In small cells like a bacterium (volume ∼1 fl) or a budding yeast (volume ∼100 fl), critical intracellular regulators are often present in small absolute numbers. In Saccharomyces cerevisiae, it is not unusual to find mRNAs expressed at concentrations of less than one molecule per cell (Ghaemmaghami et al, 2003). This means that cells will sometimes possess no molecules of some particular mRNA, sometimes one molecule, and sometimes two or more. How can cells function properly in the face of such dramatic stochastic variation? Experimental studies of the noise in synthetic gene expression networks have begun to provide important insights into this question (Elowitz et al, 2002; Raser and O'Shea, 2004; Pedraza and van Oudenaarden, 2005). Recently, Bean et al (2006) have taken an important next step in the analysis of noise, by applying modern single-cell fluorescence reporter approaches to the noise inherent in an important, ‘natural' biological phenomenon: the traversal of Start and progression of yeast cells into the cell cycle.
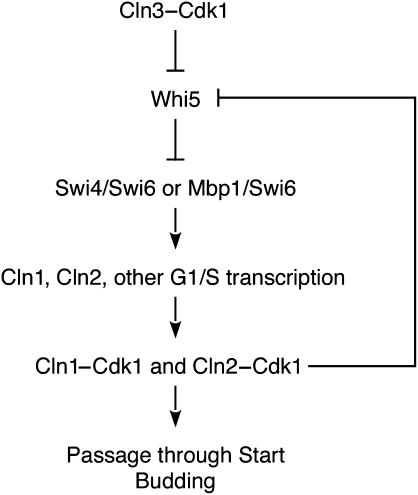
Passage through Start in budding yeast involves the transcriptional induction of a large number of G1/S-regulated genes (Spellman et al, 1998), leading to the activation of cyclin-dependent protein kinase (Cdk) complexes and the initiation of DNA replication and progression through the rest of the cell cycle (Figure 1 ). Entry into the cell cycle is initiated by the activation of Cdk1 by the G1 cyclin Cln3 (Dirick et al, 1995), promoting the transcription of G1/S genes by driving RNA polymerase II to the promoters of target genes (Cosma et al, 2001) and by driving the nuclear export of the G1-transcriptional repressor Whi5 (Costanzo et al, 2004; de Bruin et al, 2004). With the nuclear export of Whi5, the G1 transcription factors SBF (Swi4/Swi6) and MBF (Mbp1/Swi6) bind and activate transcription of many G1/S genes, including the G1 cyclins CLN1 and CLN2. The activation of Cln1,2–Cdk1 complexes initiates passage through Start, budding, and activates cell-cycle components that drive the cell cycle to completion.
Figure 1.
Schematic diagram of the regulation of Start in S. cerevisiae.
To examine the activation of the G1 transcriptional program, Bean et al developed a fluorescent biosensor that quantitatively measures the activation level of the G1/S transcriptional program. The authors constructed a yeast strain expressing a destabilized GFP under control of the G1/S transcriptional promoter of CLN2 (CLN2pr-GFP). When these cells begin to enter the cell cycle, they initiate the transcription of G1/S genes, including CLN2pr-GFP, which is expressed and then its product is rapidly degraded as G1/S transcription ceases as cells progress further into the cell cycle. This CLN2pr-GFP construct thus provides a single-cell fluorescent readout of the level of expression of the G1/S transcriptional program over the entire cell cycle.
To measure the activation of Cln1,2–Cdk1 complexes, the authors relied on the fact that full activation of Cln1,2–Cdk1 initiates bud formation, which can be easily observed microscopically. By correlating the timing of bud emergence with the timing of peak G1/S transcriptional circuit activation, as measured by peak CLN2pr-GFP expression, Bean et al measure the correlation and timing reliability of Cdk1 activation and peak transcriptional activity of the G1/S circuit. The authors refer to this as coherence: if the timing of bud emergence and that of peak CLN2pr-GFP expression are tightly linked, the coherence is high; if they are uncoupled, it is low. They then go on to analyze the role of certain known G1/S regulators in maintaining the timing and coherence of Start.
Predictably, deletion of one of the two main G1 transcription factors, SWI4, resulted in a dramatic reduction of CLN2pr-GFP fluorescence. More interestingly, swi4Δ cells showed a dramatic increase in the variation of peak CLN2pr-GFP levels and a modest decrease in the coherence between bud emergence and CLN2pr-GFP peak levels. There was also a loss of coherence between nuclear export of Whi5 and bud emergence, as cells often delayed in a Whi5-cytoplasmic, unbudded state. These findings argue that Swi4 is dispensable for Start but essential for keeping Start coherent.
Next, the authors deleted three other genes with known roles in activation of the G1/S transcriptional program, CLN3, MBP1, and RME1. In cln3Δ mbp1Δ rme1Δ cells, there was no reduction in CLN2pr-GFP induction, presumably because Swi4 is still able to activate wild-type (wt) levels of transcription. Interestingly, in this background, G1 is extended, but the coherence of Start is increased, not decreased; the variation in timing between bud emergence and peak CLN2pr-GFP expression is reduced compared to wt cells. This suggests that evolution has not maximized the coherence of Start.
