Abstract
HIV infection is initiated by the selective interaction between the cellular receptor CD4 and gp120, the external envelope glycoprotein of the virus. We used analytical ultracentrifugation, titration calorimetry, and surface plasmon resonance biosensor analysis to characterize the assembly state, thermodynamics, and kinetics of the CD4-gp120 interaction. The binding thermodynamics were of unexpected magnitude; changes in enthalpy, entropy, and heat capacity greatly exceeded those described for typical protein-protein interactions. These unusual thermodynamic properties were observed with both intact gp120 and a deglycosylated and truncated form of gp120 protein that lacked hypervariable loops V1, V2, and V3 and segments of its N and C termini. Together with previous crystallographic studies, the large changes in heat capacity and entropy reveal that extensive structural rearrangements occur within the core of gp120 upon CD4 binding. CD spectral studies and slow kinetics of binding support this conclusion. These results indicate considerable conformational flexibility within gp120, which may relate to viral mechanisms for triggering infection and disguising conserved receptor-binding sites from the immune system.
Entry of enveloped viruses into cells requires transformation of the protective envelope into a fusion-competent state. In the case of the human immunodeficiency virus (HIV-1), infection is initiated by the selective interaction between the viral exterior envelope glycoprotein, gp120, and receptors on the target cell, CD4, and obligatory chemokine receptors (CCR5 or CXCR4). Accumulating biochemical and structural evidence indicates that, in addition to contributing to viral attachment, CD4 triggers conformational alterations in the HIV envelope that promote recognition of the chemokine receptors and ultimately lead to membrane fusion (1, 2). Evidence of CD4-induced conformational changes includes enhanced protease sensitivity in gp120 variable loops (3) and release of gp120 from virus and virus-infected cells (4, 5), as well as exposure or formation of the chemokine receptor site (1, 2) and of the epitopes for neutralizing antibodies that can block chemokine receptor binding (1, 2, 6, 7). By their nature, these biochemical studies primarily probe the peripheral regions of gp120 and indicate that CD4-induced movement of variable loops occurs.
Recently, a structure was solved for a truncated core gp120 protein in a ternary complex with soluble CD4 and a Fab fragment of the neutralizing antibody 17b (8). In this complex, gp120 is organized into an inner and outer domain connected by a bridging sheet. Several elements of this structure appear to depend on association with CD4 for their stabilization. In particular, the extended conformation of the bridging sheet, which constitutes key elements of the chemokine receptor-binding site, is stabilized by direct contacts with CD4. In contrast, the structure of CD4 (D1D2) (9, 10) is virtually unchanged in the presence of gp120.
Although the crystallographic data provide an important snapshot of the structures involved in the CD4-gp120 complex, a more complete understanding of the interaction requires a description of the biophysical properties that drive complex formation. Using a series of biophysical experiments, we have examined the binding properties of soluble CD4 to soluble recombinant gp120 proteins. Our thermodynamic studies demonstrate that complex formation is accompanied by an unusually large bonding energy that is exploited to drive a substantial reordering of gp120 structure. Comparable results with a truncated gp120 protein reveal that structural rearrangement occurs within the core of gp120. Conformational flexibility of gp120 has significant implications regarding viral infection, immune evasion, and gp120-based subunit vaccine design.
Methods
Preparation of Proteins.
Soluble CD4 was produced in Chinese hamster ovarian cells (11) whereas gp120 proteins (WD61, BH10, and HXBc2 core) were from Drosophila Schneider 2 lines (12). WD61 and BH10 were used as glycosylated full-length gp120 proteins. HXBc2 core gp120 had deletions of 52 and 19 residues from the amino and carboxy termini, respectively, Gly-Ala-Gly tripeptide substitutions for 67 V1V2 loop residues and 32 V3 loop residues, and the removal of all sugar groups except for the peptide-proximal N-acetylglucosamine and fucose residues (13).
Mass Spectrometry.
Matrix-assisted laser desorption mass spectrometry data were measured with a time-of-flight instrument from Vestec LaserTec Research (Houston). Samples were prepared for analysis by mixing in phosphate buffer (pH 7.8) with sinapinic acid (saturated solution in 33% CH3CN/0.1% trifluoroacetic acid) to a final concentration of 1 pmol/μl. Phosphorylase B (97,219 Da) from rabbit muscle (Sigma) was included as an internal calibrant.
Analytical Ultracentrifugation.
