Abstract
Cellular behavior has traditionally been investigated by utilizing bulk-scale methods that measure average values for a population of cells. Such population-wide studies mask the behavior of individual cells and are often insufficient for characterizing biological processes in which cellular heterogeneity plays a key role. A unifying theme of many recent studies has been a focus on the development and utilization of single-cell experimental techniques that are capable of probing key biological phenomena in individual living cells. Recently, novel information about gene expression dynamics has been obtained from single-cell experiments that draw upon the unique capabilities of fluorescent reporter proteins.
Keywords: gene expression, noise, single cell
Introduction
Cellular heterogeneity has been observed in a wide variety of cell types ranging from simple bacterial cells (Ozbudak et al, 2002; Swain et al, 2002) to more complex mammalian cells (Ramsey et al, 2006). Any population of cells will exhibit some degree of variability, and genetic differences are one of the main factors responsible for cellular heterogeneity. However, variation is also present in genetically identical cell populations, even when the cells have been exposed to the same environment and have the same history (Elowitz et al, 2002; Rao et al, 2002; Blake et al, 2003; Raser and O'Shea, 2005). Noise, or random fluctuations, in the process of gene expression is thought to contribute to this phenotypic variation.
Analyzing gene expression at the single-cell level has provided insight into oscillatory or nonlinear behavior in asynchronous cells and has revealed the cell-to-cell variability that arises owing to the stochastic nature of gene expression. Although researchers have been interested in the stochastic nature of gene expression and its implications for many years (Benzer, 1953; Novick and Weiner, 1957; Maloney and Rotman, 1973; Spudich and Koshland, 1976; Rigney and Schieve, 1977), the techniques available for quantifying gene expression were quite limited when some of the earliest single-cell experiments were performed. For example, in 1953, cell-to-cell variability in β-galactosidase (β-gal) concentrations in Escherichia coli was investigated by using a phage whose replication depended on β-gal activity as a method for measuring the level of β-gal in each cell, because tools for directly measuring the amount of enzyme in each cell were nonexistent at that time (Benzer, 1953). The recent development of a collection of fluorescent proteins, each possessing unique biochemical characteristics, has enabled single-cell experiments in which fluorescent reporters are used to quantify protein production, examine protein localization, and monitor production of individual mRNA molecules.
Cell-to-cell variability and stochastic gene expression
Single-cell measurements are necessary for investigating the stochastic nature of gene expression because cell-to-cell variation cannot be quantified using population level measurements. Noise in gene expression arises not only from the inherent randomness of biochemical processes such as transcription and translation, but also from the fluctuations in cellular components that lead indirectly to variation in the expression of a particular gene (Swain et al, 2002). The total noise in the level of expression of a given gene can be divided into intrinsic and extrinsic components. Extrinsic noise arises from fluctuations in cellular components such as regulatory proteins and polymerases, and has a global effect (Elowitz et al, 2002). Intrinsic noise arises from the stochastic nature of the biochemical process of gene expression and causes identical copies of a gene to express at different levels (Elowitz et al, 2002).
Single-cell studies have been key in gaining insight into the stochastic nature of gene expression. Such studies often involve monitoring the level of a protein expressed from an engineered gene circuit in individual live prokaryotic or simple eukaryotic cells. For example, Ozbudak et al (2002) used point mutations to independently vary the transcriptional and translational rates of a single-gene network in Bacillus subtilis, and found that fluctuations in the level of a fluorescent reporter gene increased linearly with translational efficiency. The results were consistent with a stochastic model that predicted that noise for a single gene is determined at the translational level (Thattai and van Oudenaarden, 2001). Elowitz et al (2002) developed a method utilizing two different fluorescent proteins expressed from identical promoters to study noise in gene expression in E. coli. This study demonstrated that noise in gene expression results in fluctuations in protein levels in a clonal population and that both intrinsic and extrinsic noise contribute to total noise in gene expression. Raser and O'Shea (2004) modified the dual-reporter method to measure gene expression in the yeast Saccharomyces cerevisiae and found that gene expression variability is dominated by extrinsic noise. More recently, Bar-Even et al (2006) used 43 strains from the yeast green fluorescent protein (GFP) clone collection to analyze cell-to-cell variation in gene expression in S. cerevisiae. The study measured protein noise and mean protein abundance for each of the fusion proteins subjected to 11 different environmental conditions and discovered a strong correlation between cell-to-cell variability and mean expression level. Theoretical analysis of these results suggests that sources intrinsic to the biochemical process of gene expression make a substantial contribution to gene expression noise (Bar-Even et al, 2006). Another large-scale study of gene expression noise was recently performed by utilizing high-throughput flow cytometry to measure protein abundances in a collection of GFP-tagged yeast strains (Newman et al, 2006). This study also observed a global relationship between noise and protein abundance, suggesting that intrinsic noise dominates gene expression noise (Newman et al, 2006). The global trend observed between protein noise and mean abundance does not extend to regions of high protein abundance (Bar-Even et al, 2006; Newman et al, 2006), thus explaining the apparent discrepancy between the recent large-scale results (Bar-Even et al, 2006; Newman et al, 2006) and the results of the Raser and O'Shea study, which uses a highly abundant reporter gene.