Cln1–Cdk1 and Cln2–Cdk1 have the potential to activate their own transcription by inhibiting Whi5, and it is attractive to imagine that the accelerating tempo of this double-negative feedback loop (which, for many purposes, is equivalent to a positive feedback loop) could be critical for the synchronization of various Start-related phenomena. It is unclear how critical this feedback loop is for CLN1,2 induction in wt cells (Stuart and Wittenberg, 1995), but perhaps it does contribute enough to the normal synchronization of Start to account for the poor Start coherence in swi4Δ cells. In cln3Δ mbp1Δ rme1Δ cells, this feedback loop becomes more important as there is no Cln3 for the initial phosphorylation and inactivation of Whi5 in these cells. Here, stochastic expression of Cln1 or Cln2 might eventually culminate in the inactivation of Whi5, leading to the activation of the feedback loop where more and more Cln1 and Cln2 become active. Perhaps an increased reliance on positive feedback might explain the increased coherence of Start in the cln3Δ mbp1Δ rme1Δ cells.
Variability in the length of G1 in the cln3Δ mbp1Δ background is analogous to extrinsic noise in other studies (Elowitz et al, 2002; Swain et al, 2002; Raser and O'Shea, 2004) in that it is upstream of all events required for Start. Once Start gets rolling in cln3Δ mbp1Δ cells, it proceeds robustly and with higher coherence than wt cells. The swi4 mutant, on the other hand, constitutes a loss of reliable coherence: the initiation of one marker of Start relative to another marker is defective or mistimed. This kind of variability is loosely analogous to intrinsic noise (Elowitz et al, 2002; Swain et al, 2002; Raser and O'Shea, 2004).
The increased, rather than decreased, coherence found in the cln3Δ mbp1Δ rme1Δ background raises the question of why nature has chosen to make the events of Start less than maximally coherent. That is, does the improved coherence in the cln3Δ mbp1Δ rme1Δ strain come at some cost? Here the answer appears to be yes. The authors show that the increase in coherence is accompanied by increased variability in the length of G1. Thus, the authors speculate that the overall level of noise in the Start program may be an evolutionary compromise between requirements for both high intra-cell coherence and high inter-cell timing regularity.
Bean et al are to be commended for providing the field with a beautiful, quantitative analysis of noise and variability in an important biological context. The challenge for computational biologists now is to provide testable, mechanistic hypotheses to account for these observations, and perhaps to give us all a deeper understanding of the design principles of this biological control system.
References
- Bean JM, Siggia ED, Cross FR (2006) Coherence and timing of cell cycle start examined at single-cell resolution. Mol Cell 21: 3–14 [DOI] [PubMed] [Google Scholar]
- Cosma MP, Panizza S, Nasmyth K (2001) Cdk1 triggers association of RNA polymerase to cell cycle promoters only after recruitment of the mediator by SBF. Mol Cell 7: 1213–1220 [DOI] [PubMed] [Google Scholar]
- Costanzo M, Nishikawa JL, Tang X, Millman JS, Schub O, Breitkreuz K, Dewar D, Rupes I, Andrews B, Tyers M (2004) CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell 117: 899–913 [DOI] [PubMed] [Google Scholar]
- de Bruin RA, McDonald WH, Kalashnikova TI, Yates J III, Wittenberg C (2004) Cln3 activates G1-specific transcription via phosphorylation of the SBF bound repressor Whi5. Cell 117: 887–898 [DOI] [PubMed] [Google Scholar]
- Dirick L, Bohm T, Nasmyth K (1995) Roles and regulation of Cln–Cdc28 kinases at the start of the cell cycle of Saccharomyces cerevisiae. EMBO J 14: 4803–4813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz MB, Levine AJ, Siggia ED, Swain PS (2002) Stochastic gene expression in a single cell. Science 297: 1183–1186 [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS (2003) Global analysis of protein expression in yeast. Nature 425: 737–741 [DOI] [PubMed] [Google Scholar]
- Pedraza JM, van Oudenaarden A (2005) Noise propagation in gene networks. Science 307: 1965–1969 [DOI] [PubMed] [Google Scholar]
- Raser JM, O'Shea EK (2004) Control of stochasticity in eukaryotic gene expression. Science 304: 1811–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B (1998) Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell 9: 3273–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart D, Wittenberg C (1995) CLN3, not positive feedback, determines the timing of CLN2 transcription in cycling cells. Genes Dev 9: 2780–2794 [DOI] [PubMed] [Google Scholar]
- Swain PS, Elowitz MB, Siggia ED (2002) Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc Natl Acad Sci USA 99: 12795–12800 [DOI] [PMC free article] [PubMed] [Google Scholar]