Abstract
Genome-scale networks can now be reconstructed based on high-throughput data sets. Mathematical analyses of these networks are used to compute their candidate functional or phenotypic states. Analysis of functional states of networks shows that the activity of biochemical reactions can be highly correlated in physiological states, forming so-called co-sets representing functional modules of the network. Thus, detrimental sequence defects in any one of the genes encoding members of a co-set can result in similar phenotypic consequences. Here we show that causal single nucleotide polymorphisms in genes encoding mitochondrial components can be classified and correlated using co-sets.
Keywords: correlated reaction set, disease, human mitochondria, SNP
Introduction
Various high-throughput (HT) technologies simultaneously measure thousands of interdependent biological variables, and numerous methods are being used to reduce the complexity of HT data sets, to determine dependencies among variables, and to correlate them with biological functions (Lin et al, 2005). Dimensionality reduction is a central process that allows inference of functional principles from highly complex data sets. A conceptually simple way to reduce complexity is to identify patterns of correlation within the data. For example, correlation among mRNA levels in expression profiles can be used to identify sets of co-regulated genes (Reymond et al, 2002). Similarly, the HapMap project recently illustrated the power of ‘perfect proxy sets'—defined as sets of perfectly correlated single nucleotide polymorphisms (SNPs)—as segments of the human chromosomes and suggested their utility in identifying differences in individual genomes (Altshuler et al, 2005). Systems biology aims to reconstruct networks of cellular interactions mathematically and to compute their functional (i.e. physiological) states (Price et al, 2004). Reducing the complexity of a network by identifying modules of functionally related elements represents an important step towards the understanding of its functional properties. In the context of biochemical networks, the study of their functional states has led to the definition of correlated sets of reactions (co-sets): groups of reactions that always (Papin et al, 2004) or often (Burgard et al, 2004) function together in metabolic networks under the constraints of mass conservation, charge conservation, and thermodynamic considerations. Herein, co-sets are defined as groups of enzymatic reactions that are perfectly correlated (correlation coefficient of 1). These co-sets are often non-obvious, as the reactions within a co-set may not be adjacent on a network map. They represent mathematically functional modules of a network, and identify genes whose products are collectively required to achieve physiological states. Accordingly, perturbations affecting genes belonging to the same co-set are expected to lead to similar functional consequences.
Classifying SNPs and co-sets
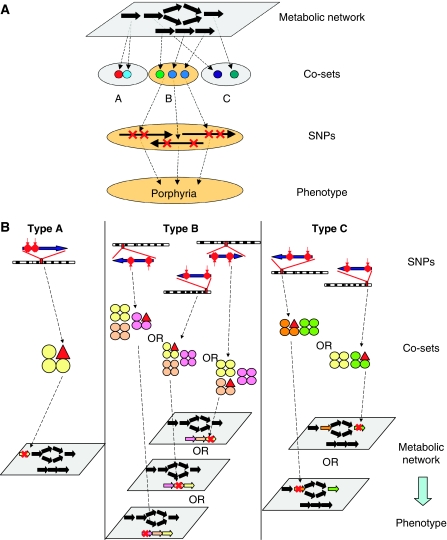
Here we use co-sets to seek dependencies among SNPs with causal implications on metabolic function, by grouping SNPs in proteins that catalyze different reactions, but are shared within the same co-set. Of course, not all SNPs will affect protein function; however, as the goal of this systems-based analysis is to study functional consequences of causal SNPs, it will be implicit throughout this work that all SNPs considered will only be those with causal implications on enzymatic function. Although the SNPs may affect different proteins with different catalytic activities, if the reactions are in the same co-set, the phenotypic consequences of such SNPs are expected to be similar. One can classify a group of genes that encode members of a co-set into three fundamental types (Figure 1). Type A describes a multimeric enzyme, where an SNP in any subunit of the multimer can thus result in the same phenotype. Type B represents a co-set of reactions in a contiguous pathway and Type C co-sets are formed by non-contiguous reactions.
Figure 1.
Relating SNPs, diseases, and correlated reaction sets. (A) Functional metabolic network analysis results in correlated reaction sets. Causal SNPs in any of the genes encoding proteins in the reaction sets are expected to have similar phenotypic states. (B) Examples of the different classes of genotype–phenotype relationships in the context of metabolic networks. Each sub-panel exemplifies a type of co-set represented by colored arrows and circles (non-black). The circles represent the protein subunits of a particular enzyme. Each enzyme is the same color. Red triangles represent altered protein subunits due to causal SNPs. Each group of colored circles (enzyme) corresponds to the same colored reaction flux in the pathway. SNPs in the gene coding for the protein subunits that catalyze the transformation from one metabolite to another will result in a phenotype that is unable to produce that particular product (Type A co-set). Type B co-sets function in an analogous manner, where causal SNPs affecting a reaction that is part of a correlated reaction set (a linear chain of reactions) will result in a phenotype characterized by an inability to produce the end point of the chain. More complex schemes may also occur in which the correlated reaction set is not a linear pathway (Type C co-set).
