Abstract
Although human epidermal growth factor receptor 2 (HER2) overexpression is implicated in tumor progression for a variety of cancer types, how it dysregulates signaling networks governing cell behavioral functions is poorly understood. To address this problem, we use quantitative mass spectrometry to analyze dynamic effects of HER2 overexpression on phosphotyrosine signaling in human mammary epithelial cells stimulated by epidermal growth factor (EGF) or heregulin (HRG). Data generated from this analysis reveal that EGF stimulation of HER2-overexpressing cells activates multiple signaling pathways to stimulate migration, whereas HRG stimulation of these cells results in amplification of a specific subset of the migration signaling network. Self-organizing map analysis of the phosphoproteomic data set permitted elucidation of network modules differentially regulated in HER2-overexpressing cells in comparison with parental cells for EGF and HRG treatment. Partial least-squares regression analysis of the same data set identified quantitative combinations of signals within the networks that strongly correlate with cell proliferation and migration measured under the same battery of conditions. Combining these modeling approaches enabled association of epidermal growth factor receptor family dimerization to activation of specific phosphorylation sites, which appear to most critically regulate proliferation and/or migration.
Keywords: cell migration, cell proliferation, epidermal growth factor receptor, signal transduction, tyrosine phosphorylation
Introduction
Human epidermal growth factor receptor 2 (HER2), also known as Erb-B2, belongs to the epidermal growth factor receptor (EGFR) family of highly regulated receptor tyrosine kinases (RTKs) composed of human epidermal growth factor receptors 1, 2, 3, and 4 (EGFR, HER2, HER3, and HER4). These receptors are differentially bound by 11 ligands, although so far no ligand has been found to bind HER2. Mutation and dysregulation of EGFR family members has been correlated with cancer development and progression (Yarden and Sliwkowski, 2001; Mendelsohn and Baselga, 2003), and overexpression of HER2 has been found in association with a variety of tumor types (Hynes and Lane, 2005). The connection between HER2 and cancer appears to be especially important in breast cancer (Lemmon, 2003), with current estimates of prevalence on the order of 25–30% of patients including metastases as well as primary tumors (Carlsson et al, 2004) and notable correlation with poor prognosis (Citri et al, 2003). For instance, one recent review has concluded that HER2/neu gene amplification or HER2 protein overexpression can predict breast cancer outcome for 90% of the studies and 92% of the patients in 81 studies including 27 161 patients (Ross et al, 2003).
Although HER2 appears to have no intrinsic ligand-binding capability, it can interact reversibly with ligand-activated EGFR or HER3 to form active heterodimers that perturb and often enhance the downstream signals that govern cell proliferation and migration (Hynes and Lane, 2005). At high concentrations, HER2 is also thought to spontaneously form signaling-competent homodimers (Harari and Yarden, 2000). In the case of HER2–EGFR heterodimers, signaling proceeds through phosphorylation events initiated from either of the activated receptors. In contrast, HER2–HER3 heterodimer signaling proceeds only through the activated HER2 receptor, as HER3 does not exhibit kinase activity itself. HER2 is considered responsible for at least the vast majority of downstream signals induced by HER3-ligand binding (Holbro et al, 2003), although there is some evidence of HER3–EGFR signaling heterodimers (Knowlden et al, 2003). Activation of HER2–HER3 heterodimers, generally considered to be the most potent signaling pair of EGFR family dimers (Pinkas-Kramarski et al, 1996), is tightly regulated (Citri et al, 2003), and HER2 overexpression has been found to mediate enhanced signaling caused by heregulin (HRG) stimulation (Aguilar et al, 1999). Various studies have shown that HRG-stimulated HER3 activation in breast cancer can induce anti-estrogen resistance, tumor progression, invasion, and metastasis (Yao et al, 2001; Atlas et al, 2003; Tsai et al, 2003). Taken together, the high correlation between EGFR family member dysregulation and cancer progression highlights the need for mechanistic understanding of the underlying cellular networks, both for improved basic knowledge of cancer and to find new and more effective drug targets.
Signal generation and resultant cellular behavior in RTK-initiated cascades is achieved through a highly coordinated series of events. Receptors are first activated, typically through ligand binding, which results in receptor autophosphorylation and leads to the dynamic phosphorylation/dephosphorylation of a variety of proteins. The dynamic state of protein phosphorylation ultimately controls cellular response, and a given protein may influence more than one response as it becomes phosphorylated at different sites or interacts with different proteins depending on timing and placement within the cell. Thus, to understand cellular signaling in the context of cell regulation, it is helpful to quantify phosphorylation dynamics of regulatory sites and their consequent association with downstream cell functions. Our work presented here offers, to our knowledge, the most detailed explanation of how signaling network phosphorylation, in the context of HER2 overexpression, governs cellular behavior such as proliferation and migration across a diverse set of ligand stimulation conditions.
To identify important epidermal growth factor (EGF)- and HRG-induced protein phosphorylation events that control cell migration and proliferation in the context of HER2 overexpression, we utilized a mass spectrometry (MS) approach that is capable of simultaneously quantifying the temporal dynamics of a large number of specific tyrosine phosphorylation sites under a given condition (Zhang et al, 2005b). Here, we extend this methodology to measure temporal dynamics of protein tyrosine phosphorylation sites following EGFR or HER3 stimulation in the presence and absence of HER2 overexpression. Protein phosphorylation sites are clustered based on similarity of dynamic response to different stimulation conditions across four time points for all four conditions (self-organizing map (SOM)), revealing co-regulated phosphorylation sites and providing potential functionality for several novel phosphorylation sites. In addition to MS analysis of protein phosphorylation, biological response data (cell migration and proliferation) were quantified for each of the four stimulation conditions. To identify signals that regulate downstream biological response to a given stimulus, computational methods were used to correlate biological responses to quantitative phosphoproteomics data. Phosphorylation sites, which correlate strongly with proliferation or migration, were identified and may be targeted in future studies to selectively inhibit a given response.
Results and discussion
Phosphotyrosine MS of parental and HER2-overexpressing HMECs in response to EGF and HRG treatment
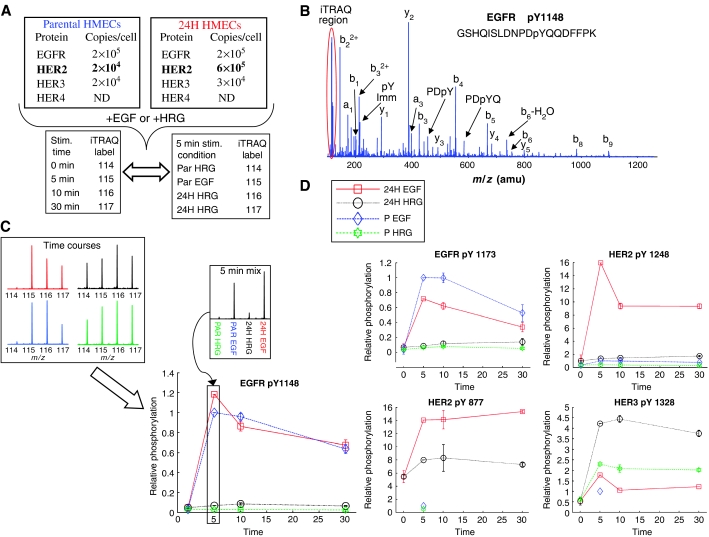
We have undertaken a quantitative investigation of the effects of HER2 overexpression in a human mammary epithelial cell (HMEC) line (184A1; Stampfer and Bartley, 1985). We compared the parental line (denoted ‘P'), which exhibits approximately 200 000 EGFR per cell, 20 000 HER2 per cell, and 20 000 HER3 per cell, to a stable retrovirally transduced clone (denoted ‘24H') that expresses HER2 at 600 000 per cell while maintaining constant EGFR levels and increased HER3 levels to about 30 000 per cell. We have previously performed combined experimental and modeling studies on this cell line enabling quantitative estimation of the various receptor dimer species under various treatment conditions (Hendriks et al, 2003a, 2005). From our previous work, we expect that 100 ng/ml EGF treatment should result in high levels of EGFR homodimers in the P HMECs (∼80 000), but lower numbers of EGFR–HER2 heterodimers (∼20 000). By comparison, treating 24H HMECs with this same concentration of EGF should drive a large increase in heterodimers (to ∼150 000) and a significant decrease in EGFR homodimers (∼10 000). HRG treatment at 80 ng/ml should yield ∼15 000 HER2–HER3 heterodimers in the P HMECs, and in the 24H HMECs this number should increase to ∼25 000. Interestingly, under both ligand treatments, 24H HMECs are expected to have large numbers of HER2 homodimers (>∼200 000), some of which may be activated through basal autophosphorylation. Here, we explore the consequences of these changing dimerization states on intracellular signaling and the subsequent control of cell proliferation and migration. We have acquired MS data describing the temporal dynamics (0, 5, 10, and 30 min stimulation) of tyrosine phosphorylation in the 184A1 cell system under four conditions: P HMECs stimulated with EGF, P HMECs stimulated with HRG, 24H HMECs stimulated with EGF, and 24H HMECs stimulated with HRG (see Figure 1).