Recent single-cell experiments have provided further insight into cell-to-cell variability by examining how noise in gene expression propagates from one gene to the next (Pedraza and van Oudenaarden, 2005), measuring the relative amplitude and timescales of intrinsic and extrinsic noise (Rosenfeld et al, 2005), analyzing the relative contribution of global noise and pathway-specific noise to cell-to-cell variation in a cell-fate decision (Colman-Lerner et al, 2005), investigating the relationship between gene circuit structure and noise frequency range (Austin et al, 2006), examining the effects of cell-cycle position on cell-to-cell variation (Colman-Lerner et al, 2005) and on nuclear protein levels and localization (Sigal et al, 2006), and investigating the source of extrinsic noise in eukaryotic gene expression (Volfson et al, 2005) (for a review of the origins and consequences of noise in gene expression, see McAdams and Arkin, 1999; Kaern et al, 2005; Raser and O'Shea, 2005).
Feedback loops play an important role in modulating gene expression noise. Single-cell studies have been useful in examining how stochastic gene expression affects the behavior of natural networks, which possess feedback loops. For example, the competence induction network in B. subtilus contains a positive feedback loop and a negative feedback loop. Because differentiation into a competent state occurs in only a small fraction of cells in an asynchronous fashion, population-level experiments are inappropriate for monitoring the differentiation process. Recently, the dynamics of this network were analyzed by using time-lapse fluorescence microscopy to monitor expression levels of two fluorescent reporter proteins, yellow fluorescent protein (YFP) and cyan fluorescent protein (CFP), in individual cells (Suel et al, 2006). The study demonstrated that the positive feedback loop and negative feedback loop can explain both the transient differentiation into a competent state and the return to vegetative growth.
Feedback loops also modulate the behavior of mammalian systems. For example, the HIV-1 life cycle in mammalian cells depends on a Tat transactivation feedback loop. A recent study examined the behavior of the Tat-positive feedback loop in Jurkat cells by utilizing a GFP reporter to observe stochastic switching between high and low expression states (Weinberger et al, 2005). Because cells with low basal expression of Tat protein can randomly produce bursts of Tat that are amplified by the positive feedback loop, bifurcating phenotypes can be generated. The study demonstrated that stochastic fluctuations in Tat expression result in two distinct expression states corresponding to latent and productive HIV-1 infection.
Quantitative dynamics of gene expression
Because the gene regulatory networks found in cells are often quite complex, designing simpler synthetic systems that are easier to model and manipulate can provide insight into the mechanisms underlying the behavior of natural gene networks (Hasty et al, 2002; Sprinzak and Elowitz, 2005). However, owing to stochastic fluctuations in network components and the cellular environment, the behavior of even simple artificial gene networks often cannot be accurately predicted using deterministic models. A better understanding of gene expression noise is essential for improving the performance of synthetic circuits and working towards the ultimate goal of programmable cells.
Simple gene modules, such as autoregulatory feedback loops (Becskei and Serrano, 2000; Becskei et al, 2001; Rosenfeld et al, 2002), toggle switches (Gardner et al, 2000; Atkinson et al, 2003), and oscillators (Elowitz and Leibler, 2000; Atkinson et al, 2003), have been engineered in model microorganisms such as E. coli and S. cerevisiae. The behavior of these simple genetic circuits has been observed in vivo by using flow cytometry or fluorescence microscopy to measure reporter protein levels in individual live cells.
Synthetic gene networks have been constructed to demonstrate the ability of negative feedback to reduce cell-to-cell fluctuations in protein concentrations, thus increasing the stability of the network (Becskei and Serrano, 2000). Positive feedback loops amplify cellular fluctuations and allow for the generation of bistability (Savageau, 1974). Bistability, or the existence of two stable states, has been observed in a synthetic positive feedback system (Becskei et al, 2001). Two distinct populations of cells were observed by fluorescence microscopy: cells that expressed low levels of GFP and cells that expressed high levels of GFP. Stochastic fluctuations in the network enabled spontaneous transitions from one expression state to the other.
