Abstract
The distribution of most genes is not random, and functionally linked genes are often found in clusters. Several theories have been put forward to explain the emergence and persistence of operons in bacteria. Careful analysis of genomic data favours the co-regulation model, where gene organization into operons is driven by the benefits of coordinated gene expression and regulation. Direct evidence that coexpression increases the individual's fitness enough to ensure operon formation and maintenance is, however, still lacking. Here, a previously described quantitative model of the network that controls the transcription factor σF during sporulation in Bacillus subtilis is employed to quantify the benefits arising from both organization of the sporulation genes into the spoIIA operon and from translational coupling. The analysis shows that operon organization, together with translational coupling, is important because of the inherent stochastic nature of gene expression, which skews the ratios between protein concentrations in the absence of co-regulation. The predicted impact of different forms of gene regulation on fitness and survival agrees quantitatively with published sporulation efficiencies.
Keywords: coupled gene expression, noise, operon, signalling networks, sporulation
Introduction
The available genome sequences demonstrate that many genes are clustered on chromosomes according to their function. Genes in bacteria are clustered but can also be organized into operons such that the expression of a group of genes is regulated by the same genetic control element. When operons were first discovered, it was assumed that the benefit of co-transcription led to operon assembly (Jacob and Monod, 1961). Other models have since been proposed, and these belong to one of three classes, the natal model, the Fisher model, or the selfish operon model (Lawrence, 1997). According to the natal model, clustering of genes is the consequence of gene duplication. However, as operons comprise genes that belong to very distant families and the majority of paralogues do not cluster, this model is insufficient to explain operon origin (Lawrence, 1997; Dandekar et al, 1998). A recast of the Fisher model, adapted to prokaryotes, proposes that clustering of genes reduces the likelihood that co-adapted genes become separated by recombination. However, this does not explain how operons can emerge, as recombination is as likely to generate clusters as to disrupt them. According to the selfish operon model, operons facilitate the horizontal transfer of functionally related genes (Lawrence and Roth, 1996). The physical proximity of genes thus does not necessarily provide a selective advantage to the individual organism but rather to the gene cluster itself, because it can be efficiently transmitted horizontally as well as vertically. Recent studies have, however, failed to observe the gene cluster pattern predicted by the model, and this strongly suggests that the selfish operon model does not explain the emergence and persistence of operons (Pal and Hurst, 2004; Price et al, 2005). So what drives operon assembly?
The idea that co-transcription of genes provides a selective advantage to the individual organism has never been contradicted. It has been questioned only because it remains unclear whether the benefits of co-transcription could be strong enough to drive the assembly of operons by rare recombination events (Lawrence and Roth, 1996; Lawrence, 1997). A genotype that confers higher fitness will dominate in a population with bounded total population size only if selection acts on a timescale that is substantially shorter than the timescale on which recombination and mutation events could negate the benefits.
There are a number of potential selective advantages given by co-transcription. In the case of operons that code for multi-protein complexes, co-transcription enables co-translational folding (Dandekar et al, 1998), it limits the half-life of toxic monomers (Pal and Hurst, 2004), and it reduces stochastic differences in gene expression (Swain, 2004). Operons that do not code for interacting proteins may be advantageous because of the co-regulation of protein expression (Price et al, 2005). Many examples of this class of operons are associated with metabolic operons (Lawrence and Roth, 1996) where co-regulated expression is likely to optimize the flux and to facilitate the regulation of functions, especially if these are required only under certain environmental conditions, or if complex regulatory structures are employed (Price et al, 2005).