Figure 1.
Data acquisition and quantification scheme with example data. (A) 184A1 parental HMEC cells and 24H HMEC cells were stimulated with either EGF or HRG for 0, 5, 10, or 30 min. For each stimulation condition, following cell lysis and tryptic digestion, peptides from each time point were labeled with iTRAQ, mixed, and analyzed by anti-phosphotyrosine peptide immunoprecipitation and immobilized metal affinity chromatography–liquid chromatography tandem mass spectrometry (IMAC–LC-MS/MS), producing temporal tyrosine phosphorylation profiles for hundreds of peptides. To normalize temporal profiles from each condition, tryptic peptides from the 5 min stimulation time point for each condition were labeled with iTRAQ, mixed, and analyzed by anti-phosphotyrosine peptide immunoprecipitation and IMAC–LC-MS/MS. (B) For each peptide, y- and b-type ions in the MS/MS spectrum provided sequence identification and site assignment, whereas the iTRAQ marker ion region (highlighted in the red oval) provided quantification. (C) iTRAQ marker ion profiles representing temporal phosphorylation profiles for EGFR pY1148 under all four conditions (top left) as well as the relative amount of phosphorylation following a 5 min stimulation for each condition (top right). The final plot of temporal response under multiple stimulation conditions (bottom) was generated by multiplying each temporal phosphorylation profile by the value obtained from the 5 min mix quantification data for the particular stimulation condition. (D) Multiple tyrosine phosphorylation sites were identified and quantified from EGFR family member proteins. Four representative phosphorylation sites are shown here, with response plots across temporal and conditional space. As can be seen by comparing HER2 pY877 and pY1248, different sites on a given protein are often differentially regulated.
As a result of these analyses, 332 phosphorylated peptides from 175 proteins were identified, including 289 singly (tyrosine) phosphorylated peptides, 42 doubly phosphorylated peptides (21 tyrosine/tyrosine, 18 serine/tyrosine, and three threonine/tyrosine), and one triply phosphorylated peptide (tyrosine/tyrosine/tyrosine) (full data set is available in Supplementary information). A total of 20 phosphorylation sites were identified on EGFR, HER2, and HER3, including nine tyrosine and two serine sites on EGFR, eight tyrosine phosphorylation sites on HER2, and one tyrosine phosphorylation site on HER3 (Figure 1B–D). Of the 20 phosphorylation sites on EGFR family members, Y1114 on EGFR and Y1005 and Y1127 on HER2 represent novel sites that have not been previously described in the literature, although synthetic phosphopeptides containing EGFR Y1114 and HER2 Y1005 have been shown to bind SHC (Schulze et al, 2005) and mutation of EGFR Y1114 has been shown to block SOCS recruitment to the receptor, regulating STAT activation (Xia et al, 2002). Downstream of the receptors, quantitative data were obtained for 36 phosphorylation sites on 15 different proteins in the EGFR canonical signaling pathway (as defined by Yarden and Sliwkowski, 2001). Coverage of the cell adhesion/migration pathway included quantitative information on 41 phosphorylation sites distributed along 16 proteins, including nine tyrosine phosphorylation sites on δ-catenin.
One of the goals in this study was to provide a more comprehensive definition of EGFR family signaling networks, including novel proteins or phosphorylation sites that may regulate differential cellular response to exogenous stimuli. To enable identification of novel phosphorylation sites, MS analyses were performed in an information-dependent acquisition mode (automated selection of the most abundant species in a given full scan mass spectrum for MS/MS analysis), rather than targeting specific peptides and known phosphorylation sites for quantification. This data-dependent mode of operation was successful at identifying novel proteins and phosphorylation sites, as 122 of the 322 phosphorylation sites have not previously been described in the literature. Unfortunately, the use of automated ion selection to discover novel phosphorylation sites often precluded selection of low-abundance precursor ions for MS/MS fragmentation, and therefore temporal phosphorylation profiles were not obtained for all conditions for many peptides. In fact, 234 of the 322 sites in this work were detected and quantified in multiple analyses, but only 68 were quantified for all four stimulation conditions and the 5 min comparison. This core group of 68 phosphopeptides, which include many of the key signaling nodes in the network, was then used for further computational analysis to obtain a mechanistic understanding of the effects of HER2 overexpression on cellular signaling networks and corresponding biological response to growth factor stimulation.
Self-organizing maps define temporal and conditionally related clusters of phosphorylation sites
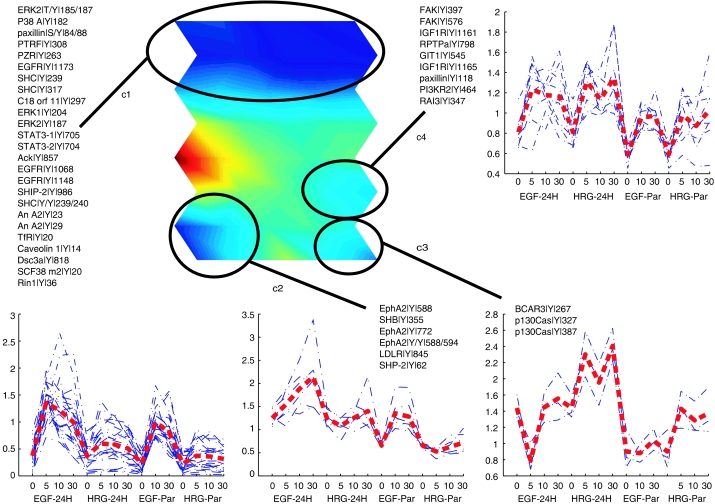
In order to identify clusters of tyrosine-phosphorylated peptides exhibiting similar temporal dynamics, as well as to globally visualize the high-dimensional information we have obtained, we used the SOM algorithm (Kohonen, 2001). The SOM is a versatile clustering algorithm that transforms high-dimensional data into lower dimensional display, in a nonlinear manner. Here, we use the unified-distance matrix (or U-matrix) approach, which allows robust identification of clusters, and the component plane representation, which facilitates comparison of peptide phosphorylation response to exogenous stimuli (Ultsch and Siemon, 1990; Kohonen, 2001). Instead of taking a single map unit as one cluster, we use U-matrix to identify groups of map units that together comprehend a cluster. The U-matrix illustrates the mean distances between neighboring map units after the SOM training phase. These distances are color-coded so that close proximity of two map units is colored with bluish colors, whereas shades of yellow and red denote dissimilarity. Clusters can be identified as continuous bluish regions (valleys) surrounded by yellow or red ‘mountains.'
We applied this SOM algorithm to 62 ‘core' phosphorylated peptides for which temporal profiles were generated under all conditions (six of the ‘core' phosphorylated peptides had minimal response across all conditions and time points and were removed from this analysis). Data were normalized with the z-score method across all time points and conditions. From the U-matrix in Figure 2, four clusters are readily identifiable. Statistical significances for these clusters were computed with a permutation test-based method (Hautaniemi et al, 2003) and the clusters were found to be statistically significant (P<0.05). The entire SOM display including the component planes is given in Supplementary information.
Figure 2.
SOM to cluster phosphorylation sites across conditional and temporal space. Phosphorylation levels for 62 core phosphorylated peptides, which were detected and quantified at four time points for each of the four stimulation conditions and the 5 min mix analysis, were normalized and placed into an SOM. Four statistically significant clusters were identified in the SOM (circled bluish valleys surrounded by yellow ‘mountains'). The components for each of the four clusters are indicated in the figure along with the phosphorylation profiles (blue dashed lines) and an average profile for each cluster (red line) across conditions and time points (16 dimensions).
By comparing the individual phosphorylation profiles (Figure 2, blue dashed lines) or average phosphorylation profile (Figure 2, red line) within each cluster, it is clear that the algorithm has clustered peptides whose phosphorylation level is increased under selected conditions, information that can be used to link phosphorylation sites within a cluster to activated receptor homo- or heterodimers. For instance, the first cluster (c1 in Figure 2) consists of 18 peptides whose phosphorylation levels, on average, are highest following EGF stimulation of P or 24H HMECs, conditions that would lead to activation of EGFR homodimers or EGFR–HER2 heterodimers, respectively. Correspondingly, most of the proteins in this cluster have been previously associated with proliferation and early response to EGF stimulation, including EGFR Y1068 and Y1148, STAT-3 Y705 (STAT-3 isoform 1) and Y704 (STAT-3 isoform 2), SHIP-2 Y986, SHC Y239, Y240 and Y317 and early effectors downstream of them, including MAP kinases ERK2 (phosphorylated at Y187 and at T185 and Y187), Erk1 Y204, and p38α Y182.
