Abstract
We mapped the transcriptional regulatory circuitry for six master regulators in human hepatocytes using chromatin immunoprecipitation and high-resolution promoter microarrays. The results show that these regulators form a highly interconnected core circuitry, and reveal the local regulatory network motifs created by regulator–gene interactions. Autoregulation was a prominent theme among these regulators. We found that hepatocyte master regulators tend to bind promoter regions combinatorially and that the number of transcription factors bound to a promoter corresponds with observed gene expression. Our studies reveal portions of the core circuitry of human hepatocytes.
Keywords: autoregulation, hepatocyte, transcriptional regulation, regulatory hierarchy, chromatin immunoprecipitation
Introduction
The liver performs a number of complex functions essential for life including the uptake and storage of glucose, synthesis of bile acids, production of plasma proteins, and drug detoxification. These functions are carried out by hepatocytes, which comprise the bulk of the liver tissue. We and others have begun to use genome-scale approaches to determine the transcriptional regulatory circuitry of hepatocytes (Friedman et al, 2004; Odom et al, 2004; Phuc Le et al, 2005; Rubins et al, 2005; Zhang et al, 2005). These studies have been limited because they focused on a small number of regulators and used low-resolution technology that explored only a subset of proximal promoters in the mammalian genome. We mapped the promoter occupancy of six master regulators in primary human hepatocytes using chromatin immunoprecipitation (ChIPs) combined with DNA microarrays representing large (10 kb) portions of promoter regions for most annotated human genes. Our results provide a high-resolution, genome-wide overview of the core transcriptional circuitry of human hepatocytes.
Results and discussion
Identification of transcription factor binding sites
To initiate mapping of the transcriptional regulatory circuitry of primary human hepatocytes, we selected regulators known to be critical to hepatocyte biology based on genetic experiments in mouse or human (HNF1α, HNF4α, FOXA2/HNF3β, HNF6/ONECUT1, CREB1, and USF1) (Table I, Supplementary Table S1) (Kuo et al, 1992; Pani et al, 1992; Cereghini 1996; Duncan et al, 1998; Zaret 2002; Costa et al, 2003; Montminy et al, 2004; Pajukanta et al, 2004; Lee et al, 2005). These liver-enriched transcription factors can also play important roles in other tissues (e.g. kidney and pancreatic islets) (Bell and Polonsky, 2001). We then determined promoter occupancy for these six regulators by combining ChIPs with microarrays that have high resolution and extensive promoter coverage (Materials and methods). We employed DNA microarrays that contain 60-mer oligonucleotide probes covering the region from −8 to +2 kb relative to the transcript start sites for almost 18 000 annotated human genes, which were compiled from five independent databases (Boyer et al, 2005). Most known transcription factor binding sites occur within –8/+2 kb of the transcription start site (Boyer et al 2005); this is also true for the transcription factors studied here, although some binding sites have been identified in other regions (Tronche et al, 1997; Rada-Iglesias et al, 2005). The sites occupied by transcription factors were represented by peaks of ChIP-enriched DNA that spanned neighboring probes (examples in Supplementary Figure S1). The coverage of promoter regions averaged one 60 mer for each 250 bases of sequence (Boyer et al, 2005).
Table 1.
Transcriptional master regulators profiled in primary human hepatocytes
| Regulator | Function | PFAM category | Genes bound |
|---|---|---|---|
| HNF1α | Metabolic control | POU homeodomain | 1016 |
| HNF4α | Development, metabolism | Nuclear receptor | 4519 |
| HNF6 (ONECUT1) | Development | CUT homeodomain | 1306 |
| HNF3β (FOXA2) | Development | Forkhead | 890 |
| CREB1 | cAMP response | bZIP | 2197 |
| USF1 | Glucose, lipid metabolism | Basic helix–loop–helix | 1632 |
Identification of transcriptional regulatory motifs
We analyzed this high-resolution data using previously reported methods (Lee et al, 2002) to identify transcriptional network regulatory motifs (the simplest units of network structure), and thus to determine how these six key hepatocyte regulators contribute to autoregulatory loops, multicomponent loops, feed-forward loops, and multi-input motifs (Figure 1, Supplementary Table S2) (Lee et al, 2002; Milo et al, 2002; Shen-Orr et al, 2002; Odom et al, 2004).
