Abstract
Cellular response to stress entails complex mRNA and protein abundance changes, which translate into physiological adjustments to maintain homeostasis as well as to repair and minimize damage to cellular components. We have characterized the response of the halophilic archaeon Halobacterium salinarum NRC-1 to 60Co ionizing gamma radiation in an effort to understand the correlation between genetic information processing and physiological change. The physiological response model we have constructed is based on integrated analysis of temporal changes in global mRNA and protein abundance along with protein–DNA interactions and evolutionarily conserved functional associations. This systems view reveals cooperation among several cellular processes including DNA repair, increased protein turnover, apparent shifts in metabolism to favor nucleotide biosynthesis and an overall effort to repair oxidative damage. Further, we demonstrate the importance of time dimension while correlating mRNA and protein levels and suggest that steady-state comparisons may be misleading while assessing dynamics of genetic information processing across transcription and translation.
Keywords: haloarchaea, iTRAQ, microarray, oxidative stress, proteomics
Introduction
Organisms of the phylogenetic domain Archaea are environmentally ubiquitous and typically represent ∼10% of the microbiota (Robertson et al, 2005). However, in environments characterized by extreme conditions, such as high temperature or salinity, archaea dominate the microbial population due to their unique physiology (Woese et al, 1990). Halophilic archaea, for example, posses a range of mechanisms (Baliga et al, 2004; Kottemann et al, 2005) to endure high levels of solar radiation, greater than 4.0 M salinity and wide temperature fluctuations, all of which contribute to intermittent desiccation/rehydration cycles. In a previous study, we showed remarkably high resistance to one factor, ultraviolet (UV) radiation, and the coordinated genome-wide response involved in survival (Baliga et al, 2004). The physiological robustness of halophiles is further evident in the extraordinary resistance of the model halophile Halobacterium salinarum strain NRC-1 (heretofore Halobacterium NRC-1) to both desiccation and gamma (γ) radiation (Kottemann et al, 2005). Both these challenges induce severe DNA damage including structural modification to nucleotide bases and DNA strand breaks (Hutchinson, 1985; Dianov et al, 2001). In case of γ radiation, most of the damage results from production of hydroxyl radicals via radiolysis of water (Riley, 1994). Therefore, mechanisms to minimize and reverse oxidative stress are also crucial components of radiation resistance.
Systems approaches enable the elucidation of global physiological responses to environmental perturbations along with underlying regulatory circuits that modulate and coordinate various cellular repair and recovery processes (Kaur et al, 2006). Ideally, a systems approach constitutes the simultaneous analyses of dynamic changes at all levels of biological information processing from DNA, RNA and protein through phenotypic responses. Owing to technological and cost limitations, extensive time-series proteomic studies have thus far have been limited and hence microarray studies are often conducted with the assumption that transcript level changes sufficiently approximate downstream physiological responses. However, several studies have indicated an uncertain correlation between mRNA levels and downstream proteomic changes (Ideker et al, 2001; Baliga et al, 2002).
We have attempted to address this issue through a systems level time-course study of Halobacterium NRC-1 response to γ radiation using whole-genome microarray analyses of mRNA transcript levels and quantitative mass spectrometry analyses of total proteins. Through these analyses, we gained an overview of the regulatory and functional aspects of cell recovery after γ irradiation. Our finding of a high level of correlation (Pearson coefficients >0.5) between mRNA and protein levels upon including an appropriate time lag demonstrates that transcriptional changes do indeed translate into protein level changes in a physiologically meaningful manner.
Results and discussion
The physiological response of Halobacterium NRC-1 during recovery from irradiation with 2500 Gy of 60Co γ rays was examined temporally at mRNA and protein levels. Unintended perturbations were discounted with identically processed but unirradiated cells (see Supplementary Figure 1 for experimental design). mRNA level changes were measured over the entire time course (240 min), whereas protein abundance changes were measured at 30, 40 and 60 min in both irradiated and control cells.