In comparison to c1, the three other clusters in the U-matrix (c2–c4) contain peptides whose phosphorylation appears to be regulated by HER2 activation, as phosphorylation levels on these peptides are highest when HER2 is overexpressed. More specifically, peptides in the second cluster (c2) are primarily phosphorylated downstream of EGFR–HER2 heterodimers, as the highest levels of phosphorylation occur in the 24H HMECs stimulated with EGF. This cluster consists of SHB Y355, SHP-2 Y62, LDL receptor Y845, and three EphA2 tyrosine sites, including the activation loop at Tyr 772 (Kinch and Carles-Kinch, 2003). Of these proteins, only SHP-2 has been previously associated with EGFR activation (Qu et al, 1999). Interestingly, each of the other proteins has been associated with the cell migration response to VEGF stimulation (Cheng et al, 2002; Holmqvist et al, 2003; Prager et al, 2004), a response that requires EGF autocrine signaling following VEGF stimulation in endothelial cells (Semino et al, 2006). Our data link these phosphorylation sites to the EGFR family signaling network, and as demonstrated below, show strong correlation between these phosphorylation sites and cell migration in response to EGF stimulation of 24H HMECs.
The third cluster (c3) contains three peptides that are primarily phosphorylated downstream of activated HER2–HER3 heterodimers, as implicated by maximal phosphorylation levels following HRG stimulation of 24H HMECs. This cluster includes phosphorylation of p130Cas at Y327 and Y387, and phosphorylation of BCAR3 at Y267. Both of these proteins are part of the same family and have been shown to regulate or be regulated by Src activity in the cell migration signaling network. Similar to c2 and c3, peptides in the fourth cluster (c4) are predominantly phosphorylated downstream of HER2, but are equally phosphorylated downstream of EGFR–HER2 or HER2–HER3 heterodimers or possibly activated through active HER2 homodimers (similar phosphorylation levels are seen in EGF- or HRG-stimulated 24H HMECs). This cluster includes phosphorylation of retinoic acid-induced protein 3 (RAI3) Y347, paxillin Y118, GIT1 Y545, FAK Y397 and Y576, receptor protein tyrosine phosphatase alpha (RPTPα) Y798, PI3K Y464, and insulin-like growth factor receptor (IGF-1R) Y1161 and Y1165. A complex containing GIT1 and paxillin has been shown to regulate cell motility, possibly through recruitment of PAK or PIX to the leading edge (Manabe et al, 2002); the role of tyrosine phosphorylation in this process has not been established. Other proteins in this cluster have also been shown to be co-regulated, as RPTPα has recently been shown to regulate integrin signaling through Src activation in a PI3K-dependent manner downstream of insulin activation (Vulin et al, 2005), and Src activation is known to regulate tyrosine phosphorylation of FAK. It is worth noting that the phosphorylation sites on IGF-1R (Y1161 and Y1165) occur within a peptide that is homologous to the insulin receptor. Although not conclusive, assignment to IGF-1R is based on the presence of a phosphorylation site on a peptide specific for IGF-1R in the larger data set and greater levels of IGF-1R as compared to insulin receptor on HMEC parental and 24H cells (HS Wiley, personal communication). Activation of IGR-1R has been found to be correlated with increased motility in a breast cancer cell line (Zhang et al, 2005a); in our system, increased phosphorylation is most likely associated with autocrine release and binding of IGF-1 following stimulation of EGFR family members.
From these clusters, it is clear that analyzing phosphorylation sites across 16 dimensions of temporal and conditional space can reveal co-regulated phosphorylation sites, and that this information can be used to connect phosphorylation of specific sites to activation of EGFR family member dimers. Although clusters 3 and 4 both contain phosphorylation sites from proteins known to regulate migration, they are statistically distinct clusters whose differential response to exogenous stimuli may be owing to protein subcellular localization, as proteins from cluster 4 are known to be associated with the membrane, whereas proteins in cluster 3 are primarily cytosolic. SOM-based data analysis can reveal interesting hypotheses and connectivity in the data, such as the potential role of GIT1 Y545 affecting cell migration or the inclusion of PTPRα, PI3K, and IGF-1R phosphorylation sites in a single cluster, but it is still necessary to relate phosphorylation and phenotypic data to solidify these hypotheses.
Cell proliferation and migration are differentially stimulated via EGFR and HER2
In order to correlate signaling data with cellular response, we quantified both cell migration and cell proliferation in the HMEC parental and 24H cell lines. Measurements were obtained under serum-free, HRG-stimulating (80 ng/ml), or EGF-stimulating (100 ng/ml) conditions.
Cell migration was measured using a wound-closure assay adapted for a 96-well plate format and automated fluorescent imaging. Under all conditions investigated, the 24H cells moved more rapidly into the induced wound, thereby reducing the original wound area at a greater rate than the parental cells (Figure 3A–C). The wound closure trajectories were fit to a line, and the slopes were the input for partial least-squares modeling. Interestingly, the greatest difference between the two cell lines occurs during EGF treatment. By comparison, HRG stimulation, while inducing a slightly higher rate of wound closure for 24H HMEC relative to P HMEC, does not seem to drive significantly enhanced migration relative to serum-free conditions.
Figure 3.
EGF and HRG drive migration and proliferation to varying extents in HMEC parental and HER2-overexpressing cells. Parental (black) and 24H (red) wound healing data shown for (A) serum-free conditions, (B) treatment with 100 ng/ml EGF, and (C) treatment with 80 ng/ml HRG. (D) Proliferation, as measured by [3H]thymidine incorporation, for both cell types under the treatment conditions shown in (A–C).
Cell proliferation was measured by [3H]thymidine uptake 25 h after ligand stimulation. Figure 3D shows that EGF treatment increased thymidine uptake to a greater extent than did HRG treatment, but both treatments induced higher amounts of proliferation than seen in serum-free conditions. In direct contrast to the migration phenotype, there was no significant difference (i.e., P≫0.05 in all cases) between the two cell types measured under any of the stimulation conditions. Thus, HER2 overexpression did not seem to facilitate enhanced proliferation in any of the conditions probed.
The fact that HER2 overexpression mediated differences in migration, particularly under EGF-stimulating conditions, but did not do so for proliferation indicates that some of the signaling molecules differentially regulated by HER2 expression levels play a role in driving higher levels of migration while at the same time remaining agnostic to cell proliferation. The linear regression modeling discussed below integrates our quantitative migration, proliferation, and signaling data to describe, among other things, a set of signaling molecules that is most relevant for the changes in migration induced by HER2 overexpression.
Modulation of EGFR signaling network by HER2
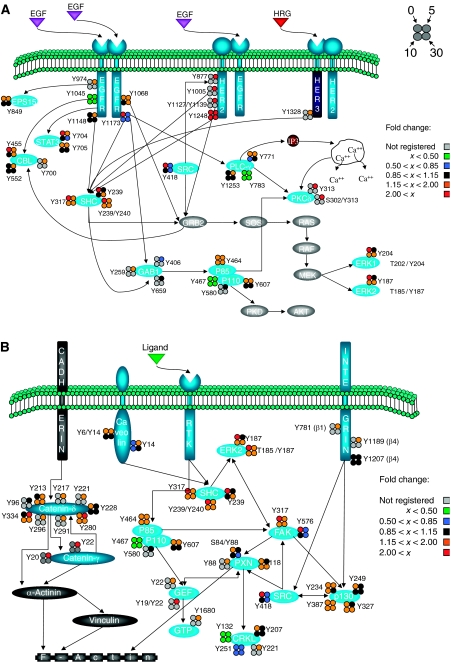
To assess the effect of increased HER2 expression levels on the canonical EGF-activated pathway, the phosphorylation level for sites observed in EGF-stimulated 24H cells was divided by the phosphorylation level for the same site and stimulation time in EGF-stimulated parental cells, producing a fold change in phosphorylation level for a given site and time. A subset of the proteins and phosphorylation sites within the canonical EGFR signaling network are shown in Figure 4A. As is evident from this figure, increased HER2 expression affects most phosphorylation sites on selected proteins in the EGFR signaling network, but not all phosphorylation sites on a given protein react equally to this perturbation. For instance, each of the multiple phosphorylation sites on EGFR exhibits different regulation at low or high HER2 expression levels, including increased phosphorylation of Y974 and decreased phosphorylation on Y1045 under HER2 overexpression as compared with basal HER2 expression. Both of these sites appear to regulate receptor internalization and degradation: Y974A or Y974F mutations have been shown to decrease receptor internalization rates (Sorkin et al, 1996), and Cbl (E3-ubiquitin ligase) binding to EGFR Y1045 is required for lysosomal sorting and receptor degradation (Grovdal et al, 2004). Decreased phosphorylation at the Y1045 site should lead to decreased ubiquitination of activated EGFR, thereby providing a mechanism for the observed increase in recycling of activated EGFR in the 24H cell line relative to parental 184A1 HMECs (Hendriks et al, 2003b).