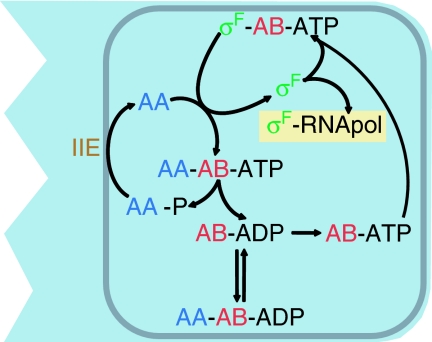
Evidence in favour of any of these proposed driving forces has so far largely been obtained from comparative genomics. Here we use a previously derived quantitative model for the network that controls the transcription factor σF during sporulation in Bacillus subtilis (Iber et al, 2006) to quantify the benefits of coexpression. Spore formation in B. subtilis is a response to nutrient deprivation at high cell density and involves asymmetric septation and compartment-specific initiation of gene expression (Hilbert and Piggot, 2004). The different gene programs in the larger mother cell and the smaller prespore are both directed by the transcription factor σF, which, although only active in the smaller prespore, affects the transcriptional programs across the septum also in the mother cell, a phenomenon that is referred to as criss-cross regulation (Losick and Stragier, 1992). Successful sporulation therefore requires the rapid septation-dependent and prespore-specific activation of σF. σF is kept inactive by binding to SpoIIAB and is released upon binding of SpoIIAA (Figure 1). SpoIIAA is phosphorylated by SpoIIAB (Min et al, 1993) and reactivated by the serine phosphatase SpoIIE (Duncan et al, 1995). The balance between kinase and phosphatase activity thus determines whether or not σF is released from its inactive complex with SpoIIAB. SpoIIE accumulates on both sides of the asymmetrically positioned septum and therefore has an increased activity in the smaller compartment (Arigoni et al, 1995). A quantitative model of the regulatory network predicts that because of the low turnover rate, most SpoIIE is bound by its substrate such that enzyme and substrate increase together in the smaller compartment (Iber et al, 2006). According to the model, this combined increase is sufficient to trigger the formation of micromolar concentrations of σF holoenzyme in the prespore.
Figure 1.
An overview of the interactions in the network that controls σF activity in B. subtilis. For details, see text. The figure is a reproduction of Figure 1 of Iber et al (2006).
It is obvious from the above that the protein concentration ratio is important. An excess of σF or SpoIIAA compared to SpoIIAB will result in free σF and σF-dependent gene expression, whereas an excess of SpoIIAB will prevent SpoIIAA-dependent σF release. In the vegetative cell, the sporulation proteins are not detectable, and septation is preceded by 90–120 min of gene expression, dependent on the exact experimental conditions (Magnin et al, 1997; Lord et al, 1999; Lucet et al, 1999). Limiting the stochastic noise inherent in protein expression can be expected to be crucial for avoiding variations in the relative protein concentrations and the resulting sporulation defects. Three of the four proteins in the network are transcribed from genes in the spoIIA operon (Figure 2A). These genes are not only co-transcribed into a single mRNA but are also most likely to be coexpressed, as the translation of the three proteins appears to be coupled, at least to some degree. This system therefore offers an excellent opportunity to analyse the influence of transcriptional and translational co-regulation of the sporulation genes on an individual's survival and fitness.
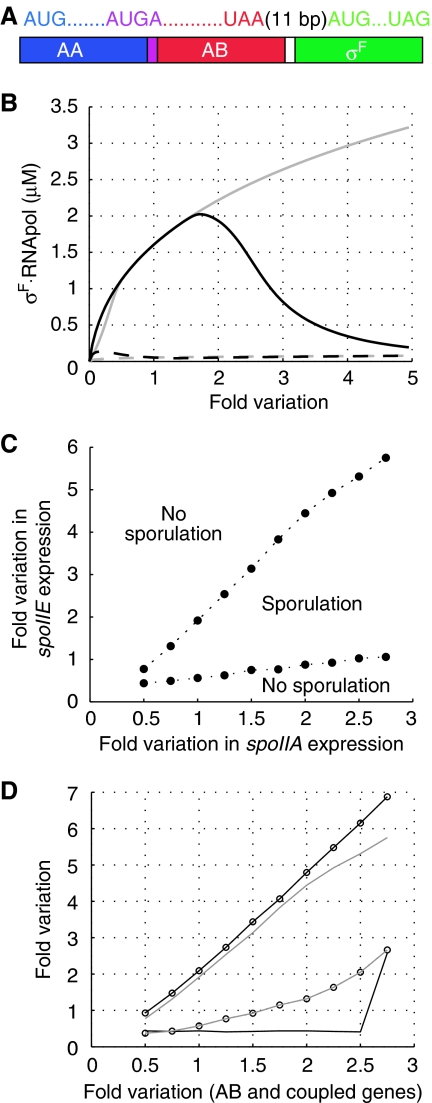
Figure 2.
