As our ability to tinker with cellular components increases, the possibility of designing and controlling the functioning of living systems seems closer than ever.
In the pioneering monograph ‘What is life?', Erwin Schrödinger pointed out the now obvious fact that living organisms obey the well-established laws of physics (Schrödinger, 1945). Yet, there is a long way from the atomic interactions at the molecular level to the behavior of even the simplest bacterium. So far, the type of predictive behavior that has been so successful in physics, chemistry, and engineering is largely missing in biology. Biological systems are not static; they evolve and adapt, grow and die. They are also intrinsically stochastic and live in a fluctuating environment. All these contingencies make quite a challenging task to apply the well-established principles of engineering to biology.
In a recent issue of Nature, Guido et al (2006) take an important step in this direction: building up networks of genes from well-characterized modules and obtaining the expected behavior in all its details. This type of challenge boils down to the core of systems biology, which aims at predicting the systemic properties in terms of the properties of the components and their interactions (cellular processes in terms of molecular interactions, organismal behavior in terms of populations of cells, and so on). If this program can be carried out at all levels of organization, building up successively on the previous one, it would be possible to design and control the functioning of living systems.
Guido et al engineered a promoter controlled by lambda cI and the lac repressor to allow simultaneous repression and activation of a gene in the bacterium Escherichia coli. The authors studied its behavior in synthetic gene networks under increasingly more involved conditions, ending with the promoter being controlled by the product of the gene it controls. The novelty with respect to previous work is the construction of a particularly complex promoter and, most remarkably, the verification of the extremely detailed predictions that were made from the behavior of the simpler components. The predicted behavior included not only the average values of protein content but also a detailed quantification of cell-to-cell variability. Such predictive power is needed to bring synthetic biology to engineering grounds.
How scalable is this type of approach? It is illustrative to take a look first at the other extreme of synthetic biology, in which many networks are randomly constructed with the hope of obtaining the wanted behavior among the resulting networks. For instance, Guet et al (2002) used a combinatorial approach to construct networks of three genes controlling their promoters in a highly interconnected way. The resulting networks displayed a myriad of different types of behavior but, remarkably, the functioning of many of these networks could not be explained in terms of the known properties of their components.
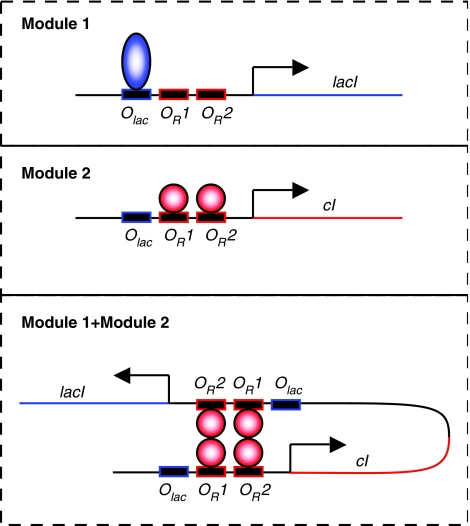
The components used by Guido et al originate from the two systems that led to the discovery of gene regulation, and we know a great deal about them. One might ask, would the approach of Guido et al work in a set up like that of Guet et al, that is, when multiple components are assembled adjacent to each other on the same DNA molecule? The answer does not follow straightforwardly. It just happens that both the lac repressor and lambda cI can loop DNA (Müller-Hill, 1996; Ptashne, 2004). Thus, placing two promoters on the same DNA strand relatively close to each other would induce the formation of DNA loops (Figure 1 ). DNA looping has been shown to have strong effects in gene regulation. It enhances the repression level about hundred times in the lac operon (Müller-Hill, 1996) and it couples two operators that are over 2 kb apart in phage lambda (Ptashne, 2004). Therefore, the formation of DNA loops would affect the behavior of the system in ways that are not present in the original components (Vilar and Saiz, 2005).
Figure 1.
Modular crosstalking in synthetic gene networks. A promoter with binding sites for lambda cI dimers (OR1 and OR2) and for the lac repressor (Olac) controls the production of the lac repressor (encoded by lacI) in module 1 and lambda cI (encoded by cI) in module 2. The lac repressor and lambda cI dimers are represented by blue ellipsoids and red circles, respectively. When the two modules are put together, two pairs of lambda cI dimers bound at different operators can loop DNA and octamerize, forming a tetramer of dimers.
This potentially emergent behavior illustrates a general theme that is present in biology at all levels: increasingly complex systems can pick up details that were hidden in simple setups. To carry on with the modular approach, crosstalking would need to be prevented. In the case of Guido et al, it is possible to devise mutant lac repressors and lambda cI proteins that do not induce DNA looping (Müller-Hill, 1996; Ptashne, 2004). Thus, in principle, it would be easy to tweak the modules to prevent them to be tangled in DNA loops.
As our ability to put together different components increases, the main challenges for a large-scale bottom-up approach to gene regulation become double-edged: how to avoid the unwanted emergent behavior in an engineered design and how to use the emergent behavior to tackle more sophisticated tasks. Naturally occurring systems seem to be placed in a middle ground where some degree of modularity is present with extensive crosstalking between different modules (Hartwell et al, 1999; Martinez Arias and Stewart, 2002). In this regard, nature seems to have followed a quasimodular approach. Perhaps, a more practical avenue to go up in complexity will be to build the core structure of the system and artificially evolve the details (Yokobayashi et al, 2002).
References
- Guet CC, Elowitz MB, Hsing W, Leibler S (2002) Combinatorial synthesis of genetic networks. Science 296: 1466–1470 [DOI] [PubMed] [Google Scholar]
- Guido NJ, Wang X, Adalsteinsson D, McMillen D, Hasty J, Cantor CR, Elston TC, Collins JJ (2006) A bottom-up approach to gene regulation. Nature 439: 856–860 [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Hopfield JJ, Leibler S, Murray AW (1999) From molecular to modular cell biology. Nature 402: C47–C52 [DOI] [PubMed] [Google Scholar]
- Martinez Arias A, Stewart A (2002) Molecular Principles of Animal Development. Oxford, New York: Oxford University Press [Google Scholar]
- Müller-Hill B (1996) The Lac Operon: A Short History of a Genetic Paradigm. Berlin, New York: Walter de Gruyter [Google Scholar]
- Ptashne M (2004) A Genetic Switch: Phage Lambda Revisited. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Schrödinger E (1945) What is Life? The Physical Aspect of the Living Cell. Cambridge: The University Press [English] [Google Scholar]
- Vilar JM, Saiz L (2005) DNA looping in gene regulation: from the assembly of macromolecular complexes to the control of transcriptional noise. Curr Opin Genet Dev 15: 136–144 [DOI] [PubMed] [Google Scholar]
- Yokobayashi Y, Weiss R, Arnold FH (2002) Directed evolution of a genetic circuit. Proc Natl Acad Sci USA 99: 16587–16591 [DOI] [PMC free article] [PubMed] [Google Scholar]