Figure 4.
Effect of increased HER2 expression on phosphorylation sites within the EGFR signaling network. Visualization of the fold change in phosphorylation level between 24H and parental cells stimulated with EGF provides a network view of the mechanistic effects of HER2 overexpression on (A) the canonical EGFR signaling cascades and (B) the cell migration-associated signaling pathway.
It is also interesting to note the higher basal phosphorylation level for selected sites in the 24H cell lines relative to parental HMECs. Increased basal phosphorylation associated with HER2 overexpression could be mediated by HER2 autophosphorylation, crossphosphorylation of EGFR in the absence of ligands, or even through increased autocrine stimulation of HER2 heterodimers. For most sites with high levels of basal phosphorylation in the 24H cells, stimulation with saturating concentrations of EGF typically resulted in a greater response in parental cells relative to 24H cells, such that after EGF stimulation for 5 min many of the sites had similar levels of phosphorylation in both cell lines. A specific example of this behavior is provided by phosphorylation of the ERK2 activation loop (Y185 and T185/Y187). Much higher levels of basal phosphorylation were detected in the 24H cells relative to parental cells, but only a slight difference in phosphorylation at these sites remained after 5 min of EGF stimulation. These results are consistent with our proliferation data, in which no significant difference between 24H and parental cells under EGF stimulation was observed, in contrast to serum-free 24H cells, which had greater proliferation than parental HMECs (P<0.1). These results are also consistent with previous literature demonstrating ERK2's role as a potent activator of proliferation (Cobb et al, 1994). In fact, many protein phosphorylation sites associated with proliferation behave similarly to ERK2 in our system, including STAT-3 Y705 (and Y704 from STAT-3 isoform 2), EGFR Y1173, and SHC Y239, Y240, and Y317.
As HER2 overexpression has been found to affect cell motility in our study as well as in previous work (Spencer et al, 2000), the effect on protein phosphorylation in a subset of the pathways related to cell migration is shown in Figure 4B. From this figure, it is clear that there is an overall increase in phosphorylation of many of these pathway proteins in the presence of HER2 overexpression. Perhaps most striking is the increase in phosphorylation for all of the phosphorylation sites detected on catenin-δ and catenin-γ. Catenins are known to interact with E-cadherin, the main cell–cell adhesion protein in epithelial cells (Davis et al, 2003). Catenin-δ, a member of the p120 catenin family, has been shown to regulate E-cadherin turnover by modulating its internalization and degradation rate (Davis et al, 2003), stabilizing it on the cell surface when bound to it. HER2 overexpression in breast carcinomas inhibits the transcription of E-cadherin (D'Souza and Taylor-Papadimitriou, 1994) and has also been found to destabilize the catenin–cadherin complex, leading to decreased adhesion (Jawhari et al, 1999). Also, EGFR has been found to directly phosphorylate p120 catenin at Y228 (Mariner et al, 2004), and EGFR inhibition was found to promote assembly of cell adhesions (Lorch et al, 2004). The accumulated evidence of these reports is consistent with our data as it argues that Src- or RTK-related phosphorylation of catenin-δ leads to separation of the catenin–cadherin complex, resulting in destabilization of E-cadherin and an increase in cell migration. Tyrosine phosphorylation of catenin-γ has also been related to loss of cell–cell adhesions in EGF-stimulated, E-cadherin-positive cervical cancer cells (Moon et al, 2001). Our data show high levels of phosphorylation of catenin-δ and catenin-γ in HER2-overexpressing cells under EGF stimulation, directly contributing to loss of cell adhesion and an increase in cell motility.
Although catenin phosphorylation and loss of E-cadherin-based cell–cell adhesion may provide part of the migratory response following EGF stimulation in 24H cells, increased phosphorylation of many additional components of the cell migration pathways is also likely to be contributing to increased migration of these cells relative to parental cells. For instance, FAK, Src, paxillin, and p130Cas have all been shown to interact with each other, to localize at the focal adhesions, and to play a fundamental role in the actin cytoskeleton reorganization and motility pathways (Webb et al, 2004). Phosphorylation of paxillin (Y31) and (Y118) is known to regulate membrane spreading and ruffling in cell migration and adhesion (Tsubouchi et al, 2002). FAK and Src are two of the major kinases involved in cell migration. FAK (Y397) is the major autophosphorylation site and acts as a docking site for Src and PI3K (Cary and Guan, 1999); this site is also necessary for both p130cas and paxillin phosphorylation in response to FAK expression (Cary and Guan, 1999). Src Y418 is a major Src autophosphorylation site, whose phosphorylation results in self-activation (Roskoski, 2004). Src can mediate FAK-related migration by further phosphorylation of paxillin or p130Cas, which may result in focal adhesion turnover and/or lamellipodia and filipodia formation via either the RAC, JNK pathway, or the RhoGAP pathway (Playford and Schaller, 2004). We conclude that the effect of increased HER2 expression levels is partly increased interaction of FAK, Src, P130Cas, and paxillin, a small but noticeable increase in ERK2 activity, and the upregulation of catenin phosphorylation and thus E-cadherin turnover, all combining to drive an increased migratory response in the 24H cell line (as in Figure 3).
HRG versus EGF stimulation in the presence of HER2
In addition to investigating the effects of HER2 on these pathways in Figure 5, our data also enabled comparison of signaling downstream of activated HER2:HER3 versus EGFR:HER2 heterodimers, resulting from stimulation of the 24H cells with HRG and EGF, respectively. For this analysis, the phosphorylation level resulting from HRG stimulation of 24H cells was divided by the phosphorylation level resulting from EGF stimulation of 24H cells to produce the fold change ratio for each phosphorylation site in the canonical signaling pathways (Figure 5A). As expected, stimulation with HRG instead of EGF caused a significant increase in HER3 phosphorylation and a large decrease in EGFR phosphorylation. Perhaps not surprisingly given the relative receptor expression levels (20 000–30 000 copies/cell for HER3 versus 200 000 copies/cell for EGFR) and the kinase-dead nature of HER3, stimulation with saturating concentrations of HRG resulted in decreased phosphorylation of almost all downstream proteins as compared to stimulation with saturating concentrations of EGF (Figure 5A). Specifically, most of the downstream effectors that lead to proliferation (STAT-3 Y705 (Y704), ERK T202/Y204, all three phosphorylation sites on SHC) were phosphorylated to a lesser degree under HRG stimulation, correlating with decreased proliferation in HRG-stimulated 24H cells as compared to EGF-stimulated 24H cells (as in Figure 3). Other sites primarily associated with migration (Src, PKC-δ, and PI3K) do not show the same decrease in phosphorylation when comparing HRG to EGF stimulation of these cells. In fact, both Src and PKC-δ show an increase in phosphorylation under HRG activation at 5 and 10 min, respectively. Together with our response data, these results indicate that signaling distinct from Src, PKC-δ, and PI3K controlled pathways may govern cell migration in our system.
Figure 5.
Effect of HRG stimulation versus EGF stimulation in high HER2-expressing cells. Visualization of the fold change in phosphorylation level between HRG-stimulated and EGF-stimulated 24H cells provides a network view of the mechanistic effects underlying differential response to distinct growth factor stimuli on (A) the canonical EGFR signaling cascades and (B) the cell migration-associated signaling pathway.
To analyze the effect of HRG stimulation on the cell migration signaling network, signaling downstream of activated HER2:HER3 versus EGFR:HER2 heterodimers was also compared for selected proteins within a subset of the pathways related to cell motility (Figure 5B). Comparing phosphorylation levels for specific sites under different stimulation conditions provides a potential hypothesis to explain the mechanism underlying the quantitative phenotypic migration measurements, in which EGF stimulation resulted in a dramatic increase in migration relative to HRG stimulation of 24H cells (see Figure 3). Given the difference in migration rates between these two cell states, it is not surprising to note that protein phosphorylation sites on almost all effectors of migration have decreased phosphorylation levels following HRG stimulation as compared to EGF stimulation. However, in contrast to most proteins in the motility network, tyrosine phosphorylation levels on FAK, p130Cas, Src, and paxillin were increased following HRG stimulation compared to EGF stimulation, indicating that the slight migratory effect of HRG stimulation of 24H cells is directed primarily through amplified phosphorylation of a very specific pathway driven through these four proteins.
