The in vivo stem cell microenvironment or niche is composed of an intricate blend of extracellular matrix proteins, soluble protein factors, immobilized protein factors, proteoglycans, small molecule signals, in some cases mineralized tissue, and numerous adjacent cell types, all of which likely vary in space and time. In concert, these many components present the cells with an ‘array' of biochemical and biomechanical signals, such that the cell is continually faced with the complex tasks of sensing these inputs, processing the signals through complex signal transduction and gene regulation networks, and executing cell behavioral or fate choices.
This native microenvironment therefore poses a daunting challenge to the fields of stem cell biology and engineering: how does one analyze the relative importance of the individual signals of this medley in regulating cell function? In addition to advancing our basic knowledge of the central question of stem cell fate choice, addressing this question will be necessary to achieve the control over cell behaviors needed for regenerative medicine efforts. Reductionist biological approaches have consistently made major strides in elucidating the importance of individual signaling factors; however, cells in general sense and respond to combinations of extracellular signals rather than any single one (Janes et al, 2005). In an article currently published in Molecular Systems Biology, Theo Palmer, Patrick Brown, and colleagues reported on a novel, high-throughput approach to address this problem (Soen et al, 2006).
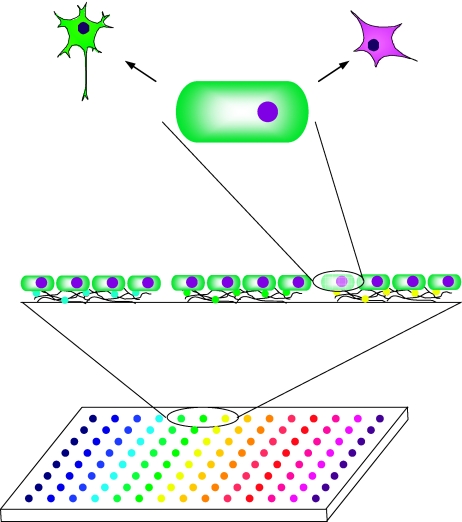
As the number of combinations to be investigated rapidly increases with the number of individual factors, high-throughput and density approaches to sample this signaling space will be of fundamental importance to this challenge. Arrays of cDNA (Ziauddin and Sabatini, 2001), biomaterials (Anderson et al, 2004), and extracellular matrix proteins (Flaim et al, 2005) have proven to be powerful approaches to exploring cell responses to varying signals and microenvironments. Soen et al (2006) have further extended array technologies by building a microarray system to systemically investigate the effects of numerous morphogens, growth factors, cell adhesion molecules, and extracellular matrix components on the fate of primary bi-potent human neural precursor cells that can differentiate into neurons or glial cells (Figure 1 ). A non-contact, piezoelectric arrayer was utilized to print combinations of factors onto an aldehyde-activated surface and thereby generate arrays of 350–500 μm diameter spots of factors covalently grafted to the surface. Human neural precursors were then isolated from tissue, seeded onto the surface, and cultured under uniform soluble media conditions. Because all cells on the array were exposed to uniform culture medium, this approach focused on the effects of the immobilized ligands on cell function, with the possible exception of local autocrine or paracrine effects that could be modulated by varying the distance between spots. Cellular behaviors (i.e. differentiation and proliferation) were then monitored by immunostaining for bromodeoxyuridine and cell fate markers, followed by high-throughput quantitative imaging analysis at the single-cell level.
Figure 1.
Schematic representation of a microenvironment array. Combinations of signals and adhesion molecules are printed onto an aldehyde-activated surface to generate an array of 350–500 μm diameter spots. Suspended cells are briefly incubated on the array and single-cell populations are captured on the spots by adherence to the printed extracellular matrix components or adhesion molecules. Automated fluorescence microscopy and computer-assisted feature extraction allows a high-throughput quantitative analysis of multiple phenotypic outcomes at the single cell level.