The impact of parallel and random variations in the expression of spoIIE and spoIIA genes on σF release. (A) The spoIIA operon comprises the genes for SpoIIAA, SpoIIAB, and σF. The genes for SpoIIAA and SpoIIAB overlap; the genes for SpoIIAB and σF are separated by 11 bp. (B) The regulatory network is robust to parallel variations in gene expression. The predicted concentration of σF-RNA polymerase holoenzyme before (dashed lines) and after septum formation (continuous lines) if either all (grey lines) or all protein expression rates except that of SpoIIE (black lines) were increased by the factor on the horizontal axis compared to the standard reference rates (6 × 10−9 M s−1 for SpoIIAA and SpoIIAB dimers and 2 × 10−9 M s−1 for σF and SpoIIE; Iber et al, 2006). (C, D) The expression rate combinations for which septation-dependent σF release is possible (between the lines) or not possible (outside the area marked by lines). (C) The impact of differential regulation of spoIIE and spoIIA expression. The vertical and horizontal axes indicate the fold variation in the spoIIE and spoIIA expression rates respectively, compared to the standard reference rates. (D) The impact of differential regulation of the expression of genes encoded in the spoIIA operon. The vertical axis indicates the fold variation in the expression of SpoIIAA (circles), σF (black lines), or SpoIIAA and σF (grey lines). The horizontal axis indicates the fold variation in the expression of SpoIIAB and of any other protein whose expression is coupled to that of SpoIIAB (which are those genes in the spoIIA operon not reported on the vertical axis). The sudden jump observed at a high SpoIIAB to σF ratio (lower black line) is the consequence of impaired σF release when the relative SpoIIAB concentration is too high.
Coupled translation is achieved when two genes are translated by the same ribosome. Reinitiation of translation at a nearby start codon after termination at the upstream gene is possible because ribosome dissociation from the mRNA is a slow and energy-dependent process (McCarthy and Gualerzi, 1990). There is currently no direct experimental evidence for coupled translation of the spoIIA operon. Such coupling can, however, be postulated based on the arrangement of genes (Fort and Piggot, 1984). The first two genes in the spoIIA operon (encoding SpoIIAA and SpoIIAB) overlap by 4 bp, whereas the genes for SpoIIAB and σF are interspaced by 11 bp (Figure 2A); coupled translation has been documented for intercistronic distances of more than 60 bp (McCarthy and Gualerzi, 1990). The majority of genes that are organized in operons are separated by distances comparable to those found in the spoIIA operon (Salgado et al, 2000), so that the studied system can be considered as representative of operons in general. The efficiency of reinitiation depends on the distance as well as the strength of the Shine–Dalgarno sequence (Adhin and van Duin, 1990; McCarthy and Gualerzi, 1990), which is, in general, located 5–13 bp upstream of a start codon and which binds to the homologous 3′ end of the 16S rRNA, a component of the 30S ribosomal subunit. Moreover, the secondary structure of the mRNA can affect lateral diffusion of the ribosomes (Adhin and van Duin, 1990).
According to the protein expression data for the spoIIA operon, it appears that the last gene in the operon which encodes σF, is expressed at much lower levels than are spoIIAA and spoIIAB, whereas SpoIIAB monomers may be expressed at equal or up to three times higher levels compared to SpoIIAA (Magnin et al, 1997; Lord et al, 1999; Lucet et al, 1999). The weaker expression of a downstream gene (as is the case for σF) can, in general, be accounted for by a weaker ribosomal binding site, which is removed far enough from the termination codon of the upstream cistron that a considerable fraction of ribosomes dissociate from the mRNA before translation can be reinitiated (McCarthy and Gualerzi, 1990). It should be noted that whereas the transcriptional and translational coupling will reduce the noise in the relative SpoIIAB to σF expression levels, the unbinding of ribosomes is necessarily a stochastic process and will therefore add a low level of noise. The stronger expression of a downstream gene (as may be the case for SpoIIAB relative to SpoIIAA) can, in general, only be observed if a strong initiation sequence for the downstream gene is occluded by mRNA secondary structure, which is melted by the ribosome that transcribes the upstream gene (McCarthy and Gualerzi, 1990). Such a condition does not seem to be met by the gene for SpoIIAB, and more accurate expression data will be necessary to establish whether more SpoIIAB than SpoIIAA is expressed.