mRNA level changes
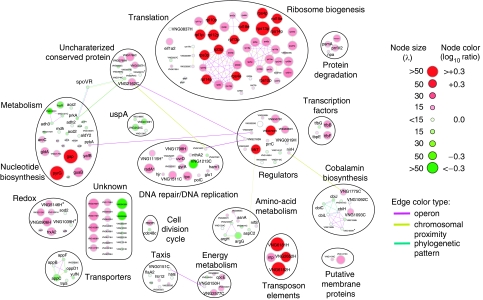
Significance of change in mRNA levels in microarray data was estimated with a maximum likelihood ratio test (Ideker et al, 2000). Comparison of identically processed biological replicates yields maximum likelihood statistic lambda (λ) values consistently below 15 for over 99% of all genes. Therefore, changes discussed heretofore are associated with λ>15.0 and correlate to >99% confidence level. Based on these statistical parameters, 216 genes (∼9% of all predicted genes) were differentially expressed of which 143 were upregulated and 73 were downregulated (Figure 1).
Figure 1.
Systems level visualization during early response to γ radiation for all genes showing systematic and significant expression changes with the layout organized by general function. The RNA changes are visualized as a network of genes (nodes) and their interactions (edges). The color of the nodes indicates upregulation (red) or downregulation (green) of that gene, whereas the size of the node relates to the significance of the observed change (λ value; see legend). Edge types are color coded to delineate the association type (see legend).
Protein level changes
We measured proteomic changes using quantitative tandem mass spectrometry analyses of four-plexed combinations of trypsinized total proteins labeled with amine-reactive isobaric iTRAQ reagents (isobaric tags for relative and absolute quantification; see Supplementary information for details on chemistry and statistical analysis). The iTRAQ chemistry was expected to yield significantly better proteome coverage relative to the isotope-coded affinity tag (ICAT) approach, as primary amines are more abundant in proteins than the cysteines targeted by ICAT. Indeed, the 1033 proteins detected using iTRAQ represented over three-fold better coverage relative to ∼300 proteins identified in a comparable ICAT analysis of Halobacterium NRC-1 (Baliga et al, 2002). Protein products for 99 of the 143 γ irradiation-induced transcripts were detected, of which 68 had significant abundance changes relative to the control (Supplementary Figure 2 and Table 2). Likewise, products for 44 proteins were detected for the 73 downregulated transcripts, and the abundances of 27 were significantly perturbed relative to the control. The lists of mRNAs and proteins that changed significantly are provided in Supplementary Tables 1 and 2.
In the sections below, we provide (1) a synthesis of the cellular response based on simultaneous analysis of transcript and protein level changes along with evolutionarily conserved functional associations and protein–DNA interactions and (2) a discussion on comparison of changes at mRNA levels to corresponding changes in protein abundance.
A systems model for physiological response to γ radiation
High-energy γ particles cause radiolysis of water, generating reactive oxygen species (ROS) (Hutchinson, 1985) resulting in oxidative stress and damage. Although the high intracellular concentration of KCl and bacterioruberins in haloarchaea have been hypothesized to provide protection by quenching ROS (Carbonneau et al, 1989), this alone is insufficient to alleviate stress induced from γ irradiation (Kottemann et al, 2005) requiring extensive repair and recovery processes. We have assimilated all transcript (Figure 1) and protein level changes during recovery from γ irradiation into a physiological model of Halobacterium NRC-1, which involves restoration of genome integrity, modulation of dehydrogenases, redoxins and cytochromes to minimize ROS reactions and inhibition of cell division (Figure 2).
Figure 2.
Graphical representation of the systems level response in Halobacterium NRC-1 to direct and indirect effects of γ radiation showing both repair and oxidative strategies for cell recovery. Text or arrow color indicates whether the cellular process or pathway was upregulated (red) or downregulated (green). Green X's indicate downregulated steps in the TCA cycle. Blue ellipses represent Htr8 and Htr12.
Physiological changes
DNA repair to restore genome integrity appears to be primarily mediated by homologous recombination and glycosylase activity. Archaeal homologous recombination proteins are structurally and functionally similar to those seen in eukaryotes (Allers and Ngo, 2003). Of the two RecA/Rad51 homologs in archaea, RadA (also called RadA1) and RadB (also called RadA2), only RadA can catalyze strand exchange (Komori et al, 2000). In Halobacterium NRC-1, RadA1 mRNA and protein levels increased during early γ response, which parallels similar DNA damage-responsive regulation of this gene in other organisms (Liu et al, 2003) (Supplementary Figures 2 and 3). Likewise, the branched structure-specific endonuclease hjr (Holliday junction resolvase) was also upregulated after γ irradiation.