Taken together, the data from Figures 4 and 5 demonstrate that 24H cell migration can be induced by either broad upregulation of the migratory pathway, including loss of cell adhesions (24H cells under EGF stimulation) or, to a much lesser extent, by more intense upregulation of a restricted subset of the migratory pathway (as in 24H cells stimulated with HRG). By comparison, 24H cell proliferation was induced to a greater extent by EGF stimulation as determined by phenotypic assay; correspondingly, phosphorylation levels were higher on almost all proliferation-associated proteins on 24H cells stimulated with EGF as compared to 24H cells stimulated with HRG. Also consistent with the phenotypic data, the effect of increasing HER2 expression levels was most dramatic for proteins in the cell migration signaling network, as most proteins displayed a sustained increase in phosphorylation levels, but was much less dramatic for proteins in the proliferation-associated network, as increased basal phosphorylation levels in 24H cells were not sustained following EGF stimulation.
Quantitative proteomics and microarray-based analyses have been used to address the question of protein interactions with cytosolic domain tyrosine phosphorylation sites from EGFR family members (Schulze et al, 2005; Jones et al, 2006; Uetz and Stagljar, 2006). Although our work is aimed at analyzing phosphorylation dynamics in the signaling network across different cellular environments, and therefore does not focus on the identification of proteins interacting with EGFR family members, several similarities can be found between our results and these recent publications. For instance, Jones et al demonstrated that HER3 interacts with a small number of downstream proteins, which is consistent with our findings that HER3 uses a very specific set of nodes to activate the migration pathway. Additionally, HER2 and EGFR were found to be the most promiscuous members of the EGFR family (Jones et al, 2006), whereas in our study EGF stimulation of HER2-overexpressing cells promoted migration via a wide range of phosphorylation events coupling several signaling cascades. Overall, the proteomics approaches described by Schultze et al and Jones et al complement the data presented in our work by highlighting proteins directly interacting with tyrosine phosphorylation sites on EGFR family members, providing additional structure to the signaling network.
PLSR modeling correlates signals with cell functional responses
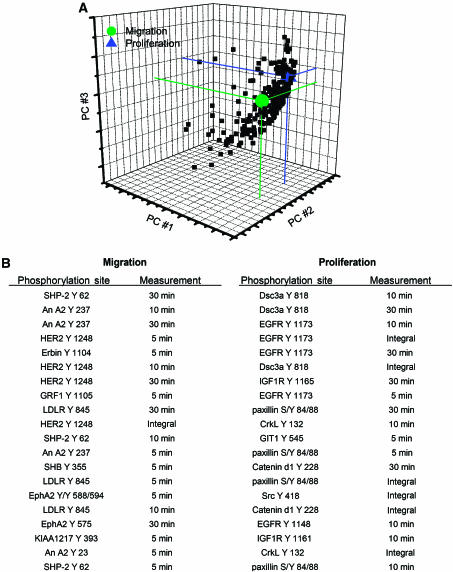
We have constructed a model using partial least-squares regression (PLSR), a method we have previously found to be effective in relating cell signaling data to cell behavioral response data in a quantitative and integrative manner (Janes et al, 2005). Information obtained through our proteomic studies was represented in an M × N matrix (the X-block), where M is the number of conditions investigated and N is the number of peptide metrics measured. An entry in the matrix with coordinates (i, j) represents the column j metric (i.e., ERK Y187 phosphorylation at 5 min) measured under the row i condition (i.e., parental cell line treated with EGF). For each condition, the metrics included in the model were phosphorylation measurements at 5, 10, and 30 min in addition to the integral of this time course (with integrals being used as a measurement for the ‘net' phosphorylation over the 30 min time course). Cell behavior measurements comprised an M × P matrix (the Y-block), where M is again the number of conditions and P is the number of behavior measurements obtained. PLSR analysis produced a vector of coefficients indicating the importance of each signaling metric with respect to cellular behavior. In addition, PLSR provided a reduced-dimension map, with axes defined as linear combinations of our original signaling metrics (Figure 6A), on which both signals and cellular behavior can be represented. Figure 6A shows that our original data set, consisting of 248 dimensions (i.e., 248 protein signal metrics), has been reduced to three dimensions using PLSR, each of which incorporates a quantitative combination of multiple signals. The projection of an individual signal in the direction of a given cellular behavior in the PLS space determines how important the phosphorylation signal is to the behavior. In Figure 6B, we list the top 20 signals that positively correlate most strongly with each cell behavior. Importantly, even though we can identify small sets of variables that correlate strongly with each cellular output, 148 out of the 248 protein metrics had a variable importance for projection (VIP) value of greater than 1, indicating that these 148 protein metrics play an important role in our model (see Supplementary information for VIP calculation). This highlights the great advantage of proteome measurements that quantitatively capture dynamic information flow through a large number of nodes. Our model was validated through crossvalidation and had a goodness of prediction (Q2) of 0.89 (see Supplementary information).
Figure 6.
PLSR correlates 248 protein metrics to cell migration and proliferation. (A) Visual representation of a reduced (3) dimension graph showing all 248 protein measurements and the cellular outputs with cell migration (green), cell proliferation (blue), and protein signals (black). (B) List of the top 20 signal–behavior covariates for both migration and proliferation.
From the top 20 signal behavior covariates for proliferation and migration in our model (Figure 6B), HER Y1248 appears to be the main upstream activator of migration, whereas proliferation seems to be activated by EGFR Y1173 and to a lesser extent EGFR Y1148. This finding correlates with literature showing that the presence of HER2 Y1248 is necessary to induce migration of breast cancer cell lines (Dittmar et al, 2002), whereas both EGFR Y1148 and Y1173 are Shc binding sites and thus are able to activate the ERK signaling pathway, thereby driving proliferation (Okabayashi et al, 1994). In this model, SHP-2 Y62 and Annexin A2 Y237, among others, also appear to be highly correlated with migration. Both of these proteins have previously been implicated in regulating migration, as SHP-2 has been shown to promote migration in MCF-7 cells by inducing loss of E-cadherin expression and production of matrix metalloproteinase 9 (Wang et al, 2005a), whereas Annexin A2 has been shown to mediate mitogenesis, cell migration, and loss of focal adhesions when activated by tenascin-c (Chung et al, 1996; Liu et al, 2003) and it has also been identified as a major substrate for EGFR phosphorylation (Rothhut, 1997). Interestingly, treating cells with withaferin, a novel inhibitor of Annexin A2, has recently been shown to decrease migration, further validating the correlation between Annexin A2 and cell migration (Falsey et al, 2006).
In addition to EGFR Y1173 and Y1148, desmocollin (Dsc3a) Y818, IGF-1R Y1165, and catenin-δ Y228 (among others) are most strongly correlated with proliferation. IGF-1R is known to induce cell proliferation and survival through activation of the MAPK and PI3K pathways (Wang et al, 2005b), and has also been associated with EGFR signaling in tamoxifen-resistant breast cancer cell lines, potentiating their mitogenic strength (Knowlden et al, 2003). The correlation of Dsc3a and catenin-δ to cellular proliferation is initially surprising, as both of these proteins are associated with cell adhesions, leading to the expectation that they should correlate to migration instead of proliferation. However, EGFR inhibition has been shown to increase the presence of desmocollin and desmoglein at cell–cell borders (Lorch et al, 2004), and EGFR has also been shown to signal to E-cadherin complexes through phosphorylation of catenin-δ Y228 (Mariner et al, 2004). As the conditional and temporal profiles for both of these sites (Dsc3a Y818 and catenin-δ Y228) display strong similarity to that observed for EGFR Y1173, it can be argued that they are either directly or closely downstream of EGFR and that phosphorylation of these sites may destabilize cell–cell adhesions, a step needed for both proliferation and migration. Further biological experiments are clearly needed to more firmly establish the functional roles of these proteins in regulating cellular proliferation under these stimulation conditions.
It is worth noting that a large proportion of the phosphorylation events contained in our regression model influence both migration and proliferation to some extent. Importantly, the model provides a quantitative metric describing the strength of correlation for each phosphorylation site relative to migration or proliferation, data that can be used to design further experiments to specifically perturb selected biological outcomes—that is, to inhibit migration one might want to target HER2, SHP-2, and EphA2. This type of data-driven modeling can also provide insight into the functionality of unknown proteins such as KIAA1217; in our model, phosphorylation of this protein at Y393 correlates strongly to migration. Interestingly, KIAA1217 is a novel protein highly homologous to p140CAP (p130Cas-associated protein), which has been shown to be tyrosine phosphorylated in response to EGF stimulation and to participate in actin cytoskeleton organization and cell spreading (Di Stefano et al, 2004).
Combining large-scale quantitative analysis of tyrosine phosphorylation with quantitative phenotypic measurements has provided the means with which to understand the relationship between these phosphorylation events and their relation to cellular responses. The findings we have discovered here illustrate how HER2 overexpression influences signaling network activities important for governing cell proliferation and migration behavior, common to and distinct between EGFR-binding ligand and HER3-binding ligand conditions. This quantitative information offers unusual opportunity for understanding prospective drug effects in a network-wide manner, and may offer novel targets for intervening in biological processes downstream of activated RTKs.