Sedimentation equilibrium experiments were performed on a Beckman XL-A analytical ultracentrifuge as described (14). Conditions were 20 mM Na2HPO4, 200 mM NaCl (pH 7.0), and a range of temperature from 15 to 40°C. Equilibrium was defined by no change in protein distribution spectra acquired 4 h apart. Partial specific volumes, calculated from amino acid and carbohydrate compositions (15) for CD4, full-length glycosylated gp120, and core gp120 were 0.738, 0.695, and 0.716 ml⋅g−1, respectively.
Isothermal Titration Calorimetry.
Isothermal titration calorimetry measurements were carried out by using a Microcal (Amherst, MA) MCS instrument with approximately 4 μM gp120 in the cell (16). CD4, full-length gp120 WD61, and core gp120 concentrations were determined by absorbance at 280 nm from extinction coefficients of 1.4, 0.74, and 1.2 ml⋅cm−1⋅mg−1, respectively. Data were analyzed with Microcal origin software using a single-site binding model. Titrations carried out in phosphate and Tris buffers yielded identical binding enthalpy change (ΔH) values, indicating that linked protonation reactions do not contribute to the observed CD4-gp120 ΔH under these analytical conditions.
The amount of apolar (ΔASAap) and polar (ΔASApol) surface area buried upon binding were calculated from the empirical relationships to the binding heat capacity (ΔC°) and enthalpy (ΔH°) changes (17, 18) as: ΔC° = 0.45⋅ΔASAap − 0.26⋅ΔASApol; ΔH°60 = −8.44⋅ΔASAap + 31.4⋅ΔASApol, where ΔH°60 is the binding enthalpy change at 60°C.
The number of residues that become ordered during binding, Rth, were predicted from the experimental association entropy change ΔS°assoc of the binding reaction (19) as: ΔS°assoc = ΔS°HE + ΔS°rt + ΔS°other, where ΔS°HE and ΔS°rt are entropy change contributions caused by the hydrophobic effect and loss of rotational and translational degrees of freedom, respectively. ΔS°HE is estimated by ΔS°HE = 1.35 ΔC ° ln(Ts/386), where Ts is the characteristic temperature for which ΔS°assoc = 0 for a given interaction. ΔS°rt has been deduced empirically to be −50 e.u. for binary protein-protein interactions. The remaining term, ΔS°other, has been shown, based on thermodynamic and high-resolution structural data, to relate to Rth as Rth = ΔS°other/−5.6 entropic units (19).
Circular Dichroism.
CD measurements were done on a JASCO J-710 spectropolarimeter in 100 mM NaCl and 3 mM Na2HPO4 (pH 7.4) by using a 0.1-cm path length water-jacketed cell. Several scans were made at a scan rate of 20 nm/min, a bandwidth of 1 nm, and a response time of 2 s. All spectra were buffer-corrected and normalized to difference molar extinction coefficient units as Δɛ = θ/(32.98·l·C), where θ is the spectral ellipticity, l is pathlength in cm, and C is the molarity of peptide bonds in the sample.
Surface Plasmon Resonance Biosensor Analysis.
Surface plasmon resonance biosensor data were collected on a BIACORE 2000 optical biosensor (Biacore AB, Uppsala). For direct binding comparisons, BH10 gp120, core gp120, and deglycosylated core gp120 were immobilized onto separate flow cells within the same sensor chip using amine coupling chemistry (20). Flow cell four was left blank as a control for nonspecific binding and refractive index changes. With the instrument operating in parallel sensing mode, CD4 was injected over all four flow cells at a concentration of 1 μM and at a flow rate of 100 μl/min. To perform a kinetic study, CD4 was immobilized onto a low charge (B1) sensor chip using carbodiimide coupling (21). Coupling in this manner produced a homogeneous surface because the glycosylation sites in domains 3 and 4 of CD4 are distant from the gp120 binding site in domain 1. The binding capacity of the CD4 surface was kept low to avoid mass transport effects and steric hindrance. Sensor data were prepared for kinetic analysis by subtracting binding responses collected from a blank reference surface. The association and dissociation phase data were fitted simultaneously to a single-site binding model by using the nonlinear data analysis program clamp (22, 23).
Results
Assembly State of CD4-gp120 Interaction.