Communication between DNA replication, repair and cell cycle progression is imperative to maintain genomic stability (Sancar et al, 2004). An inverse relationship was observed between mRNA changes of mcm (upregulated) and cdc48c (downregulated), a CdcH ortholog putatively involved in cell division (Supplementary Figure 2B), implying a pause in the cell division cycle as has been observed in other organisms (Rieger and Chu, 2004; Sancar et al, 2004) putatively to ensure completion of DNA repair before cell division. In accordance with a pause in cell cycle, a transient global downregulation was observed similar to that seen after UV irradiation (Baliga et al, 2004) (Supplementary Figure 4). However, unlike the UV response, key recovery-related pathways such as nucleotide biogenesis (PyrG), protein degradation (VNG0557H, PsmA), ribosome biogenesis (43 genes) and DNA repair were upregulated (Supplementary Figure 4).
Upregulation of nucleotide biosynthesis genes including pyrG (CTP synthase) and cmk (cytidydylate kinase) indicates increased de novo synthesis of nucleotides, which is consistent with damage responses observed in both higher eukaryotes (Rieger and Chu, 2004) and D. radiodurans (Liu et al, 2003). Increased nucleotide production may be necessary to accommodate increases in transcription and for DNA replication and repair. In line with this, a deletion of the ura3 homolog pyrF, which encodes an enzyme central to pyrimidine biosynthesis, significantly reduced resistance of Halobacterium NRC-1 to γ irradiation (see Supplementary information). An increased nucleotide pool would also serve to support enhanced ribosome genesis (43 ribosomal genes were upregulated).
Electron transport systems are especially sensitive to increased ROS production (Imlay, 2003). Therefore, the observed downregulation of eight dehydrogenases (e.g. adh2, adh3, sdh and mdh) may reflect depletion of intracellular reducing equivalents during severe oxidative stress (Golden and Ramdath, 1987) and an attempt to minimize ROS production by subsequent auto-oxidation reactions (Imlay, 2003). Likewise, increased protein abundances for superoxide dismutase Sod2 and redox-related functions such as thioredoxin (trxA2; Supplementary Figure 2A) might serve to scavenge free radicals (Cannio et al, 2000). Modulation of general cell metabolism may indirectly stem from these attempts to minimize oxidative stress; for example, as in D. radiodurans, several TCA cycle-related dehydrogenases in Halobacterium NRC-1were also downregulated (Liu et al, 2003; Ghosal et al, 2005).
The simultaneous analysis of transcriptional and translational changes has provided functional relevance for our observations enabling the synthesis of a coherent overview of physiological adjustments necessary for withstanding extreme levels of γ irradiation. In the next section, we will discuss aspects of transcriptional regulation for coordinating these various processes.
Transcriptional control
The construction of a gene regulatory map provides a framework to hypothesize and further investigate which transcription factors and regulators might directly mediate the observed transcriptional changes during a stress response. In Halobacterium NRC-1, transcription is mediated by an ∼12-subunit eukaryotic-like RNA polymerase II enzyme (RNAP), and two families of general transcription factors (GTFs): six TATA-binding proteins (TBPs) and seven Transcription Factor IIB orthologs (TFBs) (Baliga et al, 2000; Geiduschek and Ouhammouch, 2005). Transcription is further modulated by approximately 130 additional proteins orthologous to bacterial regulators (Sivaraman et al, 2005). During the γ response, at least nine regulators were upregulated along with one TBP (tbpE) and two TFBs (tfbB and tfbF). Of the five regulators that were downregulated, two were of unknown function and have been newly annotated (Supplementary Table 3). Besides these five regulators, tfbG was also repressed during early stages of the response.