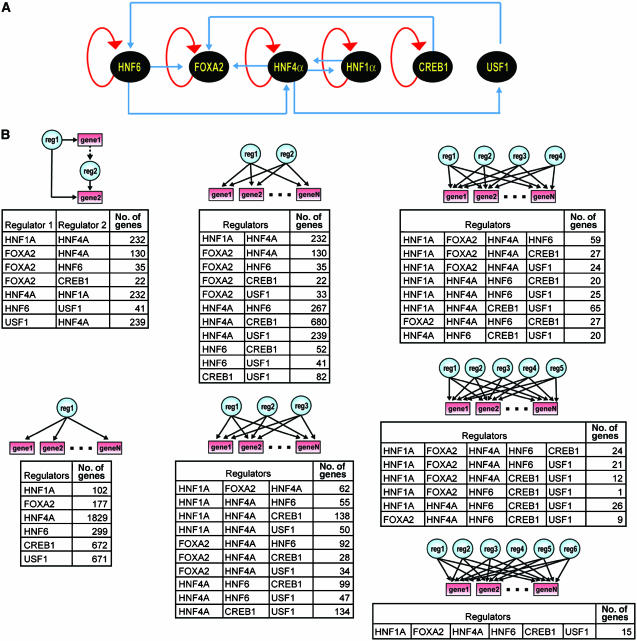
Figure 1.
(A) Transcription factor crosstalk and autoregulation in the core regulatory circuitry of human hepatocytes. Regulators are shown as black ovals, with genomic occupancy of promoter regions indicated by blue or red (for autoregulatory loops) lines. (B) Frequency of network motifs in core hepatocyte regulatory network. For clarity, only combinations potentially controlling 20 or more genes are shown for the two-, three-, and four-factor multi-input motifs. (See also Supplementary Table S2).
Several aspects of the regulatory circuitry present among these master regulators have been noted or suggested previously. The network is highly interconnected (Figure 1A), and combinatorial control plays an important role in directing gene expression (Krivan and Wasserman, 2001). HNF4α and HNF1α bound each others promoters; the FOXA2 promoter was bound by both HNF6 and FOXA2; and the HNF4α promoter is occupied by multiple HNF factors (Figure 1A, Supplementary Figure S1) (Pani et al, 1992; Duncan et al, 1998; Bailly et al, 2001; Costa et al, 2003; Briancon et al, 2004; Odom et al, 2004). Our data further showed the promoter of transthyretin, an archetypic hepatocyte gene, is occupied by HNF1α, HNF4α, FOXA2, and HNF6 as predicted from extensive site-specific and sequence-based analysis (Supplementary Figure S1) (reviewed in Costa et al, 2003).
Feed-forward loops were observed for seven combinations of transcriptional regulators (Figure 1). FOXA2 is involved with three separate feed-forward motifs, and potentially acts as a master regulator via the feed-forward motif for over 180 genes. This is consistent with previous suggestions that FOXA2 is at the top of regulatory hierarchies within the liver as well as other tissues (Lee et al, 2005). Single input motifs are present for all six regulators, although it is likely that many of the genes presently classed as single input are probably coordinately regulated and cobound by other as-yet-uncharacterized factors. Multi-input motifs were present for most combinations of transcriptional regulators (Figure 1B, Supplementary Table S2).