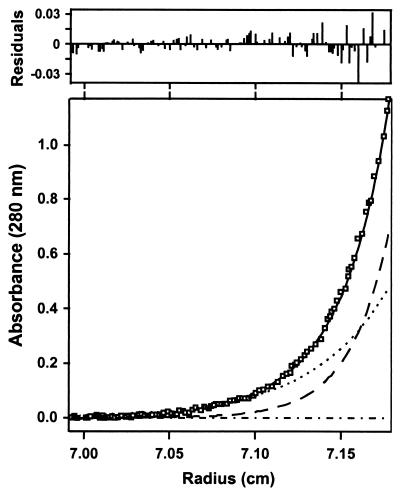
Our analysis of the CD4-gp120 binding interaction involved binding of soluble CD4 (external domains D1-D4) with recombinant gp120 proteins derived from HIV-1 isolates WD61 and BH10, and a loop-deleted and termini-truncated HXBc2 gp120 protein referred to as “core gp120” (8). A prerequisite to interpreting binding data for these reactions was to characterize the assembly states of the reactants and product. This was done by comparing their molecular masses determined by matrix-assisted laser desorption ionization mass spectrometry to those obtained in solution using analytical ultracentrifugation. Fig. 1 shows sedimentation equilibrium data for the complex of CD4 and core gp120. CD4 is seen to form a 1:1 complex with core gp120 and shows no evidence of higher-order aggregation, consistent with the crystal structure of the core gp120 protein complexed with CD4(D1D2) (8). Similar studies on the various proteins from 15–40°C showed that both core gp120 and CD4 are monomeric under the conditions studied and that the complexes are formed in 1:1 molar ratios (Table 1).
Figure 1.
Analysis of a mixture of CD4 and core gp120 by analytical ultracentrifugation. The main figure shows the sedimentation equilibrium data and the best-fit curve (solid) for a mixture of CD4 and core gp120. Under the fit, the theoretical sum is deconvoluted into contributions from the core gp120-CD4 complex (long dashed line), excess CD4 (short dashed line), and baseline (dashed-dotted line).
Table 1.
Assembly state of the CD4-gp120 complex by mass by matrix-assisted laser desorption ionization (MALDI) and analytical ultracentrifugation (AUC)
| Mass by
|
||
|---|---|---|
| MALDI | AUC | |
| CD4 | 44,600 | 45,000 ± 300 |
| WD61 full-length gp120 | 99,600 | 97,000 ± 300 |
| Core gp120 | 39,000 | 35,000 ± 1,000 |
| CD4-WD61 full-length gp120 complex | 144,100* | 138,000 ± 800 |
| CD4-core gp120 complex | 83,600* | 83,000 ± 200 |
Predicted mass for 1:1 complex.
Thermodynamics of CD4-gp120 Interaction.
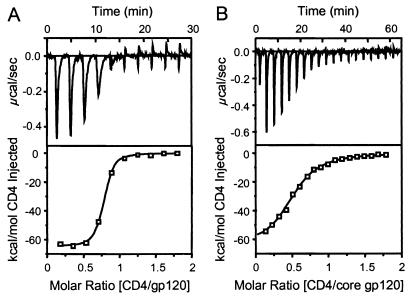
The thermodynamics of the CD4-gp120 interaction (Table 2) were measured by titration microcalorimetry. Fig. 2 shows the titration data for CD4 binding to both full-length and core gp120 proteins. A striking feature of both interactions is the enormously favorable binding enthalpy (ΔH ° ≈ −63 kcal/mol), which demonstrates that a large number of bonding interactions (e.g., hydrogen bonds and van der Waals interactions) occur during complex formation. However, the binding entropy change for both proteins is also extremely large (−TΔS ° ≈ 52 kcal/mol), but unfavorable, demonstrating a substantial loss in the degrees of freedom upon binding. Thus, the modest KD37°C values of 5 nM and 190 nM observed for full-length and core gp120, respectively, result from a balance between highly favorable bonding interactions and highly unfavorable molecular ordering. The 40-fold difference in affinity between the gp120 proteins, although experimentally distinguishable, reflects a difference of only 2.2 kcal/mol in Gibbs free energy change and is a small perturbation to the overall energetics. The similar thermodynamics for core and full-length gp120 proteins indicates they bind with similar overall mechanisms.
Table 2.
Thermodynamics of the CD4-gp120 interaction at 37°C by isothermal titration calorimetry
| ΔG°, kcal/mol | ΔH°, kcal/mol | −TΔS°, kcal/mol | ΔC°, kcal/mol/K | KD, nM | |
|---|---|---|---|---|---|
| WD61 full-length gp120 | −11.8 ± 0.3 | −63 ± 3 | 51.2 ± 3 | −1.2 ± 0.2 | 5 ± 3 |
| Core gp120 | −9.5 ± 0.1 | −62 ± 3 | 52.5 ± 3 | −1.8 ± 0.4 | 190 ± 30 |
Figure 2.
CD4-gp120 thermodynamics. Calorimetry data for the titration of WD61 full-length (A) and core (B) gp120 with CD4 in 10 mM Na2HPO4, 200 mM NaCl, and 0.5 mM EDTA (pH 7.4). The top panels show raw data in power versus time. The area under each spike is proportional to the heat produced at each injection. The lower panels show integrated areas normalized to the number of moles of CD4 injected at each injection step. Best-fit curves represent binding enthalpy changes of −63 and −62 kcal/mol CD4 for full-length and core gp120, respectively. Equilibrium binding KD values were determined as 5 nM and 190 nM, respectively.
