliswaran I , Charlab R , Chaturvedi K , Deng Z , Di Francesco V , Dunn P , Eilbeck K , Evangelista C , Gabrielian AE , Gan W , Ge W , Gong F , Gu Z , Guan P , Heiman TJ , Higgins ME , Ji RR , Ke Z , Ketchum KA , Lai Z , Lei Y , Li Z , Li J , Liang Y , Lin X , Lu F , Merkulov GV , Milshina N , Moore HM , Naik AK , Narayan VA , Neelam B , Nusskern D , Rusch DB , Salzberg S , Shao W , Shue B , Sun J , Wang Z , Wang A , Wang X , Wang J , Wei M , Wides R , Xiao C , Yan C , Yao A , Ye J , Zhan M , Zhang W , Zhang H , Zhao Q , Zheng L , Zhong F , Zhong W , Zhu S , Zhao S , Gilbert D , Baumhueter S , Spier G , Carter C , Cravchik A , Woodage T , Ali F , An H , Awe A , Baldwin D , Baden H , Barnstead M , Barrow I , Beeson K , Busam D , Carver A , Center A , Cheng ML , Curry L , Danaher S , Davenport L , Desilets R , Dietz S , Dodson K , Doup L , Ferriera S , Garg N , Gluecksmann A , Hart B , Haynes J , Haynes C , Heiner C , Hladun S , Hostin D , Houck J , Howland T , Ibegwam C , Johnson J , Kalush F , Kline L , Koduru S , Love A , Mann F , May D , McCawley S , McIntosh T , McMullen I , Moy M , Moy L , Murphy B , Nelson K , Pfannkoch C , Pratts E , Puri V , Qureshi H , Reardon M , Rodriguez R , Rogers YH , Romblad D , Ruhfel B , Scott R , Sitter C , Smallwood M , Stewart E , Strong R , Suh E , Thomas R , Tint NN , Tse S , Vech C , Wang G , Wetter J , Williams S , Williams M , Windsor S , Winn-Deen E , Wolfe K , Zaveri J , Zaveri K , Abril JF , Guigo R , Campbell MJ , Sjolander KV , Karlak B , Kejariwal A , Mi H , Lazareva B , Hatton T , Narechania A , Diemer K , Muruganujan A , Guo N , Sato S , Bafna V , Istrail S , Lippert R , Schwartz R , Walenz B , Yooseph S , Allen D , Basu A , Baxendale J , Blick L , Caminha M , Carnes-Stine J , Caulk P , Chiang YH , Coyne M , Dahlke C , Mays A , Dombroski M , Donnelly M , Ely D , Esparham S , Fosler C , Gire H , Glanowski S , Glasser K , Glodek A , Gorokhov M , Graham K , Gropman B , Harris M , Heil J , Henderson S , Hoover J , Jennings D , Jordan C , Jordan J , Kasha J , Kagan L , Kraft C , Levitsky A , Lewis M , Liu X , Lopez J , Ma D , Majoros W , McDaniel J , Murphy S , Newman M , Nguyen T , Nguyen N , Nodell M , Pan S , Peck J , Peterson M , Rowe W , Sanders R , Scott J , Simpson M , Smith T , Sprague A , Stockwell T , Turner R , Venter E , Wang M , Wen M , Wu D , Wu M , Xia A , Zandieh A , Zhu X (2001) The sequence of the human genome. Science 291: 1304–1351 [DOI] [PubMed] [Google Scholar]
- Vinson CR , Hai T , Boyd SM (1993) Dimerization specificity of the leucine zipper-containing bZIP motif on DNA binding: prediction and rational design. Genes Dev 7: 1047–1058 [DOI] [PubMed] [Google Scholar]
- Vinson C , Myakishev M , Acharya A , Mir AA , Moll JR , Bonovich M (2002) Classification of human B-ZIP proteins based on dimerization properties. Mol Cell Biol 22: 6321–6335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walshaw J , Woolfson DN (2001) Socket: a program for identifying and analysing coiled-coil motifs within protein structures. J Mol Biol 307: 1427–1450 [DOI] [PubMed] [Google Scholar]
- Westman BJ , Perdomo J , Matthews JM , Crossley M , Mackay JP (2004) Structural studies on a protein-binding zinc-finger domain of Eos reveal both similarities and differences to classical zinc fingers. Biochemistry 43: 13318–13327 [DOI] [PubMed] [Google Scholar]
- Westman BJ , Perdomo J , Sunde M , Crossley M , Mackay JP (2003) The C-terminal domain of Eos forms a high order complex in solution. J Biol Chem 278: 42419–42426 [DOI] [PubMed] [Google Scholar]
- Wolfe SA , Grant RA , Elrod-Erickson M , Pabo CO (2001) Beyond the ‘recognition code': structures of two Cys2His2 zinc finger/TATA box complexes. Structure (Cambridge) 9: 717–723 [DOI] [PubMed] [Google Scholar]
- Wolfe SA , Greisman HA , Ramm EI , Pabo CO (1999) Analysis of zinc fingers optimized via phage display: evaluating the utility of a recognition code. J Mol Biol 285: 1917–1934 [DOI] [PubMed] [Google Scholar]
- Wolfe SA , Nekludova L , Pabo CO (2000) DNA recognition by Cys2His2 zinc finger proteins. Annu Rev Biophys Biomol Struct 29: 183–212 [DOI] [PubMed] [Google Scholar]
- Zeng X , Zhu H , Lashuel HA , Hu JC (1997) Oligomerization properties of GCN4 leucine zipper e and g position mutants. Protein Sci 6: 2218–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2A
Supplementary Figure 2B
Supplementary Figure 3A
Supplementary Figure 3B
Supplementary Table 1
Supplementary Table 2
Supplemental Figure and Table Legends