Prevalence of autoregulation among hepatocyte regulators
A remarkable feature of the portions of hepatocyte regulatory circuitry we studied here is the frequency of autoregulatory loops: five of the six regulators (83%) occupied their own promoters (Figure 1A, Supplementary Figure S2). Consistent with this, we use hypothesis-driven binding sequence analysis (MacIsaac et al, 2006) to determine that optimized binding sequences for each transcription factor, and their presence close to each autoregulatory binding event (Supplementary information). Most bacterial transcription factors form autoregulatory loops (Thieffry et al, 1998; Shen-Orr et al, 2002). However, an analysis of nearly all yeast transcription factors has shown that only 10% have autoregulatory loops, suggesting that this form of regulation occurs much more rarely in eukaryotes (Lee et al, 2002; Harbison et al, 2004). These observations prompted us to consider the possibility that autoregulatory motifs are general features of eukaryotic transcription factors at the top of regulatory hierarchies or that play key roles in major cellular processes. When we inspected data for all transcription factors in yeast (Harbison et al, 2004; Supplementary information), we found that master regulators of key cellular processes were significantly more likely to autoregulate than other regulators (Supplementary Table S3 and S4, Supplementary Material). For instance, STE12 and TEC1 form autoregulatory loops: STE12 is the master regulator of mating and TEC1 is a key regulator of pseudohyphal growth (Zeitlinger et al, 2003).
In mammalian cells, transcription factors that have autoregulatory loops are frequently considered master regulators of tissues or key processes. These include, for example, OCT4 in embryonic stem cells (Boyer et al, 2005; Okumura-Nakanishi et al, 2005), MyoD and MyoG in muscle (Tapscott and Weintraub, 1991; Blais et al, 2005), FOXA2 in hepatocytes (Pani et al, 1992; Lee et al, 2005), and PU.1 in myeloid cells (Chen et al, 1995) (Supplementary Table S5). Importantly, most characterized mammalian transcription factors have been investigated because they play key roles in a particular tissue or cellular process. However, there are transcription factors (such as USF1) that do not occupy their own promoters (Supplementary Table S6), indicating that autoregulation is not simply a universal feature of mammalian transcription factors (Bateman, 1998).
Autoregulatory motifs may be a general feature of transcription factors that play key roles in major cellular processes because they impart stability to regulatory networks (Becskei and Serrano, 2000; Brandman et al, 2005). Master regulators of hepatocytes, like those of other tissues, are known to play both positive and negative regulatory roles (Briancon et al, 2004); both positive and negative feedback loops allow systems to be resistant to noise (Becskei and Serrano, 2000; Rosenfeld et al, 2002; Brandman et al, 2005). These characteristics may be crucial for regulators that are responsible for tissue-specific programs in higher eukaryotes.
Statistical enrichment of multiple binding events and correlation of promoter occupancy with gene expression
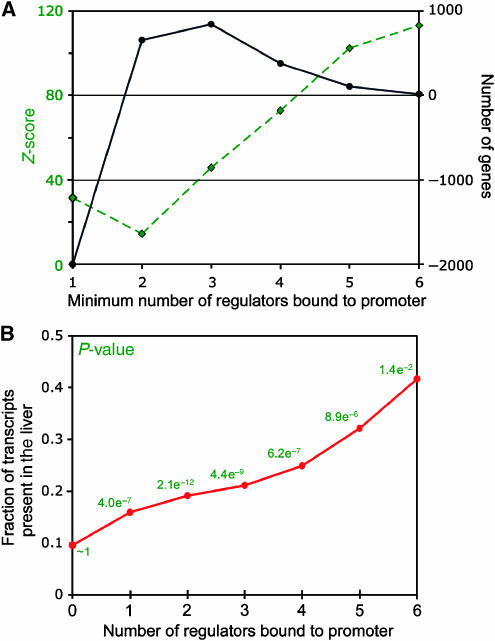
Well-characterized liver promoters are often controlled combinatorially by multiple liver-enriched transcription factors (Cereghini, 1996; Krivan and Wasserman, 2001; Costa et al, 2003; Friedman et al, 2004; Phuc Le et al, 2005; Rada-Iglesias et al, 2005). This prompted us to inspect our genome-wide binding data for evidence of enrichment in combinatorial promoter occupancy. Comparison of the experimental data against randomized binding data (randomized by assuming all factors bind independently) revealed a statistically significant enrichment in the number of promoters bound by two or more transcriptional regulators. The enrichment generally increases with the number of transcriptional regulators bound (Figure 2A, Supplementary Figure S3). For instance, 1188 genes are bound by three or more regulators, whereas we would expect by random chance to see 345 genes cobound by three or more factors (z-score 45.8). Similar analyses of four, five, and six bound regulator combinations yield z-scores that increase with increasing number of bound regulators (Figure 2B). These results are consistent with previous suggestions that liver transcriptional regulation is controlled by multiple transcription factors acting in concert.