Available expression data can best be captured by an expression rate for SpoIIAB dimers and SpoIIAA of 6 × 10−9 M s−1 and 2 × 10−9 M s−1 for σF and SpoIIE (Iber et al, 2006); it should be noted that the simulation yields qualitatively similar results if SpoIIAB monomers and SpoIIAA are expressed at equal rates (6 × 10−9 M s−1), as long as the σF and SpoIIE expression rate is then reduced to 10−9 M s−1 (Iber, 2006). As discussed by Iber (2006), the linear increase in the protein concentration assumed here does not fully match the experimental observations. There are, nonetheless, two good reasons to use a linear model. First of all, the data are too inaccurate and, in parts, contradictory to be modelled exactly. Secondly, the chosen rates correspond to the protein concentrations measured at the time of septation (Magnin et al, 1997; Lord et al, 1999; Lucet et al, 1999), the critical time point to judge sporulation success. This is because, in the cell, the IIE concentration increases more slowly than the other protein concentrations and only increases sharply immediately before septation (Feucht et al, 2002). As a consequence, the greatest danger of spontaneous uncompartmentalized activation of σF is just before septation, and this risk is fully assessed by the linear expression model. As our analysis focuses mainly at what happens minutes before and after septation, individual fluctuations in the global expression rates during the 2 h preceding septation are not important and the linear protein expression rates used should be considered as an averaged protein expression rate per bacterium.
Our quantitative ordinary differential equation model is very detailed—it comprises 50 dependent variables and 150 kinetic constants to describe the dynamics of only four proteins; the reader is referred to a detailed discussion of the model in Supplementary information of Iber et al (2006). Given its high level of detail and accuracy, the model predicts the phenotypes of essentially all mutants for which the biochemical effect is known. We can therefore expect that the predicted sporulation efficiencies in response to changes in parameter values are realistic. In the following, we employ the model to quantify how far different levels of stochastic noise in gene expression, as modulated by different degrees of coupling of protein expression (that is by the coupling of both transcription and translation), affect the sporulation efficiency, that is the survival chances.
Results and discussion
In addressing how variations in the protein expression rates affect the sporulation efficiency we will look at the effect of parallel changes in all protein expression rates as well as at the effects of independent changes that skew the ratios of protein concentrations. As the standard, ‘wild-type' protein expression rates, we use 6 × 10−9 M s−1 for SpoIIAA and SpoIIAB dimers and 2 × 10−9 M s−1 for σF and SpoIIE (Iber et al, 2006). After 120 min of protein expression, the septum forms and SpoIIE accumulates on both sides of this septum. This is modelled by a four-fold increase in the concentration of SpoIIE, together with its associated substrate (phosphorylated SpoIIAA) in the prespore. As before, we define a successful sporulation event by the requirement that before septation the concentration of σF. RNA polymerase holoenzyme does not exceed 0.4 μM, whereas after septation the concentration exceeds 1 μM (Iber et al, 2006).