We integrated all significant mRNA changes with a physical map of genome-wide protein–DNA interactions for seven TFBs (Facciotti et al, unpublished data) to investigate whether some of them may have specialized roles in the γ stress response. Of the 216 γ-responsive genes, transcription binding sites could be identified for 39% of them (84 genes) including 12 transcription regulators and kinases (Supplementary Table 4, gray boxes). An important point to note is that during the response to γ radiation, tfbB, tfbF and tfbG were differentially regulated and binding sites for at least one of these are observed in all but 10 of the 84 genes, representing a statistically significant enrichment of these binding sites (P-value=0.002). In fact, binding sites for TFBb and TFBf, the two TFBs upregulated by γ irradiation, were the most prevalent (>1.3-fold enrichment, P-value <0.009) upstream to genes that were also transcriptionally modulated during the response (Supplementary Table 5). In other words, although the presence of other TFB binding sites and possible condition-specific promoter binding by these GTFs complicate inference of exclusive stress-specific control by these TFBs, our data suggest statistically significant association between distribution of TFBb, TFBf and TFBg binding sites and transcriptional modulation of downstream genes in response to γ irradiation. Further, the differential regulation of tfbB, tfbF and tfbG in response to several stressors (Baliga et al, 2004; Kaur et al, 2006) motivates the hypothesis that these three TFBs coordinate various aspects of physiology that together constitute complex cellular responses during adjustment to diverse stress agents (Facciotti et al, unpublished data). This hypothesis will be tested in future experiments for a mechanistic understanding of stress response and its regulation.
Comparison of changes in mRNA levels and their corresponding protein abundances
The relationship between changes in a given mRNA and a corresponding change in its protein abundance is a function of their synthesis and degradation rates as well as their stability. Although there is no evidence for ubiquitin-ligase-mediated targeted protein degradation in halophilic archaea, a recent study has demonstrated likely involvement of C-terminal degradation signals for proteasomes in protein stability (Reuter and Maupin-Furlow, 2004). Furthermore, transient up- or downregulation of some genes (Kaur et al, 2006), differing rates of protein synthesis and post-transcriptional protein modifications all lend further complexity in correlating transcription and translation.
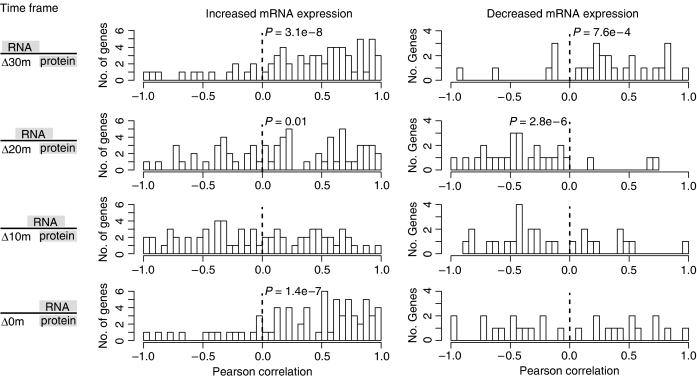
We have addressed the sequential nature and therefore likely temporal separation of these processes by calculating Pearson correlations (PC) for each gene by temporally shifting the protein level changes with respect to mRNA level changes; for example, protein levels at 30, 40 and 60 min compared with mRNA levels at 10, 20 and 40 min represent a time lag of 20 min (Δ20m). This analysis was conducted for all 95 genes with significant changes in both mRNA and protein abundances. The resulting correlations for each gene were binned (bin size=0.05) and are presented as histograms with P-values indicating the significance of the distribution of correlation values (Figure 3).
Figure 3.
Time-lagged Pearson correlations between mRNA and protein abundance for genes with significant changes in both. The time lag is given on the side ranging from 30 min (Δ30m) to no shift (Δ0m). P-values indicate the statistical significance of the distribution of Pearson coefficients and are only stated when a significant skew is observed. P-values on the right of the dashed line indicate a skew towards positive correlations, whereas those on the left indicate a negative correlation.