Conclusions
To investigate the effect of HER2 overexpression on signaling networks and corresponding cell behavior, HMECs were stimulated by EGF or HRG and temporal tyrosine phosphorylation profiles were quantified by MS. To correlate signals with cell response, proliferation and migration rates were also quantified for these same cell states and stimulation conditions. Phenotypically, HER2 overexpression promoted increased cell migration, but had minimal effect on cell proliferation. More specifically, EGF stimulation of HER2-overexpressing cells promoted migration by the phosphorylation of proteins from multiple pathways (e.g., PI3K, MAPK, catenins, and FAK), whereas HRG stimulation of HER2-overexpressing cells activated only a very specific subset of proteins in the canonical migration pathway, in particular FAK, Src, paxillin, and p130Cas. In contrast, proliferation was primarily driven by EGF stimulation, and was not affected by HER2 expression levels.
From a mechanistic point of view, computational analysis of the phosphoproteomic data set resulted in the identification of clusters of phosphorylation sites that are highly responsive to particular sets of cellular stimulation conditions, linking specific phosphorylation sites with activated receptor homo- or heterodimers. In addition, PLSR analysis of the phosphoproteomic and phenotypic data sets provided quantitative weighting to describe the relationship between specific phosphorylation sites and downstream biological behavior. From this list, we were able to identify the phosphorylation sites that are most strongly correlated with migration and proliferation. Although some of these sites are intuitive (e.g., HER2 Y1248 with migration or EGFR Y1173 with proliferation), the presence of unexpected phosphorylation sites (e.g., KIAA1217 Y393 with migration) in these lists gives a new level of insight into the nature of EGFR family-related signaling networks. Overall, the identification of specific phosphorylation sites coupled to their relative weighting toward biological outcome (proliferation and migration) enables a suite of additional experiments in which specific proteins may be targeted to inhibit particular biological responses to stimulation.
Materials and methods
Cell culture and stimulation
184A1 HMECs (Stampfer and Bartley, 1985) (HMEC parental) were a kind gift from Martha Stampfer (Lawrence Berkeley Laboratory, Berkeley, CA) via Steve Wiley (Pacific Northwest National Laboratory, Richland, WA) and were maintained in DFCI-1 medium supplemented with 12.5 ng/ml EGF, as described by Hendriks et al (2003a). 184A1 HMECs clone 24H (HMECs overexpressing HER2 30-fold; Hendriks et al, 2003a) (HMEC 24H) were a kind gift from Steve Wiley (Pacific Northwest National Laboratories, Richland, WA) and were maintained in DFCI-1 medium supplemented with 12.5 ng/ml EGF and 150 μg/ml of geneticin. Cells were washed with PBS and incubated for 12 h in serum-free media (DFCI-1 without EGF, bovine pituitary extract, or fetal bovine serum) after 80% confluence was reached in 15 cm plates (∼2 × 107 cells). Synchronized cells were washed with PBS after removal of media. Cells were then stimulated with 100 ng/ml EGF or 80 ng/ml HRG in serum-free media for 5, 10, or 30 min, or left untreated with serum-free media for 5 min as control.
Sample preparation, peptide immunoprecipitation, IMAC, and MS analysis
Samples were prepared and analyzed as described previously (Zhang et al, 2005b). Detailed description is available in Supplementary information.
Self-organizing maps
The idea behind the SOM is to nonlinearly transform high-dimensional input data into a lower dimensional display that consists of several map units. Each map unit consists of a reference vector whose dimension is the same as the dimension of an input pattern (Kohonen, 2001). The clustering is carried out in two steps. In the training phase, the SOM algorithm computes distance between an input pattern and all reference vectors. The map unit consisting of reference vector that correlates the best with the input pattern is declared as the winner map unit and is updated with maximal strength, whereas topologically close map units are updated with gradually decreasing strength. This process is executed using all 62 peptide profiles and repeated a few times leading to self-organization. After the training phase, each peptide is assigned to the map unit whose reference vector is most similar to the input pattern.
The parameters for the SOM used in this study were as follows. Topology of the map was chosen to be sheet, distance measure was correlation, and the number of map units was chosen to be
, where p is the number of input vectors as suggested by Vesanto et al (2000). We used the batch learning algorithm, and the neighborhood function was chosen to be Gaussian with the parameters given by Vesanto et al (2000). The SOM analysis was performed in MATLAB with the publicly available SOM Toolbox (Vesanto et al, 2000).
We used the U-matrix method to identify a group of map units that comprehend a cluster (Ultsch and Siemon, 1990). The U-matrix illustrates Euclidean distances between the map units with colors. Adjacent map units colored with blue shades constitute a cluster (valleys) and the clusters are separated by red and yellow colors that represent high distance between two map units (mountains). For each cluster, we computed statistical significance with the permutation test-based method given by Hautaniemi et al (2003) as follows. First, we computed correlation distances for all combinations of the peptide profiles in a cluster. If the two profiles correlate perfectly, their distance is zero and perfect negative correlation results in the distance value two. Then, we computed the mean of these pairwise comparisons. This was followed by choosing randomly the same number of profiles as there are in the original cluster and computing all combinations of the pairwise correlation distances. For example, if a cluster consisted of 18 peptides, we randomly chose 18 peptides from the set of 62 peptides and computed the mean of 153 correlation distance values (18 choose 2), which was compared to the mean of the peptide correlations in the original cluster. If the mean distance of a randomly chosen set is less than, or equal to, the original, we added one to counter. For each cluster, we created 5000 samples and the final P-value is computed by dividing the counter by 5000. Large values suggest that the original cluster may be owing to chance.
ELISA protocol for ErbB3 receptor quantification
Reagents for the ErbB3 ELISA were purchased from R&D Systems as a Duoset DY348 (capture antibody, biotinylated detection antibody, recombinant human Erb3, and streptavidin-HRP). The ELISA conditions were those recommended by the manufacturer; additional protocol details are available in Supplementary information.
Proliferation
HMECs were plated on 24-well tissue culture plastic plates (∼1 × 104 cells/cm2) and grown for 24 h to ∼60–70% confluence. The medium was then removed and cells were serum-starved as previously described for 12–16 h. Starved cells were then treated with new serum-free media, serum-free media containing EGF (100 ng/ml), or serum-free media containing HRG (80 ng/ml). The cells were grown for 15 h at which time [3H]thymidine (10 μCi/ml) was added. After 10 h, the cells were washed with cold PBS and treated with trichloroacetic acid (5% w/v) at 4°C. The resulting precipitate was dissolved in NaOH (0.5 N) and quantified using liquid scintillation counting.
Migration
HMECs were seeded in 96-well tissue culture plastic plates (Packard View Plate Black, ref. 6005225) at confluence (∼50 000 cells/cm2) and allowed to adhere for 4–6 h. Media were then removed and cells were serum starved for 12–16 h as described previously. Starved cells were treated with 5-chloromethylfluorescein diacetate (CMFDA, 9 μM in serum-free media) for 30 min. CMFDA-containing media were removed and cells were then treated with new serum-free media, serum-free media containing EGF (100 ng/ml), or serum-free media containing HRG (80 ng/ml). A wound width ∼700 μm was scraped in each well and cell movement imaged every 15 min for 12 h using Cellomics KineticScan. Kinetics of wound closure were quantified using an in-house analysis software that calculated the wound area at each time point normalized by the initial wound area. A time averaging algorithm was used to average wound closure in four wells of equal treatment into a single trajectory at 30 min intervals. The trajectories shown here have been additionally normalized to their 2 h time points for the purpose of comparing treatments. The wound closure data used in the described PLSR model are the linearly fitted slope (2–5.5 h) of each trajectory shown.
Partial least-squares regression
Computational analysis was performed using the SIMCA-P 10.0 (Umetrics) software suite, as detailed elsewhere (Janes et al, 2004; Gaudet et al, 2005). The software uses the nonlinear iterative partial least-squares algorithm to perform decompositions and regressions. The model was evaluated for goodness of fit (R2Y) and goodness of prediction (Q2), and was validated against overfitting via response permutation (response matrix was randomly permuted 50 times and Q2 values were obtained for each run). All matrices were mean centered and unit variance scaled (z-score normalized) before partial least-squares analysis.
Supplementary Material
Supplementary materials and methods
Supplementary Self Organizing Map Figure
Supplementary Table with Full Data Set
Acknowledgments
We thank Steve Wiley at Pacific Northwest National Laboratory and members of the White and Lauffenburger groups for helpful discussions. This work was supported by NCI grant CA96504, NCI grant CA112967, NIH grant P50-GM68762, the David Koch Research Fund, Pirkanmaan Cultural Foundation (SH), Academy of Finland (SH), the Ludwig Foundation (AW-Y), and a grant from AstraZeneca.