Figure 2.
(A) Combinatorial occupancy of promoter regions is significantly enriched, using expectations calculated assuming a binomial distribution of binding events. The difference in numbers between expected and actual binding events is shown (black) with the z-score of the difference (green) (see also Supplementary Figure S3). (B) Comparison of the number of bound transcription factors to gene expression. P-values were determined using a hypergeometric calculation, and transcripts present were established by analysis of publicly available compendiums of tissue-specific gene expression (see also Supplementary Figure S4).
It might be expected that the presence of larger numbers of liver-specific transcription factors at a promoter region would increase the probability that the associated gene was expressed. We tested this hypothesis by classifying genes by the presence or absence of transcripts in the liver (Su et al, 2004), and comparing these categories with the number of regulators bound to corresponding promoters. We found a strong correspondence between these two values (Figure 2B), although the statistical significance drops at higher multi-input motifs due to small numbers of genes in the higher multi-input motifs. This correspondence is independent of the stringency used to call a transcript present (Supplementary Figure S4). Nevertheless, there are transcripts expressed in the absence of binding by these six master regulators, and there are genes bound by multiple factors, which are not appreciably expressed in human hepatocytes. These observations highlight the complexity of the hepatocyte transcriptional program.
Conclusions
We have used a systematic approach to identify the set of human promoters bound by six master regulators that are essential for proper liver development and function. The results show that these regulators form a highly interconnected core circuitry in human hepatocytes, identify the local regulatory network motifs created by regulator–gene interactions, and reveal that autoregulation is a predominant theme among liver-enriched transcription factors. The data support previous predictions that these factors co-occupy many genes to control hepatocyte gene expression, and there exists a direct correspondence between the number of regulators bound to a promoter region and the probability that a gene is expressed. This initial analysis of a portion of the regulatory circuitry in human liver should lay the foundation for future efforts to more fully elucidate the hepatic transcriptional program.
Data Accession Number
Data accession number at ArrayExpress is E-WMIT-9.
Supplementary Material
Supplementary Material
Supplementary Information 1
Supplementary Information 2
Supplementary Information 3
Supplementary Information 4
Supplementary Information 5
Supplementary Information 6
Acknowledgments
We thank the Whitehead Center for Microarray Technology and Whitehead Bioinformatics and Research Computing; E Herbolsheimer, S Strom, and W Gordon for experimental and computational support; FC Wardle, G Gerber, and C Harbison for helpful discussions. This work was supported by NIH Grants DK68766 and DK20595 (GIB), DK070813 (DTO), DK68655, and HG002668 (RAY). RAY and DKG are consultants for Agilent Technologies.