Disease-associated SNP co-sets in the mitochondria
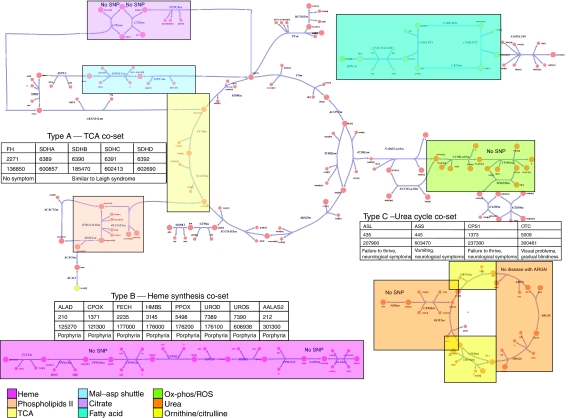
We mapped the human mitochondrial metabolic co-sets (Thiele et al, 2005) to various diseases in the Online Mendelian Inheritance in Man (OMIM; Hamosh et al, 2005) database and then identified those cases in which SNPs have been described in the literature (Figure 2). The succinate dehydrogenase (SDH) forms a Type A co-set of genes. A series of SNPs in the different subunits of SDH have been found to have similar phenotypic consequences.
Figure 2.
Map of mitochondrial metabolism with SNP-associated co-sets. The co-sets are color-coded according to the legend at the bottom of the figure. An example of each type of co-set (Type A: TCA cycle; Type B: Heme biosynthesis; Type C: Urea cycle) has a summary chart that includes the following information: gene abbreviation, gene IDs, MIMs, and SNP-associated signs or symptoms, respectively column-wise.
The genes that encode the enzymes leading to heme biosynthesis constitute a Type B co-set (Figure 2). Many SNPs in this set of genes result in various manifestations of porphyria. There is a range of severity and symptoms for a given enzyme and across the different enzymes in this gene set. These variations may be attributable to the specific location of particular SNPs, the presence or absence of other SNPs across the genome, differential tissue expression, the specific metabolic by-products that accumulate or diminish based on the specific reaction, or mitochondrial heteroplasmy.
A Type C co-set is found in the urea cycle (Figure 2). There is clinical coherence between SNPs in three of the four reactions in this set. Type C co-sets are perhaps the most interesting of the three classifications because they are the most non-obvious; consequently, they may have the greatest effect on revising previous views of interactions and classifications of disease. Another particularly interesting case is the citrulline/ornithine co-set. There is only SNP-related disease information for one of the two reactions in the co-set, the SLC25A15 transporter, whose deficiency results in the hyperornithinemia, hyperammonemia, and homocitrullinuria syndrome (HHH). Although SNP-related diseases have not been described for the other reaction in the co-set, overexpression of SLC25A2 can rescue patients with HHH due to SLC25A15 deficiency (Camacho et al, 2003). This example presents implications for therapeutic strategies in disease treatment with enzymes capable of binding a range of substrates. If two reactions with overlapping substrate utilization are in the same co-set, then overexpression of one enzyme can compensate for the deficiency or lack of expression of the other.
The complete set of SNP-associated co-sets identified in the mitochondria can be found in Supplementary Tables 1–9 online. The majority of identified co-sets are Type B. Although there is a significant amount of overlap between these co-sets, there is variability among them. Indeed even for a particular disease type, there can be a remarkably broad range of resulting phenotypes. As referred to above, this can be due to a range of factors including differential expression and genomic differences in other regions of the genome. The appropriate manner to resolve many of these issues and to increase the predictive power of these approaches is to incrementally increase the level of detail by accounting for more detailed biological information such as intracellular regulation, intercellular interactions, and different tissue expression states.