A high degree of correlation was observed for upregulated genes and their protein abundance at each time lag interval except at Δ10m (Figure 3). Transcript and protein levels for DNA repair genes (e.g. RadA1 and UvrD) had highest correlations (PC>0.6) over virtually all time lags perhaps due to a continuous increase in both over the entire time series. Further, whereas ribosomal proteins encoded within the two major operons had highly correlated changes in transcript and protein, those encoded by genes elsewhere in the genome were less correlated. Likewise, whereas TfbB and TfbF mRNA and protein level changes were highly correlated to one another in the absence of any time lag, TbpE protein abundance change was manifested after a 30 min time lag (Figure 3). In fact, transcript and protein changes for most downregulated genes were significantly correlated only after a time lag of 30 min. Thus, our analysis confirms that transcript and protein level changes vary gene-by-gene and on a temporal scale, and further that simplistic global correlations of mRNA and protein level changes at steady state might sometimes be misleading.
Conclusion
In this study, we have identified the cooperative physiological mechanisms that render Halobacterium NRC-1 resistant to γ radiation and have shown that these are reflected at both the transcript and protein level. This study further supports the view that transcript level changes might indeed be truly reflective of a significant fraction of protein level changes and therefore the physiological manifestations of those changes. Further, as general discordance between steady-state mRNA and protein levels are generally attributed to post-transcriptional regulation (Gygi et al, 1999; Ideker et al, 2001), our observation of relatively high degree of correlation between temporally shifted mRNA and protein levels demonstrates the potential importance of the time dimension while interpreting the mechanisms of genetic information processing.
Materials and methods
See supplementary information.
Data Accession Number
Data accession number at Gene Expression Omnibus (GEO) is GSE5557.
Supplementary Material
Supplementary Tables
Supplementary Information
Acknowledgments
We thank MT Facciotti for providing early access to ChIP-chip (protein–DNA interaction) data, R Bonneau for help with protein function annotation, A Schmid for helpful comments, MP Krebs for the Δura3 strain and V Adams for his technical assistance in irradiating the cells. This work was supported by research grants from NSF (EIA-0220153, EF-0313754), NIH (P50 GM076547) and DoE (DE-FG02-04ER63807 and DE-AC02-05CH11231) to NSB, MCB-0425825 to NSB and JDR, HFSP grant #RG522002 to JDR and an NSF OPP Postdoctoral Fellowship to KW.
References
- Allers T, Ngo HP (2003) Genetic analysis of homologous recombination in Archaea: Haloferax volcanii as a model organism. Biochem Soc Trans 31: 706–710 [DOI] [PubMed] [Google Scholar]
- Baliga NS, Bjork SJ, Bonneau R, Pan M, Iloanusi C, Kottemann MC, Hood L, DiRuggiero J (2004) Systems level insights into the stress response to UV radiation in the halophilic archaeon Halobacterium NRC-1. Genome Res 14: 1025–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baliga NS, Goo YA, Ng WV, Hood L, Daniels CJ, DasSarma S (2000) Is gene expression in Halobacterium NRC-1 regulated by multiple TBP and TFB transcription factors? Mol Microbiol 36: 1184–1185 [DOI] [PubMed] [Google Scholar]
- Baliga NS, Pan M, Goo YA, Yi EC, Goodlett DR, Dimitrov K, Shannon P, Aebersold R, Ng WV, Hood L (2002) Coordinate regulation of energy transduction modules in Halobacterium sp. analyzed by a global systems approach. Proc Natl Acad Sci USA 99: 14913–14918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannio R, Fiorentino G, Morana A, Rossi M, Bartolucci S (2000) Oxygen: friend or foe?. Archaeal superoxide dismutases in the protection of intra- and extracellular oxidative stress. Front Biosci 5: D768–D779 [DOI] [PubMed] [Google Scholar]
- Carbonneau MA, Melin AM, Perromat A, Clerc M (1989) The action of free radicals on Deinococcus radiodurans carotenoids. Arch Biochem Biophys 275: 244–251 [DOI] [PubMed] [Google Scholar]