References
- Bailly A, Torres-Padilla ME, Tinel AP, Weiss MC (2001) An enhancer element 6 kb upstream of the mouse HNF4alpha1 promoter is activated by glucocorticoids and liver-enriched transcription factors. Nucleic Acids Res 29: 3495–3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman E (1998) Auto-regulation of eukaryotic transcription factors. Prog Nucleic Acid Res Mol Biol 60: 133–168 [DOI] [PubMed] [Google Scholar]
- Becskei A, Serrano L (2000) Engineering stability in gene networks by auto-regulation. Nature 405: 590–593 [DOI] [PubMed] [Google Scholar]
- Bell GI, Polonsky KS (2001) Diabetes mellitus and genetically programmed defects in beta-cell function. Nature 414: 788–791 [DOI] [PubMed] [Google Scholar]
- Blais A, Tsikitis M, Acosta-Alvear D, Sharan R, Kluger Y, Dynlacht BD (2005) An initial blueprint for myogenic differentiation. Genes Dev 19: 553–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA (2005) Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122: 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O, Ferrell JE, Li R, Meyer T (2005) Interlinked fast and slow positive feedback loops drive reliable cell decisions. Science 310: 496–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briancon N, Bailly A, Clotman F, Jacquemin P, Lemaigre FP, Weiss MC (2004) Expression of the alpha7 isoform of hepatocyte nuclear factor (HNF) 4 is activated by HNF6/OC-2 and HNF1 and repressed by HNF4alpha1 in the liver. J Biol Chem 279: 33398–33408 [DOI] [PubMed] [Google Scholar]
- Cereghini S (1996) Liver-enriched transcription factors and hepatocyte differentiation. FASEB J 10: 267–282 [PubMed] [Google Scholar]
- Chen H, Ray-Gallet D, Zhang P, Hetherington CJ, Gonzalez DA, Zhang DE, Moreau-Gachelin F, Tenen DG (1995) PU.1 (Spi-1) auto-regulates its expression in myeloid cells. Oncogene 11: 1549–1560 [PubMed] [Google Scholar]
- Costa RH, Kalinichenko VV, Holterman AX, Wang X (2003) Transcription factors in liver development, differentiation, and regeneration. Hepatology 38: 1331–1347 [DOI] [PubMed] [Google Scholar]
- Duncan SA, Navas MA, Dufort D, Rossant J, Stoffel M (1998) Regulation of a transcription factor network required for differentiation and metabolism. Science 281: 692–695 [DOI] [PubMed] [Google Scholar]
- Friedman JR, Larris B, Le PP, Peiris TH, Arsenlis A, Schug J, Tobias JW, Kaestner KH, Greenbaum LE (2004) Orthogonal analysis of C/EBPbeta targets in vivo during liver proliferation. Proc Natl Acad Sci USA 101: 12986–12991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, Jennings EG, Zeitlinger J, Pokholok DK, Kellis M, Rolfe PA, Takusagawa KT, Lander ES, Gifford DK, Fraenkel E, Young RA (2004) Transcriptional regulatory code of a eukaryotic genome. Nature 431: 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivan W, Wasserman WW (2001) A predictive model for regulatory sequences directing liver-specific transcription. Genome Res 11: 1559–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CJ, Conley PB, Chen L, Sladek FM, Darnell JE Jr, Crabtree GR (1992) A transcriptional hierarchy involved in mammalian cell-type specification. Nature 355: 457–461 [DOI] [PubMed] [Google Scholar]
- Lee CS, Friedman JR, Fulmer JT, Kaestner KH (2005) The initiation of liver development is dependent on Foxa transcription factors. Nature 435: 944–947 [DOI] [PubMed] [Google Scholar]
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, Zeitlinger J, Jennings EG, Murray HL, Gordon DB, Ren B, Wyrick JJ, Tagne JB, Volkert TL, Fraenkel E, Gifford DK, Young RA (2002) Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298: 799–804 [DOI] [PubMed] [Google Scholar]
- Macisaac KD, Gordon DB, Nekludova L, Odom DT, Schreiber J, Gifford DK, Young RA, Fraenkel E (2006) A hypothesis-based approach for identifying the binding specificity of regulatory proteins from chromatin immunoprecipitation data. Bioinformatics 22: 423–429 [DOI] [PubMed] [Google Scholar]
- Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U (2002) Network motifs: simple building blocks of complex networks. Science 298: 824–827 [DOI] [PubMed] [Google Scholar]