If the protein expression rates are all varied in parallel, that is by a common factor as denoted on the horizontal axis in Figure 2B, we find that the predicted sporulation efficiency is not affected as long as a minimal expression rate is kept to provide sufficient σF for binding to the RNA polymerase (Figure 2B, grey lines). If the expression of SpoIIE is kept constant (in order to reflect that this protein is transcribed from a different locus and may therefore vary independently), then an independent 2.5-fold increase in the other sporulation proteins can still be tolerated before the relative activity of the phosphatase becomes too weak (Figure 2B, black lines). An even higher independent increase in the expression of the spoIIA genes can be tolerated if we assume that the expression of the spoIIA and spoIIE genes is at least weakly correlated such that a large increase in the expression of the spoIIA genes is accompanied by a small increase in the expression of the spoIIE genes (Figure 2C). Such a correlation is not unexpected, considering that variations in gene expression are the result of both intrinsic and extrinsic noise. The latter, which reflects cell-to-cell variation in the concentration of other molecular species such as the RNA polymerase, will affect all genes similarly. We can conclude that the independent regulation of the spoIIA and spoIIE genes is unlikely to generate a major risk of failed sporulation. Separation of the spoIIA and spoIIE genes on the bacterial chromosome, on the other hand, has benefits because it ensures that, upon septation, each compartment retains one copy of spoIIE while initially (for the first 10–15 min) two copies of spoIIA are in the mother cell but none in the prespore (Frandsen et al, 1999). This initial transient genetic imbalance may protect the mother cell from a relative increase of spoIIE to spoIIA gene products (Iber, 2006).
If the expression levels of the genes in the spoIIA operon are varied independently of each other, the tolerance of the network to variations in gene expression drops substantially. In particular, if expression of SpoIIAB and SpoIIAA is no longer co-regulated, the network is sensitive to rather small changes (Figure 2D, grey lines and circles). Thus, if the SpoIIAA expression rate remains fixed and the SpoIIAB expression rate increases by 60% (corresponding to the factor 1.6 on the horizontal axis in Figure 2D), then sporulation is predicted to fail; 60% variation from the mean is a noise level observed in bacterial (Escherichia coli) expression systems (Elowitz et al, 2002). On the other hand, if expression of SpoIIAA and SpoIIAB remains co-regulated but σF expression is regulated independently (Figure 2D, black lines), the network is rather robust to variations in gene expression as long as the expression of SpoIIAB is increased more than the expression of σF and the overall σF concentration remains high enough to form micromolar concentrations of the holoenzyme. The transcriptional coupling together with a strong translational coupling of SpoIIAA and SpoIIAB therefore substantially increases the robustness of the network to fluctuations in gene expression. Stochastic variations in the relative rate of σF translation, on the other hand, are not as detrimental as long as the translation efficiency for σF is lower than for SpoIIAA and SpoIIAB, as can be achieved by a weaker ribosomal binding site and the resulting (stochastic) dissociation of ribosomes. An advantage of preferential dissociation of the ribosomes before translating the gene for σF is that the bacterium saves the energy that would otherwise be required to translate, and subsequently degrade, unnecessary (harmful) copies of σF. Considering that σF comprises 255 amino acids and linkage of each amino acid requires the equivalent of four ATPs, the energy by not translating and degrading 10 μM σF corresponds to more than 10 mM ATP, which is a considerable amount considering that the bacterial ATP concentration is 1–3 mM (Jolliffe et al, 1981; Guffanti et al, 1987; Hecker et al, 1988) and sporulation is a response to starvation, that is energy deprivation.