References
- Aguilar Z, Akita RW, Finn RS, Ramos BL, Pegram MD, Kabbinavar FF, Pietras RJ, Pisacane P, Sliwkowski MX, Slamon DJ (1999) Biologic effects of heregulin/neu differentiation factor on normal and malignant human breast and ovarian epithelial cells. Oncogene 18: 6050–6062 [DOI] [PubMed] [Google Scholar]
- Atlas E, Cardillo M, Mehmi I, Zahedkargaran H, Tang C, Lupu R (2003) Heregulin is sufficient for the promotion of tumorigenicity and metastasis of breast cancer cells in vivo. Mol Cancer Res 1: 165–175 [PubMed] [Google Scholar]
- Carlsson J, Nordgren H, Sjostrom J, Wester K, Villman K, Bengtsson NO, Ostenstad B, Lundqvist H, Blomqvist C (2004) HER2 expression in breast cancer primary tumours and corresponding metastases. Original data and literature review. Br J Cancer 90: 2344–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cary LA, Guan JL (1999) Focal adhesion kinase in integrin-mediated signaling. Front Biosci 4: D102–D113 [DOI] [PubMed] [Google Scholar]
- Cheng N, Brantley DM, Liu H, Lin Q, Enriquez M, Gale N, Yancopoulos G, Cerretti DP, Daniel TO, Chen J (2002) Blockade of EphA receptor tyrosine kinase activation inhibits vascular endothelial cell growth factor-induced angiogenesis. Mol Cancer Res 1: 2–11 [PubMed] [Google Scholar]
- Chung CY, Murphy-Ullrich JE, Erickson HP (1996) Mitogenesis, cell migration, and loss of focal adhesions induced by tenascin-C interacting with its cell surface receptor, annexin II. Mol Biol Cell 7: 883–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citri A, Skaria KB, Yarden Y (2003) The deaf and the dumb: the biology of ErbB-2 and ErbB-3. Exp Cell Res 284: 54–65 [DOI] [PubMed] [Google Scholar]
- Cobb MH, Hepler JE, Cheng M, Robbins D (1994) The mitogen-activated protein kinases, ERK1 and ERK2. Semin Cancer Biol 5: 261–268 [PubMed] [Google Scholar]
- D'Souza B, Taylor-Papadimitriou J (1994) Overexpression of ERBB2 in human mammary epithelial cells signals inhibition of transcription of the E-cadherin gene. Proc Natl Acad Sci USA 91: 7202–7206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MA, Ireton RC, Reynolds AB (2003) A core function for p120-catenin in cadherin turnover. J Cell Biol 163: 525–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Stefano P, Cabodi S, Boeri Erba E, Margaria V, Bergatto E, Giuffrida MG, Silengo L, Tarone G, Turco E, Defilippi P (2004) P130Cas-associated protein (p140Cap) as a new tyrosine-phosphorylated protein involved in cell spreading. Mol Biol Cell 15: 787–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar T, Husemann A, Schewe Y, Nofer JR, Niggemann B, Zanker KS, Brandt BH (2002) Induction of cancer cell migration by epidermal growth factor is initiated by specific phosphorylation of tyrosine 1248 of c-erbB-2 receptor via EGFR. FASEB J 16: 1823–1825 [DOI] [PubMed] [Google Scholar]
- Falsey RR, Marron MT, Gunaherath GM, Shirahatti N, Mahadevan D, Gunatilaka AA, Whitesell L (2006) Actin microfilament aggregation induced by withaferin A is mediated by annexin II. Nat Chem Biol 2: 33–38 [DOI] [PubMed] [Google Scholar]
- Gaudet S, Janes KA, Albeck JG, Pace EA, Lauffenburger DA, Sorger PK (2005) A compendium of signals and responses triggered by Prodeath and Prosurvival cytokines. Mol Cell Proteomics 4: 1569–1590 [DOI] [PubMed] [Google Scholar]
- Grovdal LM, Stang E, Sorkin A, Madshus IH (2004) Direct interaction of Cbl with pTyr 1045 of the EGF receptor (EGFR) is required to sort the EGFR to lysosomes for degradation. Exp Cell Res 300: 388–395 [DOI] [PubMed] [Google Scholar]
- Harari D, Yarden Y (2000) Molecular mechanisms underlying ErbB2/HER2 action in breast cancer. Oncogene 19: 6102–6114 [DOI] [PubMed] [Google Scholar]
- Hautaniemi S, Yli-Harja O, Astola J, Kauraniemi P, Kallioniemi A, Wolf M, Ruiz J, Mousses S, Kallioniemi O-P (2003) Analysis and visualization of gene expression microarray data in human cancer using self-organizing maps. Mach Learn 52: 45–66 [Google Scholar]
- Hendriks BS, Opresko LK, Wiley HS, Lauffenburger D (2003a) Coregulation of epidermal growth factor receptor/human epidermal growth factor receptor 2 (HER2) levels and locations: quantitative analysis of HER2 overexpression effects. Cancer Res 63: 1130–1137 [PubMed] [Google Scholar]
- Hendriks BS, Opresko LK, Wiley HS, Lauffenburger D (2003b) Quantitative analysis of HER2-mediated effects on HER2 and epidermal growth factor receptor endocytosis: distribution of homo- and heterodimers depends on relative HER2 levels. J Biol Chem 278: 23343–23351 [DOI] [PubMed] [Google Scholar]
- Hendriks BS, Orr G, Wells A, Wiley HS, Lauffenburger DA (2005) Parsing ERK activation reveals quantitatively equivalent contributions from epidermal growth factor receptor and HER2 in human mammary epithelial cells. J Biol Chem 280: 6157–6169 [DOI] [PubMed] [Google Scholar]
- Holbro T, Beerli RR, Maurer F, Koziczak M, Barbas CF III, Hynes NE (2003) The ErbB2/ErbB3 heterodimer functions as an oncogenic unit: ErbB2 requires ErbB3 to drive breast tumor cell proliferation. Proc Natl Acad Sci USA 100: 8933–8938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmqvist K, Cross M, Riley D, Welsh M (2003) The Shb adaptor protein causes Src-dependent cell spreading and activation of focal adhesion kinase in murine brain endothelial cells. Cell Signal 15: 171–179 [DOI] [PubMed] [Google Scholar]
- Hynes NE, Lane HA (2005) ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer 5: 341–354 [DOI] [PubMed] [Google Scholar]
- Janes KA, Albeck JG, Gaudet S, Sorger PK, Lauffenburger DA, Yaffe MB (2005) A systems model of signaling identifies a molecular basis set for cytokine-induced apoptosis. Science 310: 1646–1653 [DOI] [PubMed] [Google Scholar]
- Janes KA, Kelly JR, Gaudet S, Albeck JG, Sorger PK, Lauffenburger DA (2004) Cue-signal-response analysis of TNF-induced apoptosis by partial least squares regression of dynamic multivariate data. J Comput Biol 11: 544–561 [DOI] [PubMed] [Google Scholar]
- Jawhari AU, Farthing MJ, Pignatelli M (1999) The E-cadherin/epidermal growth factor receptor interaction: a hypothesis of reciprocal and reversible control of intercellular adhesion and cell proliferation. J Pathol 187: 155–157 [DOI] [PubMed] [Google Scholar]
- Jones RB, Gordus A, Krall JA, MacBeath G (2006) A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature 439: 168–174 [DOI] [PubMed] [Google Scholar]
- Kinch MS, Carles-Kinch K (2003) Overexpression and functional alterations of the EphA2 tyrosine kinase in cancer. Clin Exp Metast 20: 59–68 [DOI] [PubMed] [Google Scholar]
- Knowlden JM, Hutcheson IR, Jones HE, Madden T, Gee JM, Harper ME, Barrow D, Wakeling AE, Nicholson RI (2003) Elevated levels of epidermal growth factor receptor/c-erbB2 heterodimers mediate an autocrine growth regulatory pathway in tamoxifen-resistant MCF-7 cells. Endocrinology 144: 1032–1044 [DOI] [PubMed] [Google Scholar]
- Kohonen T (2001) Self-organizing Maps. Berlin, Germany: Springer [Google Scholar]
- Lemmon MA (2003) The EGF receptor family as therapeutic targets in breast cancer. Breast Dis 18: 33–43 [DOI] [PubMed] [Google Scholar]
- Liu JW, Shen JJ, Tanzillo-Swarts A, Bhatia B, Maldonado CM, Person MD, Lau SS, Tang DG (2003) Annexin II expression is reduced or lost in prostate cancer cells and its re-expression inhibits prostate cancer cell migration. Oncogene 22: 1475–1485 [DOI] [PubMed] [Google Scholar]
- Lorch JH, Klessner J, Park JK, Getsios S, Wu YL, Stack MS, Green KJ (2004) Epidermal growth factor receptor inhibition promotes desmosome assembly and strengthens intercellular adhesion in squamous cell carcinoma cells. J Biol Chem 279: 37191–37200 [DOI] [PubMed] [Google Scholar]