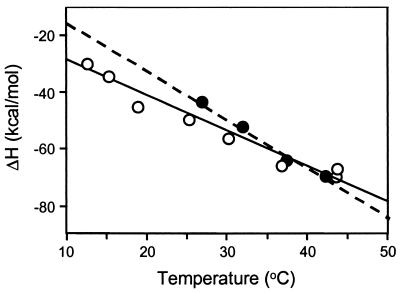
Further evidence of a common CD4 binding mechanism for full-length and core gp120 comes from the similar temperature dependencies of their binding enthalpies (Fig. 3). The slopes of the lines in Fig. 3 yield ΔC° values of −1.2 ± 0.2 and −1.8 ± 0.4 kcal/mol/deg for full-length and core gp120, respectively. These values are significantly greater than the −0.2 to −0.7 kcal/mol/deg typically observed for protein-protein interactions (24), suggesting that extensive apolar surface area is buried upon CD4-gp 120 complexation (25).
Figure 3.
Temperature dependence of the CD4 binding enthalpy change for WD61 full-length gp120 (open circles) and HXBc2 core gp120 (filled circles). Slopes of the plots, obtained from the best-fit lines shown, yielded the binding heat capacity changes (ΔC°) in Table 2.
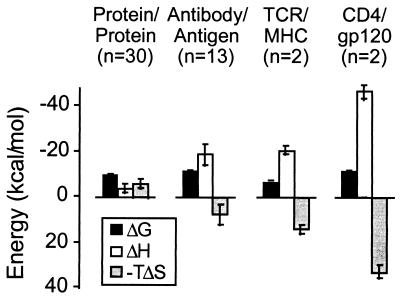
The magnitudes of the ΔH° and −TΔS° terms for the CD4-gp120 interaction are unusually large relative to other binary protein interactions (24, 26). Typical protein interactions characterized to date show a small favorable entropy of association (Fig. 4), whereas the entropy for both the antibody and T cell receptor interactions is substantially more unfavorable. As discussed by Wilcox et al. (26), this unfavorable entropy most likely results from reduction of conformational flexibility in the antibody or T cell receptor upon complex formation. Constriction of water molecules at the interfaces can also contribute unfavorably to the binding entropy change of protein interactions. In the case of the CD4-gp120 complex, only 15 water molecules are within the van der Waals radius + 1 Å of both molecules (8). This value is near the average for the 36 x-ray structures that have been analyzed (27) and suggests that the exceptionally large ΔH° and −TΔS° values for the CD4-gp120 interaction do not arise from entrapment of water. Instead, these values point to substantial conformational rearrangement during binding.
Figure 4.
Protein-protein binding thermodynamics at 25°C for several classes of protein interactions. Sample numbers here are n = 30, 13, 2, and 2 for protein-protein, antibody-antigen, T cell receptor/MHC peptide, and CD4-gp120 interactions, respectively. Average contributions to the binding free energy from enthalpic and entropic driving forces for protein-protein binding reactions are shown as bars. All values except those for CD4-gp120 were taken from ref. 24.
Interpretation of Binding Thermodynamics.
The extent of structural rearrangement that occurs during complex formation can be estimated from relationships that correlate structure with binding and folding energetics (19, 28). One approach is derived from the correlation that exists between protein folding thermodynamics and the amount of water-accessible surface area buried during the reaction (17, 25). By using empirical relationships developed by Freire and colleagues (see Methods), the large ΔH° and ΔC° values observed for the CD4-gp120 interaction infer that 10,000 (±2,000) Å2 of surface area are buried during complex formation. This value is much greater than the 1,500 Å2 observed at the CD4-gp120 interface (8), suggesting that complex formation includes burial of surface area outside the observed binding interface. In an independent approach, Spolar and Record have related the binding ΔC° and ΔS° of interactions to the number of residues that reorganize during binding by comparing thermodynamics to high-resolution structures (19). Application of this method to the CD4-gp120 binding thermodynamics suggests the reorganizing of 94 (±15) and 126 (±30) residues upon CD4 binding to full-length and core gp120, respectively. Within experimental error, these values are the same and are among the largest reported for protein-protein interactions (19).
CD of CD4-gp120 Complex.