- Dianov GL, O'Neill P, Goodhead DT (2001) Securing genome stability by orchestrating DNA repair: removal of radiation-induced clustered lesions in DNA. BioEssays 23: 745–749 [DOI] [PubMed] [Google Scholar]
- Geiduschek EP, Ouhammouch M (2005) Archaeal transcription and its regulators. Mol Microbiol 56: 1397–1407 [DOI] [PubMed] [Google Scholar]
- Ghosal D, Omelchenko MV, Gaidamakova EK, Matrosova VY, Vasilenko A, Venkateswaran A, Zhai M, Kostandarithes HM, Brim H, Makarova KS, Wackett LP, Fredrickson JK, Daly MJ (2005) How radiation kills cells: survival of Deinococcus radiodurans and Shewanella oneidensis under oxidative stress. FEMS Microbiol Rev 29: 361–375 [DOI] [PubMed] [Google Scholar]
- Golden MH, Ramdath D (1987) Free radicals in the pathogenesis of kwashiorkor. Proc Nutr Soc 46: 53–68 [DOI] [PubMed] [Google Scholar]
- Gygi SP, Rochon Y, Franza BR, Aebersold R (1999) Correlation between protein and mRNA abundance in yeast. Mol Cell Biol 19: 1720–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson F (1985) Chemical changes induced in DNA by ionizing radiation. Prog Nucleic Acid Res Mol Biol 32: 115–154 [DOI] [PubMed] [Google Scholar]
- Ideker T, Thorsson V, Ranish JA, Christmas R, Buhler J, Eng JK, Bumgarner R, Goodlett DR, Aebersold R, Hood L (2001) Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science 292: 929–934 [DOI] [PubMed] [Google Scholar]
- Ideker T, Thorsson V, Siegel AF, Hood LE (2000) Testing for differentially-expressed genes by maximum-likelihood analysis of microarray data. J Comput Biol 7: 805–817 [DOI] [PubMed] [Google Scholar]
- Imlay JA (2003) Pathways of oxidative damage. Annu Rev Microbiol 57: 395–418 [DOI] [PubMed] [Google Scholar]
- Kaur A, Pan M, Meislin M, Facciotti MT, El-Geweley R, Baliga NS (2006) A systems view of haloarchaeal strategies to withstand stress from transition metals. Genome Res 16: 841–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori K, Miyata T, DiRuggiero J, Holley-Shanks R, Hayashi I, Cann IK, Mayanagi K, Shinagawa H, Ishino Y (2000) Both RadA and RadB are involved in homologous recombination in Pyrococcus furiosus. J Biol Chem 275: 33782–33790 [DOI] [PubMed] [Google Scholar]
- Kottemann M, Kish A, Iloanusi C, Bjork S, Diruggiero J (2005) Physiological responses of the halophilic archaeon Halobacterium sp. strain NRC1 to desiccation and gamma irradiation. Extremophiles 9: 219–227 [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhou J, Omelchenko MV, Beliaev AS, Venkateswaran A, Stair J, Wu L, Thompson DK, Xu D, Rogozin IB, Gaidamakova EK, Zhai M, Makarova KS, Koonin EV, Daly MJ (2003) Transcriptome dynamics of Deinococcus radiodurans recovering from ionizing radiation. Proc Natl Acad Sci USA 100: 4191–4196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter CJ, Maupin-Furlow JA (2004) Analysis of proteasome-dependent proteolysis in Haloferax volcanii cells, using short-lived green fluorescent proteins. Appl Environ Microbiol 70: 7530–7538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieger KE, Chu G (2004) Portrait of transcriptional responses to ultraviolet and ionizing radiation in human cells. Nucleic Acids Res 32: 4786–4803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley PA (1994) Free radicals in biology: oxidative stress and the effects of ionizing radiation. Int J Radiat Biol 65: 27–33 [DOI] [PubMed] [Google Scholar]
- Robertson CE, Harris JK, Spear JR, Pace NR (2005) Phylogenetic diversity and ecology of environmental Archaea. Curr Opin Microbiol 8: 638. [DOI] [PubMed] [Google Scholar]
- Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S (2004) Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem 73: 39–85 [DOI] [PubMed] [Google Scholar]
- Sivaraman K, Seshasayee AS, Swaminathan K, Muthukumaran G, Pennathur G (2005) Promoter addresses: revelations from oligonucleotide profiling applied to the Escherichia coli genome. Theor Biol Med Model 2: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Kandler O, Wheelis ML (1990) Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 87: 4576–4579 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables
Supplementary Information