- Montminy M, Koo SH, Zhang X (2004) The CREB family: key regulators of hepatic metabolism. Ann Endocrinol (Paris) 65: 73–75 [DOI] [PubMed] [Google Scholar]
- Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, Volkert TL, Schreiber J, Rolfe PA, Gifford DK, Fraenkel E, Bell GI, Young RA (2004) Control of pancreas and liver gene expression by HNF transcription factors. Science 303: 1378–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura-Nakanishi S, Saito M, Niwa H, Ishikawa F (2005) Oct-3/4 and Sox2 regulate Oct-3/4 gene in embryonic stem cells. J Biol Chem 280: 5307–5317 [DOI] [PubMed] [Google Scholar]
- Pajukanta P, Lilja HE, Sinsheimer JS, Cantor RM, Lusis AJ, Gentile M, Duan XJ, Soro-Paavonen A, Naukkarinen J, Saarela J, Laakso M, Ehnholm C, Taskinen MR, Peltonen L (2004) Familial combined hyperlipidemia is associated with upstream transcription factor 1 (USF1). Nat Genet 36: 371–376 [DOI] [PubMed] [Google Scholar]
- Pani L, Quian XB, Clevidence D, Costa RH (1992) The restricted promoter activity of the liver transcription factor hepatocyte nuclear factor 3 beta involves a cell-specific factor and positive autoactivation. Mol Cell Biol 12: 552–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phuc Le P, Friedman JR, Schug J, Brestelli JE, Parker JB, Bochkis IM, Kaestner KH (2005) Glucocorticoid receptor-dependent gene regulatory networks. PLoS Genet 1: e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Wallerman O, Koch C, Ameur A, Enroth S, Clelland G, Wester K, Wilcox S, Dovey OM, Ellis PD, Wraight VL, James K, Andrews R, Langford C, Dhami P, Carter N, Vetrie D, Ponten F, Komorowski J, Dunham I, Wadelius C (2005) Binding sites for metabolic disease related transcription factors inferred at base pair resolution by chromatin immunoprecipitation and genomic microarrays. Hum Mol Genet 14: 3435–3447 [DOI] [PubMed] [Google Scholar]
- Rosenfeld N, Elowitz MB, Alon U (2002) Negative auto-regulation speeds the response times of transcription networks. J Mol Biol 323: 785–793 [DOI] [PubMed] [Google Scholar]
- Rubins NE, Friedman JR, Le PP, Zhang L, Brestelli J, Kaestner KH (2005) Transcriptional networks in the liver: hepatocyte nuclear factor 6 function is largely independent of Foxa2. Mol Cell Biol 25: 7069–7077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen-Orr SS, Milo R, Mangan S, Alon U (2002) Network motifs in the transcriptional regulation network of Escherichia coli. Nat Genet 31: 64–68 [DOI] [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, Cooke MP, Walker JR, Hogenesch JB (2004) A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA 101: 6062–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapscott SJ, Weintraub H (1991) MyoD and the regulation of myogenesis by helix–loop–helix proteins. J Clin Invest 87: 1133–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieffry D, Huerta AM, Perez-Rueda E, Collado-Vides J (1998) From specific gene regulation to genomic networks: a global analysis of transcriptional regulation in Escherichia coli. Bioessays 20: 433–440 [DOI] [PubMed] [Google Scholar]
- Tronche F, Ringeisen F, Blumenfeld M, Yaniv M, Pontoglio M (1997) Analysis of the distribution of binding sites for a tissue-specific transcription factor in the vertebrate genome. J Mol Biol 266: 231–245 [DOI] [PubMed] [Google Scholar]
- Zaret KS (2002) Regulatory phases of early liver development: paradigms of organogenesis. Nat Rev Genet 3: 499–512 [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, Simon I, Harbison CT, Hannett NM, Volkert TL, Fink GR, Young RA (2003) Program-specific distribution of a transcription factor dependent on partner transcription factor and MAPK signaling. Cell 113: 395–404 [DOI] [PubMed] [Google Scholar]
- Zhang X, Odom DT, Koo SH, Conkright MD, Canettieri G, Best J, Chen H, Jenner R, Herbolsheimer E, Jacobsen E, Kadam S, Ecker JR, Emerson B, Hogenesch JB, Unterman T, Young RA, Montminy M (2005) Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc Natl Acad Sci USA 102: 4459–4464 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Supplementary Information 1
Supplementary Information 2
Supplementary Information 3
Supplementary Information 4
Supplementary Information 5
Supplementary Information 6