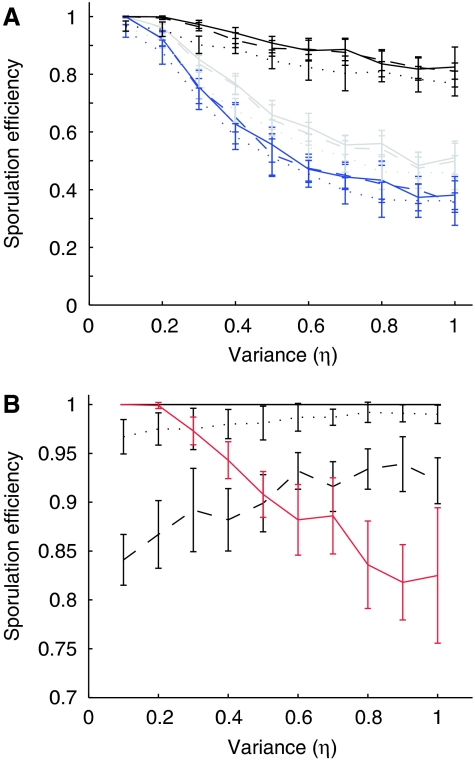
In a last step, we can now quantify the impact of gene organization on sporulation efficiency, and therefore fitness. For this, we assume that the gene expression levels in the cell population follow a normal distribution with variance η around the mean value. Given the complex regulation pattern of gene expression, gene expression levels are unlikely to be distributed exactly normally. A normal distribution is, however, still likely to provide an approximation no worse than what could be obtained with a detailed model of the regulatory process in the absence of sufficient data to determine all required parameter values (Swain, 2004). Sporulation efficiency is determined as the fraction of simulation runs for which the concentration of σF. RNA polymerase holoenzyme does not exceed 0.4 μM before septation and exceeds one micromolar after septation (Iber et al, 2006). For each condition, the mean sporulation efficiency and standard deviation are calculated from 100 independent runs that are carried out 10 times. In each run, the protein expression rates were set randomly such that overall the respective distributions of the protein expression rates were obtained. Determination of the sporulation efficiency for η∈[0,1] shows that as long as the sporulation genes are translationally coupled, even high variances hardly affect the sporulation efficiency (Figure 3A, black lines). The sporulation efficiency is even higher at high noise level, η, if spoIIE expression co-varies with spoIIA expression, at least weakly (Figure 3B). A lengthening of the transcription time (that is a delay in septation), when transcription levels are too low to generate sufficient σF until septation, will further increase robustness to fluctuations in the rate of protein expression. Such a dependency of the time point of septation on the protein (and in particular the SpoIIE) concentration is in agreement with experiments (Khvorova et al, 1998; Ben-Yehuda and Losick, 2002) and might explain the large variance in the delay between the onset of sporulation and septation that is observed under different sporulation conditions. Low levels of additional stochastic noise in σF expression (broken lines), as may arise because of the stochastic dissociation of ribosomes, also has rather little impact and confirms that the weak coupling of SpoIIAB and σF translation does not substantially reduce sporulation efficiency. If on the other hand, spoIIAB is removed from the operon and controlled independently by the same promoter, then the sporulation efficiency drops rapidly (Figure 3A, blue lines). This is in good quantitative agreement with experiments, which find that the sporulation efficiency drops to 40–80% of wild-type levels (Dworkin and Losick, 2001), especially when considering that η∼[0.3, 0.6] for these expression levels (Elowitz et al, 2002). If spoIIAA is moved instead, then the effect is reduced (J Clarkson, personal communication), as also predicted by the model (Figure 3A, grey lines).
Figure 3.
The impact of stochastic variation in gene expression on sporulation efficiency. (A) The fraction of successful sporulation events (as defined in the text) dependent on the variance in gene expression if expression of the spoIIA genes is either coupled (black lines), the expression of SpoIIAB and σF is coupled (grey lines), or the expression of SpoIIAA and σF is coupled (blue lines). SpoIIE is expressed throughout at the standard rate of 2 × 10−9 M s−1. The broken lines show the effect of an additional independent normal variation in the rate of σF expression with ηS=0.1 (dashed lines) or ηS=0.3 (dotted lines) from the coupled rates. If σF is one of the coupled rates, then σF expression is varied both together with its coupling partner and additionally independently to reflect the additive levels of noise acting at the initiation of translation and the re-initiation/dissociation step. (B) The fraction of successful sporulation events (as defined in the text) dependent on the variance in gene expression if expression of the spoIIA and spoIIE genes is coupled (to assess the benefits of correlated expression), and an additional noise term ηE is added to the expression of spoIIE with ηE=0.1 (black continuous line), ηE=0.3 (dotted line), or ηE=0.6 (dashed line); ηE assesses the effects of independent promoters and spatial heterogeneity in the concentration of transcription and translation factors. The red line is identical to the continuous black line in panel A (noise in coupled spoIIA expression, SpoIIE expressed at 2 × 10−9 M s−1). Mean and standard deviation are based on 10 times 100 independent runs.