- Manabe R, Kovalenko M, Webb DJ, Horwitz AR (2002) GIT1 functions in a motile, multi-molecular signaling complex that regulates protrusive activity and cell migration. J Cell Sci 115: 1497–1510 [DOI] [PubMed] [Google Scholar]
- Mariner DJ, Davis MA, Reynolds AB (2004) EGFR signaling to p120-catenin through phosphorylation at Y228. J Cell Sci 117: 1339–1350 [DOI] [PubMed] [Google Scholar]
- Mendelsohn J, Baselga J (2003) Status of epidermal growth factor receptor antagonists in the biology and treatment of cancer. J Clin Oncol 21: 2787–2799 [DOI] [PubMed] [Google Scholar]
- Moon HS, Choi EA, Park HY, Choi JY, Chung HW, Kim JI, Park WI (2001) Expression and tyrosine phosphorylation of E-cadherin, beta- and gamma-catenin, and epidermal growth factor receptor in cervical cancer cells. Gynecol Oncol 81: 355–359 [DOI] [PubMed] [Google Scholar]
- Okabayashi Y, Kido Y, Okutani T, Sugimoto Y, Sakaguchi K, Kasuga M (1994) Tyrosines 1148 and 1173 of activated human epidermal growth factor receptors are binding sites of Shc in intact cells. J Biol Chem 269: 18674–18678 [PubMed] [Google Scholar]
- Pinkas-Kramarski R, Soussan L, Waterman H, Levkowitz G, Alroy I, Klapper L, Lavi S, Seger R, Ratzkin BJ, Sela M, Yarden Y (1996) Diversification of Neu differentiation factor and epidermal growth factor signaling by combinatorial receptor interactions. EMBO J 15: 2452–2467 [PMC free article] [PubMed] [Google Scholar]
- Playford MP, Schaller MD (2004) The interplay between Src and integrins in normal and tumor biology. Oncogene 23: 7928–7946 [DOI] [PubMed] [Google Scholar]
- Prager GW, Breuss JM, Steurer S, Olcaydu D, Mihaly J, Brunner PM, Stockinger H, Binder BR (2004) Vascular endothelial growth factor receptor-2-induced initial endothelial cell migration depends on the presence of the urokinase receptor. Circ Res 94: 1562–1570 [DOI] [PubMed] [Google Scholar]
- Qu CK, Yu WM, Azzarelli B, Feng GS (1999) Genetic evidence that Shp-2 tyrosine phosphatase is a signal enhancer of the epidermal growth factor receptor in mammals. Proc Natl Acad Sci USA 96: 8528–8533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roskoski R Jr (2004) Src protein-tyrosine kinase structure and regulation. Biochem Biophys Res Commun 324: 1155–1164 [DOI] [PubMed] [Google Scholar]
- Ross JS, Fletcher JA, Linette GP, Stec J, Clark E, Ayers M, Symmans WF, Pusztai L, Bloom KJ (2003) The Her-2/neu gene and protein in breast cancer 2003: biomarker and target of therapy. Oncologist 8: 307–325 [DOI] [PubMed] [Google Scholar]
- Rothhut B (1997) Participation of annexins in protein phosphorylation. Cell Mol Life Sci 53: 522–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze WX, Deng L, Mann M (2005) Phosphotyrosine interactome of the ErbB-receptor kinase family. Mol Syst Biol 1: 2005.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semino CE, Kamm RD, Lauffenburger DA (2006) Autocrine EGF receptor activation mediates endothelial cell migration and vascular morphogenesis induced by VEGF under interstitial flow. Exp Cell Res 312: 289–298 [DOI] [PubMed] [Google Scholar]
- Sorkin A, Mazzotti M, Sorkina T, Scotto L, Beguinot L (1996) Epidermal growth factor receptor interaction with clathrin adaptors is mediated by the Tyr974-containing internalization motif. J Biol Chem 271: 13377–13384 [DOI] [PubMed] [Google Scholar]
- Spencer KS, Graus-Porta D, Leng J, Hynes NE, Klemke RL (2000) ErbB2 is necessary for induction of carcinoma cell invasion by ErbB family receptor tyrosine kinases. J Cell Biol 148: 385–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stampfer MR, Bartley JC (1985) Induction of transformation and continuous cell lines from normal human mammary epithelial cells after exposure to benzo[a]pyrene. Proc Natl Acad Sci USA 82: 2394–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MS, Shamon-Taylor LA, Mehmi I, Tang CK, Lupu R (2003) Blockage of heregulin expression inhibits tumorigenicity and metastasis of breast cancer. Oncogene 22: 761–768 [DOI] [PubMed] [Google Scholar]
- Tsubouchi A, Sakakura J, Yagi R, Mazaki Y, Schaefer E, Yano H, Sabe H (2002) Localized suppression of RhoA activity by Tyr31/118-phosphorylated paxillin in cell adhesion and migration. J Cell Biol 159: 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P, Stagljar I (2006) The interactome of human EGF/ErbB receptors. Mol Syst Biol 2: 2006.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ultsch A, Siemon HP (1990) Kohonen's self-organizing feature maps for exploratory data analysis. Presented at Proceedings of INNC'90, International Neural Network Conference, Dordrecht, The Netherlands
- Vesanto J, Himberg J, Alhoniemi E, Parhankangas J (2000) SOM toolbox for Matlab 5. In Technical Report A57. Finland: Helsinki University of Technology [Google Scholar]
- Vulin AI, Jacob KK, Stanley FM (2005) Integrin activates receptor-like protein tyrosine phosphatase alpha, Src, and Rho to increase prolactin gene expression through a final phosphatidylinositol 3-kinase/cytoskeletal pathway that is additive with insulin. Endocrinology 146: 3535–3546 [DOI] [PubMed] [Google Scholar]
- Wang FM, Liu HQ, Liu SR, Tang SP, Yang L, Feng GS (2005a) SHP-2 promoting migration and metastasis of MCF-7 with loss of E-cadherin, dephosphorylation of FAK and secretion of MMP-9 induced by IL-1beta in vivo and in vitro. Breast Cancer Res Treat 89: 5–14 [DOI] [PubMed] [Google Scholar]
- Wang Y, Hailey J, Williams D, Wang Y, Lipari P, Malkowski M, Wang X, Xie L, Li G, Saha D, Ling WL, Cannon-Carlson S, Greenberg R, Ramos RA, Shields R, Presta L, Brams P, Bishop WR, Pachter JA (2005b) Inhibition of insulin-like growth factor-I receptor (IGF-IR) signaling and tumor cell growth by a fully human neutralizing anti-IGF-IR antibody. Mol Cancer Ther 4: 1214–1221 [DOI] [PubMed] [Google Scholar]
- Webb DJ, Donais K, Whitmore LA, Thomas SM, Turner CE, Parsons JT, Horwitz AF (2004) FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat Cell Biol 6: 154–161 [DOI] [PubMed] [Google Scholar]
- Xia L, Wang L, Chung AS, Ivanov SS, Ling MY, Dragoi AM, Platt A, Gilmer TM, Fu XY, Chin YE (2002) Identification of both positive and negative domains within the epidermal growth factor receptor COOH-terminal region for signal transducer and activator of transcription (STAT) activation. J Biol Chem 277: 30716–30723 [DOI] [PubMed] [Google Scholar]
- Yao J, Xiong S, Klos K, Nguyen N, Grijalva R, Li P, Yu D (2001) Multiple signaling pathways involved in activation of matrix metalloproteinase-9 (MMP-9) by heregulin-beta1 in human breast cancer cells. Oncogene 20: 8066–8074 [DOI] [PubMed] [Google Scholar]
- Yarden Y, Sliwkowski MX (2001) Untangling the ErbB signalling network. Nat Rev Mol Cell Biol 2: 127–137 [DOI] [PubMed] [Google Scholar]
- Zhang X, Lin M, van Golen KL, Yoshioka K, Itoh K, Yee D (2005a) Multiple signaling pathways are activated during insulin-like growth factor-I (IGF-I) stimulated breast cancer cell migration. Breast Cancer Res Treat 93: 159–168 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wolf-Yadlin A, Ross PL, Pappin DJ, Rush J, Lauffenburger DA, White FM (2005b) Time-resolved mass spectrometry of tyrosine phosphorylation sites in the epidermal growth factor receptor signaling network reveals dynamic modules. Mol Cell Proteomics 4: 1240–1250 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials and methods
Supplementary Self Organizing Map Figure
Supplementary Table with Full Data Set