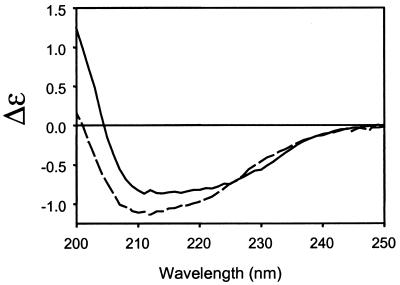
Given the strong indication of a conformational change from the CD4-gp120 binding thermodynamics, circular dichroism (CD) was used to investigate whether changes in secondary structure could be observed during complex formation. As shown in Fig. 5, a substantial change in the CD spectrum was observed for the experimental mixture of CD4 and gp120 in comparison to the predicted mixture. A similar, but smaller, change in the spectrum was observed for CD4 binding to core gp120 (data not shown). The large positive increase in ellipticity at 200 nm is consistent with a reduction in random coil (29) and suggests that complexation induces structural rigidity.
Figure 5.
Secondary structural changes detected by circular dichroism spectroscopy. CD spectra for a stoichiometric mixture of CD4 (D1D2) with WD61 full-length gp120 at 5 μM shown as difference molar extinction coefficient versus wavelength. Experimental data (solid) are compared with theoretical mixtures computed as the sum of the individual component spectra (dashed). Both full-length and core gp120 proteins exhibited this spectral shift over the temperature range 12–37°C. The shift in spectral intensity to higher wavelengths is consistent with an increase in secondary structure upon binding.
Kinetic Analysis of CD4-gp120 Interaction.
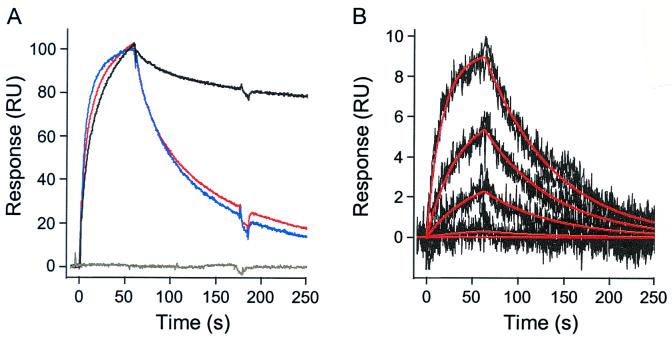
The binding kinetics of soluble CD4 to the full-length BH10 and core HXBc2 gp120 proteins were compared by surface plasmon resonance (Fig. 6). There is little difference in the association rates, demonstrating that both forms of gp120 bind to CD4 through similar forward reaction mechanisms (Table 3). In agreement with previous studies (30), the association rate constant for these gp120 proteins is slow (ka37°C ≈ 6 × 104 M−1⋅s−1), which is consistent with conformational adaptation during binding, as was suggested by our previous observation that mutation of the critical phenylalanine 43 in the gp120 binding site of CD4 primarily impacted this association rate (30). CD4 dissociates more rapidly from core than from full-length gp120. Because the HXBc2 core protein differs from the corresponding region in full-length BH10 gp120 at only four residues, the 9.3-fold increase in dissociation rate for core gp120 likely stems from a loss of favorable CD4- or self-contacts in the deleted segments. Together, the kinetics predict a 10-fold decrease in affinity for core versus full-length gp120 (220 vs. 22 nM, respectively, at 37°C) (Fig. 6B), which is in agreement with the affinities determined by titration calorimetry. Glycosylation had little effect on CD4 binding kinetics (Fig. 6A) and is consistent with the observation that the CD4 binding site on core gp120 is devoid of residues modified by carbohydrate addition (8).
Figure 6.
Kinetic analysis of the CD4-gp120 interaction. (A) Normalized binding responses for CD4 (1 μM) over a full-length BH10 gp120 (black), glycosylated HXBc2 core gp120 (red), core-gp120 (blue), and an unmodified sensor surface (gray). Responses were normalized to 100% bound at the start of the dissociation phase. (B) Kinetic data set collected for core gp120 binding to a CD4 surface. Core gp120 was injected over the CD4 surface at concentrations of 975, 325, 108, 36, and 0 nM. The injections were done at a flow rate of 100 μl/min in 10 mM Na2HPO4, 200 mM NaCl, and 0.5 mM EDTA (pH 7.4) at 37°C. Each injection was replicated to demonstrate that the binding responses were reproducible. The experimental data (black lines) were globally fit to a single site interaction model (red lines) to determine the binding kinetics.
Table 3.