Implications of SNP co-set network analysis
There are two points worth highlighting in this new conceptual framework and the resulting analysis. First, the approach taken to network reconstruction is a ‘bottom-up' approach (Reed et al, 2006). In this approach, network reconstruction is based on documented physical interactions and biochemical knowledge, rather than inferred interactions from HT data sets. Such reconstructions are a biochemically, genomically, and genetically structured (BiGG) database that represents an integration of all of our knowledge about the network being analyzed (Reed et al, 2006). Consequently, the co-set predictions made are a direct result of a network-wide analysis reflecting fundamental properties of the reconstructed biochemical network. The use of co-sets to detect functionally related reactions is but one approach to analyzing reconstructed biological networks (Papin et al, 2004) and a number of others are emerging (Hatzimanikatis et al, 2004; Price et al, 2004; Sauer, 2004; Borodina and Nielsen, 2005). This type of analysis of bottom-up networks can be used in conjunction with top-down analysis of HT data sets to help elucidate functional biological relationships.
Second, the ability to map similarly causal SNPs to co-sets represents a new dimension in SNP analysis that is enabled by systems biology. Like ‘perfect-proxy sets' in the HapMap (Altshuler et al, 2005), co-sets take a step beyond trying to track individual components independently, towards appreciating how a number of cellular components interact to produce biological functionality and thus how their malfunction to result in pathophysiological states can correlate. Therefore, by adopting a systems approach, we can hopefully begin to define some of the ‘general underlying principles' of biological functions, and in doing so, can impact the classification of diseases, the mechanistic understanding of the genotype–phenotype relationship, and the potential identification of therapeutic targets and strategies for disease treatment.
Supplementary Material
Supplementary Online Material
Acknowledgments
This work was supported in part by an NIH Training Grant.
Competing Interest Statements:The authors declare that they have no competing financial interests.
References
- Altshuler D, Brooks LD, Chakravarti A, Collins FS, Daly MJ, Donnelly F (2005) A haplotype map of the human genome. Nature 437: 1299–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodina I, Nielsen J (2005) From genomes to in silico cells via metabolic networks. Curr Opin Biotechnol 16: 350–355 [DOI] [PubMed] [Google Scholar]
- Burgard AP, Nikolaev EV, Schilling CH, Maranas CD (2004) Flux coupling analysis of genome-scale metabolic network reconstructions. Genome Res 14: 301–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho JA, Rioseco-Camacho N, Andrade D, Porter J, Kong J (2003) Cloning and characterization of human ORNT2: a second mitochondrial ornithine transporter that can rescue a defective ORNT1 in patients with the hyperornithinemia–hyperammonemia–homocitrullinuria syndrome, a urea cycle disorder. Mol Genet Metab 79: 257–271 [DOI] [PubMed] [Google Scholar]
- Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA (2005) Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res 33 (Database issue): D514–D517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzimanikatis V, Li C, Ionita JA, Broadbelt LJ (2004) Metabolic networks: enzyme function and metabolite structure. Curr Opin Struct Biol 14: 300–306 [DOI] [PubMed] [Google Scholar]
- Lin B, White JT, Lu W, Xie T, Utleg AG, Yan X, Yi EC, Shannon P, Khrebtukova I, Lange PH, Goodlett DR, Zhou D, Vasicek TJ, Hood L (2005) Evidence for the presence of disease-perturbed networks in prostate cancer cells by genomic and proteomic analyses: a systems approach to disease. Cancer Res 65: 3081–3091 [DOI] [PubMed] [Google Scholar]
- Papin JA, Reed JL, Palsson BO (2004) Hierarchical thinking in network biology: the unbiased modularization of biochemical networks. Trends Biochem Sci 29: 641–647 [DOI] [PubMed] [Google Scholar]
- Price ND, Reed JL, Palsson BO (2004) Genome-scale models of microbial cells: evaluating the consequences of constraints. Nat Rev Microbiol 2: 886–897 [DOI] [PubMed] [Google Scholar]
- Reed JL, Famili I, Thiele I, Palsson BO (2006) Towards multidimensional genome annotation. Nat Rev Genet 7: 1–12 [DOI] [PubMed] [Google Scholar]
- Reymond A, Marigo V, Yaylaoglu MB, Leoni A, Ucla C, Scamuffa N, Caccioppoli C, Dermitzakis ET, Lyle R, Banfi S, Eichele G, Antonarakis SE, Ballabio A (2002) Human chromosome 21 gene expression atlas in the mouse. Nature 420: 582–586 [DOI] [PubMed] [Google Scholar]
- Sauer U (2004) High-throughput phenomics: experimental methods for mapping fluxomes. Curr Opin Biotechnol 15: 58–63 [DOI] [PubMed] [Google Scholar]
- Thiele I, Price ND, Vo TD, Palsson BO (2005) Candidate metabolic network states in human mitochondria. Impact of diabetes, ischemia, and diet. J Biol Chem 280: 11683–11695 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Online Material