It should be noted that this drop in sporulation efficiency has previously been accounted for by the loss of the transient genetic imbalance when spoIIAB is moved to a chromosomal position close to the origin of replication (Dworkin and Losick, 2001). The transient lack of SpoIIAB expression in the prespore together with accelerated degradation of unbound SpoIIAB (Pan et al, 2001) had been suggested to enable σF release (Dworkin and Losick, 2001). However, we have shown previously that the transient genetic imbalance does not affect σF release on the timescale on which it persists (Iber, 2006), and stochastic effects are therefore a much more likely explanation for the observed phenotype of the mutants.
We conclude from the analysis of this well-studied model system that the protection from stochastic variation in the expression rate of interacting proteins can substantially increase viability, and therefore constitutes a driving force for gene clustering and co-regulation. Although the importance of gene dosage had been recognized before (Veitia, 2002), and underexpression and overexpression of protein complex subunits in yeast had been shown to lower fitness (Papp et al, 2003), this study reveals that much smaller variances, as can result from stochastic effects, can already have substantial detrimental effects. The detailed analysis of the expression of the sporulation proteins therefore demonstrates the optimized character of gene regulation and suggests that co-regulation of genes serves to optimize cellular network dynamics in spite of the inherent noise in all biological processes.
Acknowledgments
I thank Iain D Campbell, Joanna Clarkson, and Michael D Yudkin for many valuable discussions, and Iain D Campbell for his critical reading of the manuscript. The work was supported by a DTA EPSRC studentship as well as by a Junior Research fellowship held at St John's College, University of Oxford.
References
- Adhin MR, van Duin J (1990) Scanning model for translational reinitiation in eubacteria. J Mol Biol 213: 811–818 [DOI] [PubMed] [Google Scholar]
- Arigoni F, Pogliano K, Webb C, Stragier P, Losick R (1995) Localization of protein implicated in establishment of cell type to sites of asymmetric division. Science 270: 637–640 [DOI] [PubMed] [Google Scholar]
- Ben-Yehuda S, Losick R (2002) Asymmetric cell division in B. subtilis involves a spirallike intermediate of the cytokinetic protein FtsZ. Cell 109: 257–266 [DOI] [PubMed] [Google Scholar]
- Dandekar T, Snel B, Huynen M, Bork P (1998) Conservation of gene order: a fingerprint of proteins that physically interact. Trends Biochem Sci 23: 324–328 [DOI] [PubMed] [Google Scholar]
- Duncan L, Alper S, Arigoni F, Losick R, Stragier P (1995) Activation of cell-specific transcription by a serine phosphatase at the site of asymmetric division. Science 270: 641–644 [DOI] [PubMed] [Google Scholar]
- Dworkin J, Losick R (2001) Differential gene expression governed by chromosomal spatial asymmetry. Cell 107: 339–346 [DOI] [PubMed] [Google Scholar]
- Elowitz MB, Levine AJ, Siggia ED, Swain PS (2002) Stochastic gene expression in a single cell. Science 297: 1183–1186 [DOI] [PubMed] [Google Scholar]
- Feucht A, Abbotts L, Errington J (2002) The cell differentiation protein SpoIIE contains a regulatory site that controls its phosphatase activity in response to asymmetric septation. Mol Microbiol 45: 1119–1130 [DOI] [PubMed] [Google Scholar]
- Fort P, Piggot PJ (1984) Nucleotide sequence of sporulation locus spoIIA in Bacillus subtilis. J Gen Microbiol 130: 2147–2153 [DOI] [PubMed] [Google Scholar]
- Frandsen N, Barak I, Karmazyn-Campelli C, Stragier P (1999) Transient gene asymmetry during sporulation and establishment of cell specificity in Bacillus subtilis. Genes Dev 13: 394–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guffanti AA, Clejan S, Falk LH, Hicks DB, Krulwich TA (1987) Isolation and characterization of uncoupler-resistant mutants of Bacillus subtilis. J Bacteriol 169: 4469–4478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecker M, Heim C, Volker U, Wolfel L (1988) Induction of stress proteins by sodium chloride treatment in Bacillus subtilis. Arch Microbiol 150: 564–566 [DOI] [PubMed] [Google Scholar]