Kinetic rate constants at 37°C by surface plasmon resonance
| ka, M−1⋅s−1 | kd, s−1 | KD, nM | |
|---|---|---|---|
| BH10 full-length gp120 | 6.72 × 104 ± 1 × 102 | 1.5 × 10−3 ± 4 × 10−4 | 22 ± 6 |
| Core gp120 | 6.27 × 104 ± 9 × 102 | 1.4 × 10−2 ± 2 × 10−3 | 220 ± 40 |
Discussion
The unusual nature of the CD4 binding thermodynamics of gp120 indicates that a substantial conformational change accompanies this interaction. The comparable observations with the core gp120 protein, and the invariance of the CD4 protein structure between unbound (9, 10) and bound states (8), demonstrate that these changes are associated with the core structural region of gp120 (6, 31). The slow kinetics of binding are also consistent with structural adaptation during binding, as are the significant changes in CD spectral properties upon complex formation. Although localized conformational changes have been demonstrated previously (1–7), they have been based on methods that probe the periphery of gp120, rather than the restructuring of the core.
The present findings reinforce the importance of the crystallographic analysis of the core gp120-CD4 structure as a model for the CD4-gp210 interaction. The structural and biophysical data dovetail to provide unique insights into the mechanism of complex formation. From the structural viewpoint, the CD4-gp120 interface is suggestive of induced conformational changes because CD4 binds at the nexus of three different gp120 domains, with the gp120 binding surface comprising six discrete sequence elements (8).
The thermodynamic data imply that much of the core of gp120 undergoes structural rearrangement during complex formation with the consequent burial of extensive surface area outside the interface. Our observations can be fitted either by a model in which 100 residues reorganize or by a model in which 10,000 Å2 are buried. Inspection of the co-crystal structure in the context of the present findings suggests that CD4 binding drives the folding of the bridging sheet, as well as the pulling together of the bridging sheet, inner domain, and outer domains of gp120. The bridging sheet encompasses 54 residues, and its folding would account for about half of the structural rearrangement implicated from the thermodynamics. The remainder of the implicated structural change could be well explained by the amount of surface area that is buried when loosely associated domains are pulled together by CD4. Surface areas buried within CD4-complexed gp120 from the inner domain (1,311 Å2), the outer domain (1,103 Å2) and the folded bridging sheet (591 Å2), plus the buried area at the gp120-CD4 interface (1,544 Å2), account for a total over 4,500 Å2 in area that might be free in the absence of CD4. The thermodynamic, kinetic, and spectral findings reported here are consistent with complex formation being a process of induced fit, but do not exclude the possibility that CD4 may constrain gp120 into one of two or more conformations that may preexist in the unbound state.
HIV is a tenacious infectious agent, able to evade and overcome host immune defenses. The gp120 protein that is shed from virus and infected cells is strongly immunogenic but elicits predominantly non-neutralizing antibodies. This “decoy” strategy may help the virus evade the humoral immune response (32–35). The flexibility of the gp120 protein revealed in our studies may relate to the inefficiency with which monomeric gp120 elicits broadly neutralizing antibodies. The epitopes of most broadly neutralizing antibodies map to discontinuous segments of gp120 that overlap the receptor-binding regions, which are on a contiguous surface that spans the interface between domains of core gp120 (36). In conjunction with structural analysis, our results indicate that this region is disordered or in a substantially different conformation in the absence of CD4. This masking or instability of the CD4- and chemokine-receptor-binding sites on gp120 may explain the poor immunogenicity of these regions on gp120. This insight suggests that unmasking and conformational stabilization of the receptor-binding regions on gp120 may be useful in vaccine development.
How the flexibility in monomeric gp120 relates to the state of gp120 in the viral envelope complex is not yet clear. The conformation of gp120 may be constrained in its assembly with gp41 into a trimeric complex. Such constraints as do exist must not be highly restrictive, however. Many conformationally sensitive monoclonal antibodies recognize both soluble and membrane-associated forms of gp120 (37), and CD4 binding induces recognition of the CCR5 chemokine co-receptor similarly in both states (1). On the other hand, primary HIV isolates are differentiated from laboratory-adapted isolates in reduced affinity for CD4 whereas irrespectively the monomeric gp120 forms have comparable CD4 affinities (38). These properties correlate with a reduced susceptibility to neutralizing antibodies (39). It may be that oligomeric contacts or steric hindrance restrict conformational changes differently in the trimers of primary isolates than in those of laboratory-adapted strains. Changes at only a few key residues can have dramatic effects, as found with mutations selected to enable CD4-independent infection in cell culture (40, 41).