Utilizing strong experimental analysis, they found numerous interesting phenotypic outcomes. Some factors that bias cell differentiation into glia exerted additive effects when utilized in combination (e.g. the Notch ligand Jagged-1 and ciliary neurotrophic factor). However, others that would be anticipated to exert conflicting effects on cell differentiation (e.g. Wnt-3A versus Notch ligands) unexpectedly resulted in enhanced cell proliferation with low expression of cell fate markers. Intriguingly, bone morphogenetic protein (BMP)-4 yielded numerous cells that coexpressed markers of both fates in an apparently ‘androgynous' cell state.
In addition, it is likely that information is conveyed to cells not only in the identity of signals but in their quantity as a function of time (Zandstra et al, 2000; Saha and Schaffer, 2006). The arrayer was utilized to vary signal dosage, and with increasing density of the Notch ligand Jagged-1 the fraction of glia and neurons monotonically increased or decreased, respectively. Moreover, whereas the ratio of cells expressing the glial versus neuronal rapidly reached a plateau on BMP-4, it progressively increased over time on Jagged-1.
Besides extending cellular microarray technologies to additional factors and cell types, there are numerous other areas for expansion. Microarrays in general provide large, valuable data sets that can subsequently be explored in further mechanistic detail, particularly the observed ‘nonlinear' interactions among multiple factors. In addition, mechanical signals regulate stem cell function (McBeath et al, 2004), so it will be valuable for future systems to offer the ability to modulate both the biochemical and the biomechanical components of the cellular microenvironment. Likewise, cell–cell contacts in three dimensions may be important aspects of the stem cell niche. Finally, this system can be extended to numerous biomedical applications, including developing platforms for protein and drug screening, spatial patterning of cell fates for tissue engineering efforts, or diagnostic analysis of small cell samples such as tumors. Cellular microarrays can also offer economic benefits for such applications as they can utilize minimal quantities of proteins (∼10 s of pg/spot) and cells (∼100 s/spot).
Taken together, a system of high-density, microarrayed, immobilized microenvironments provides a robust method to investigate the direct, integrated effects of multiple cooperative or even conflicting signals on cell behavior. The approach developed by Soen et al thus provides new capabilities for the study of basic stem cell biology and the advancement of regenerative medicine.
References
- Anderson DG, Levenberg S, Langer R (2004) Nanoliter-scale synthesis of arrayed biomaterials and application to human embryonic stem cells. Nat Biotechnol 22: 863–866 [DOI] [PubMed] [Google Scholar]
- Flaim CJ, Chien S, Bhatia SN (2005) An extracellular matrix microarray for probing cellular differentiation. Nat Methods 2: 119–125 [DOI] [PubMed] [Google Scholar]
- Janes KA, Albeck JG, Gaudet S, Sorger PK, Lauffenburger DA, Yaffe MB (2005) A systems model of signaling identifies a molecular basis set for cytokine-induced apoptosis. Science 310: 1646–1653 [DOI] [PubMed] [Google Scholar]
- McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS (2004) Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell 6: 483–495 [DOI] [PubMed] [Google Scholar]
- Saha K, Schaffer DV (2006) Signal dynamics in Sonic hedgehog tissue patterning. Development 133: 889–900 [DOI] [PubMed] [Google Scholar]
- Soen Y, Mori A, Palmer TD, Brown PO (2006) Exploring the regulation of human neural precursor cell differentiation using arrays of signaling microenvironments. Mol Syst Biol 37. doi:10.1038/msb4100076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandstra PW, Lauffenburger DA, Eaves CJ (2000) A ligand-receptor signaling threshold model of stem cell differentiation control: a biologically conserved mechanism applicable to hematopoiesis. Blood 96: 1215–1222 [PubMed] [Google Scholar]
- Ziauddin J, Sabatini DM (2001) Microarrays of cells expressing defined cDNAs. Nature 411: 107–110 [DOI] [PubMed] [Google Scholar]