- Hilbert DW, Piggot PJ (2004) Compartmentalization of gene expression during Bacillus subtilis spore formation. Microbiol Mol Biol Rev 68: 234–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iber D (2006) A computational analysis of the impact of the transient genetic imbalance on compartmentalized gene expression during sporulation in Bacillus subtilis. J Mol Biol 360: 15–20 [DOI] [PubMed] [Google Scholar]
- Iber D, Clarkson J, Yudkin MD, Campbell ID (2006) The mechanism of cell differentiation in Bacillus subtilis. Nature 441: 371–374 [DOI] [PubMed] [Google Scholar]
- Jacob F, Monod J (1961) On the regulation of gene activity. Cold Spring Harbor Symp Quant Biol 26: 193–211 [DOI] [PubMed] [Google Scholar]
- Jolliffe LK, Doyle RJ, Streips UN (1981) The energized membrane and cellular autolysis in Bacillus subtilis. Cell 25: 753–763 [DOI] [PubMed] [Google Scholar]
- Khvorova A, Zhang L, Higgins ML, Piggot PJ (1998) The spoIIE locus is involved in the Spo0A-dependent switch in the location of FtsZ rings in Bacillus subtilis. J Bacteriol 180: 1256–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JG (1997) Selfish operons and speciation by gene transfer. Trends Microbiol 5: 355–359 [DOI] [PubMed] [Google Scholar]
- Lawrence JG, Roth JR (1996) Selfish operons: horizontal transfer may drive the evolution of gene clusters. Genetics 143: 1843–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord M, Barilla D, Yudkin MD (1999) Replacement of vegetative σF by sporulation-specific σF as a component of the RNA polymerase holoenzyme in sporulating Bacillus subtilis. J Bacteriol 181: 2346–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick R, Stragier P (1992) Crisscross regulation of cell-type-specific gene expression during development in B. subtilis. Nature 355: 601–604 [DOI] [PubMed] [Google Scholar]
- Lucet I, Borriss R, Yudkin MD (1999) Purification, kinetic properties, and intracellular concentration of SpoIIE, an integral membrane protein that regulates sporulation in Bacillus subtilis. J Bacteriol 181: 3242–3245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnin T, Lord M, Yudkin MD (1997) Contribution of partner switching and SpoIIAA cycling to regulation of σF activity in sporulating Bacillus subtilis. J Bacteriol 179: 3922–3927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy JE, Gualerzi C (1990) Translational control of prokaryotic gene expression. Trends Genet 6: 78–85 [DOI] [PubMed] [Google Scholar]
- Min KT, Hilditch CM, Diederich B, Errington J, Yudkin MD (1993) σF, the first compartment-specific transcription factor of B. subtilis, is regulated by an anti-sigma factor that is also a protein kinase. Cell 74: 735–742 [DOI] [PubMed] [Google Scholar]
- Pal C, Hurst LD (2004) Evidence against the selfish operon theory. Trends Genet 20: 232–234 [DOI] [PubMed] [Google Scholar]
- Pan Q, Garsin DA, Losick R (2001) Self-reinforcing activation of a cell-specific transcription factor by proteolysis of an anti-sigma factor in B. subtilis. Mol Cell 8: 873–883 [DOI] [PubMed] [Google Scholar]
- Papp B, Pal C, Hurst LD (2003) Dosage sensitivity and the evolution of gene families in yeast. Nature 424: 194–197 [DOI] [PubMed] [Google Scholar]
- Price MN, Huang KH, Arkin AP, Alm EJ (2005) Operon formation is driven by co-regulation and not by horizontal gene transfer. Genome Res 15: 809–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado H, Moreno-Hagelsieb G, Smith TF, Collado-Vides J (2000) Operons in Escherichia coli: genomic analyses and predictions. Proc Natl Acad Sci USA 97: 6652–6657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain PS (2004) Efficient attenuation of stochasticity in gene expression through post-transcriptional control. J Mol Biol 344: 965–976 [DOI] [PubMed] [Google Scholar]
- Veitia RA (2002) Exploring the etiology of haploinsufficiency. BioEssays 24: 175–184 [DOI] [PubMed] [Google Scholar]