Besides the implications of flexibility for avoidance of immune responses by HIV, CD4-induced conformational rigidity of the kind observed here may also have direct importance for the mechanism of viral entry into targeted cells. Trimeric gp120:gp41 complexes on virions are stable and inert; however, once fixed in a chemokine-receptor binding conformation by CD4, the association of gp41 with gp120 is metastable (4, 5) and predisposed to fusion (42, 43). Binding to CD4 brings the trimeric complex into proximity with the chemokine receptor, and it properly times the induction of metastability in gp41 for the triggering of fusion between the viral envelope and the cellular membrane. Thus, conformational flexibility in gp120 provides an attractive hypothesis for understanding of both immune evasion and cell invasion.
Acknowledgments
We thank Ian Brooks and Mark Hemling for mass spectrometry measurements and Shahzad Majeed for help with deglycosylation and purification of core gp120. This work was supported by grants from the National Institutes of Health (AI 31783, AI 39420, and AI 40895) and by Center for AIDS Research grants to the Dana-Farber Cancer Institute (AI 28691) and to Columbia-Rockefeller (AI 42848). The Dana-Farber and Huntsman Cancer Institutes are recipients of Cancer Center grants from the National Institutes of Health (CA 06516 and CA 42014, respectively). This work was made possible by gifts from the late William McCarty-Cooper, the G. Harold and Leila Y. Mathers Charitable Foundation, the Aaron Diamond Foundation, the Friends 10, and Douglas and Judi Krupp. R.W. was a fellow of the American Foundation for AIDS research. P.D.K. was a recipient of a Burroughs Wellcome career development award.
References
- 1.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardins E, Newman W, et al. Nature (London) 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 2.Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore J P, Katinger H. J Virol. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sattentau Q J, Moore J P. J Exp Med. 1991;174:407–415. doi: 10.1084/jem.174.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moore J P, McKeating J A, Weiss R A, Sattentau Q J. Science. 1990;250:1139–1142. doi: 10.1126/science.2251501. [DOI] [PubMed] [Google Scholar]
- 5.Hart T K, Kirsh R, Ellens H, Sweet R W, Lambert D M, Petteway S R, Jr, Leary J, Bugelski P J. Proc Natl Acad Sci USA. 1991;88:2189–2193. doi: 10.1073/pnas.88.6.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thali M, Moore J P, Furman C, Charles M, Ho D D, Robinson J, Sodroski J. J Virol. 1993;67:3978–3988. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang W, Canziani G, Plugariu C, Wyatt R, Sodroski J, Sweet R, Kwong P, Hendrickson W, Chaiken I. Biochemistry. 1999;38:9405–9416. doi: 10.1021/bi990654o. [DOI] [PubMed] [Google Scholar]
- 8.Kwong P D, Wyatt R, Robinson J, Sweet R W, Sodroski J, Hendrickson W A. Nature (London) 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ryu S E, Kwong P D, Truneh A, Porter T G, Arthos J, Rosenberg M, Dai X P, Xuong N H, Axel R, Sweet R W, Hendrickson W A. Nature (London) 1990;348:419–426. doi: 10.1038/348419a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J H, Yan Y W, Garrett T P, Liu J H, Rodgers D W, Garlick R L, Tarr G E, Husain Y, Reinherz E L, Harrison S C. Nature (London) 1990;348:411–418. doi: 10.1038/348411a0. [DOI] [PubMed] [Google Scholar]
- 11.Deen K C, McDougal S, Inacker R, Folena-Wasserman G, Arthos J, Rosenberg J, Maddon P J, Axel R, Sweet R W. Nature (London) 1988;331:82–84. doi: 10.1038/331082a0. [DOI] [PubMed] [Google Scholar]
- 12.Culp J S, Johansen H, Hellmig B, Beck J, Matthews T J, Delers A, Rosenberg M. Bio/Technology. 1991;9:173–177. doi: 10.1038/nbt0291-173. [DOI] [PubMed] [Google Scholar]
- 13.Kwong P D, Wyatt R, Desjardins E, Robinson J, Culp J S, Hellmig B D, Sweet R W, Sodroski J, Hendrickson W A. J Biol Chem. 1999;274:4115–4123. doi: 10.1074/jbc.274.7.4115. [DOI] [PubMed] [Google Scholar]
- 14.Hensley P. Curr Biol. 1996;4:367–373. [Google Scholar]
- 15.Laue T M, Shah B D, Ridgeway T M, Pelletier S L. In: Analytical Ultracentrifugation in Biochemistry and Polymer Science. Harding S E, Rowe A J, Horton J C, editors. Cambridge, U.K.: R. Soc. Chem.; 1992. pp. 90–125. [Google Scholar]
- 16.Wiseman T, Williston S, Brandts J F, Lin L N. Anal Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 17.Xie D, Freire E. Proteins. 1994;19:291–301. doi: 10.1002/prot.340190404. [DOI] [PubMed] [Google Scholar]
- 18.Murphy K P, Freire E. Adv Protein Chem. 1992;43:313–361. doi: 10.1016/s0065-3233(08)60556-2. [DOI] [PubMed] [Google Scholar]
- 19.Spolar R S, Record M T. Science. 1994;263:777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- 20.Johnsson B, Lofas S. Anal Biochem. 1991;198:268–277. doi: 10.1016/0003-2697(91)90424-r. [DOI] [PubMed] [Google Scholar]
- 21.O'Shannessy D J, Brigham-Burke M, Peck K. Biochemistry. 1992;205:132–136. doi: 10.1016/0003-2697(92)90589-y. [DOI] [PubMed] [Google Scholar]
- 22.Myszka D G, Morton T A. Trends Biochem Sci. 1998;23:149–150. doi: 10.1016/s0968-0004(98)01183-9. [DOI] [PubMed] [Google Scholar]
- 23.Morton T A, Myszka D G. Methods Enzymol. 1998;295:268–294. doi: 10.1016/s0076-6879(98)95044-3. [DOI] [PubMed] [Google Scholar]
- 24.Stites W E. Chem Rev. 1997;97:1233–1250. doi: 10.1021/cr960387h. [DOI] [PubMed] [Google Scholar]
- 25.Spolar R S, Ha J-H, Record M T. Proc Natl Acad Sci USA. 1989;86:8382–8385. doi: 10.1073/pnas.86.21.8382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilcox B E, Gao G F, Wyer J R, Ladbury J E, Bell J I, Jakobsen B K, van der Merwe P A. Immunity. 1999;10:357–365. doi: 10.1016/s1074-7613(00)80035-7. [DOI] [PubMed] [Google Scholar]
- 27.Lo Conte L, Chothia C, Janin J. J Mol Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 28.Freire E. Annu Rev Biophys Biomol Struct. 1995;24:141–165. doi: 10.1146/annurev.bb.24.060195.001041. [DOI] [PubMed] [Google Scholar]
- 29.Woody R W. Methods Enzymol. 1995;246:34–71. doi: 10.1016/0076-6879(95)46006-3. [DOI] [PubMed] [Google Scholar]
- 30.Wu H, Myszka D G, Tendian S W, Brouillette C G, Sweet R W, Chaiken I M, Hendrickson W A. Proc Natl Acad Sci USA. 1996;93:15030–15035. doi: 10.1073/pnas.93.26.15030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wyatt R, Moore J, Accola M, Desjardins E, Robinson J, Sodroski J. J Virol. 1995;69:5723–5733. doi: 10.1128/jvi.69.9.5723-5733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore J P, Sattentau Q J, Wyatt R, Sodroski J. J Virol. 1994;68:469–484. doi: 10.1128/jvi.68.1.469-484.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moore J P, Sodroski J. J Virol. 1996;70:1863–1872. doi: 10.1128/jvi.70.3.1863-1872.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burton D R, Montefiori D C. AIDS. 1997;11, Suppl. A:S87–S98. [PubMed] [Google Scholar]
- 35.Burton D R, Moore J P. Nat Med. 1998;4:495–498. doi: 10.1038/nm0598supp-495. [DOI] [PubMed] [Google Scholar]
- 36.Wyatt R, Kwong P D, Desjardins E, Sweet R W, Robinson J, Hendrickson W A, Sodroski J G. Nature (London) 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 37.Sattentau Q J, Moore J P. J Exp Med. 1995;182:185–196. doi: 10.1084/jem.182.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brightly D W, Rosenberg M, Chen I S, Ivey-Hoyle M. Proc Natl Acad Sci USA. 1991;88:7802–7805. doi: 10.1073/pnas.88.17.7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moore J P, Ho D D. AIDS. 1995;9, Suppl. A:5117–5136. [Google Scholar]
- 40.Kolchinsky P, Mirzabekov T, Farzan M, Kiprilov E, Cayabyab M, Mooney L, Choe H, Sodroski J. J Virol. 1999;73:8120–8126. doi: 10.1128/jvi.73.10.8120-8126.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoffman T L, LaBranche C C, Zhang W, Canziani G, Robinson J, Chaiken I, Hoxie J A, Doms R W. Proc Natl Acad Sci USA. 1999;96:6359–6364. doi: 10.1073/pnas.96.11.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Furuta R A, Wild C T, Weng Y, Weiss C D. Nat Struct Biol. 1998;5:276–279. doi: 10.1038/nsb0498-276. [DOI] [PubMed] [Google Scholar]
- 43.Chan D C, Kim P S. Cell. 1998;93:681–684. doi: 10.1016/s0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]