Simple, well-characterized gene modules can be linked to form more complex networks. A recent study constructed a repressor-only system, an activator-only system, and a system that combines the activator and repressor modules and (Guido et al, 2006). A stochastic model accurately predicted the behavior of the modular system. The study demonstrated that the properties of individual regulatory modules can be used to predict the behavior of more complex gene regulatory networks, setting the stage for the systematic construction of synthetic gene networks of increasing complexity, which mimic the behavior of naturally occurring systems.
For some synthetic gene network studies, the circuit can be characterized by obtaining distributions of cellular reporter protein levels at various time points. For instance, Gardner et al (2000) used this approach to analyze a synthetic genetic toggle switch. The bistability of the synthetic network was demonstrated by showing that transient chemical or thermal induction could switch E. coli cells from one stable state corresponding to high expression of a GFP reporter to a second stable state corresponding to low GFP expression levels. Flow cytometry was used to measure GFP expression levels from individual cells and GFP distributions were obtained to show the existence of a stable low GFP expressing state and a stable high GFP expressing state. A bimodal distribution appears as switching begins, and the return to a unimodal distribution occurs when switching is complete.
Although obtaining distributions of reporter protein levels may be sufficient for understanding the behavior of synthetic networks such as the genetic toggle switch, monitoring the dynamics of gene expression in individual live cells is needed for fully characterizing many gene circuits. For example, Elowitz and Leibler (2000) used three repressors to build a synthetic oscillatory network called ‘the repressilator', and they characterized the oscillations by quantifying expression levels of a GFP reporter protein in individual E. coli cells. Individual repressilator-containing cells are not synchronized. Thus, observing oscillations in gene expression required tracking expression levels from single cells at multiple time points. This was accomplished by using fluorescence microscopy to monitor temporal oscillations in GFP expression in individual cells.
The networks responsible for generating oscillations in natural systems tend to be quite complicated, and many of the mechanisms governing the behavior of natural oscillators are still unknown. Synthetic clocks such as ‘the repressilator' have been developed with the objective of understanding the molecular design principles responsible for generating oscillations in natural systems (Elowitz and Leibler, 2000; Atkinson et al, 2003; Fung et al, 2005). The synthetic biology approach, coupled with recent advances in single-cell imaging, will help to advance our understanding of cellular rhythms.
Naturally occurring oscillations in single cells
Cellular rhythms occur at all levels of biological organization from gene networks to animal populations (Goldbeter, 2002). Because oscillations in individual cells are often asynchronous, the dynamics of many biological rhythms must be observed at the single-cell level. Single-cell studies have advanced our current understanding of natural oscillators by revealing information about cellular behavior that could not be obtained using traditional population-based methods.
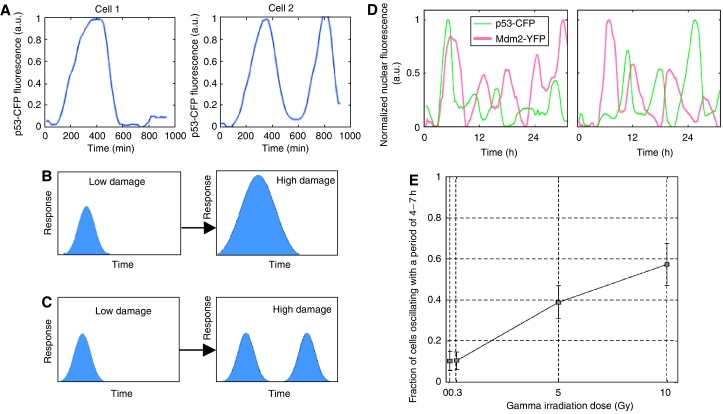
The p53–Mdm2 negative feedback loop is an example of a system that has been studied extensively primarily using population-based methods (Michael and Oren, 2003). Experiments performed at the population level have found that damped oscillations in p53 and Mdm2 occur following strong DNA damage (Lev Bar-Or et al, 2000). Recent studies have observed p53 and Mdm2 oscillations in individual cells in response to DNA damage caused by gamma irradiation (Lahav et al, 2004; Geva-Zatorsky et al, 2006). Two fusion proteins, p53-CFP and Mdm2-YFP, were used to follow the dynamics of p53 and Mdm2 in individual cells using time-lapse fluorescence microscopy. Using this system to monitor cells for approximately 16 h, Lahav et al (2004) demonstrated that individual cells can show repeated pulses of p53 and Mdm2 (see Figure 1). This study reported a distinct correlation between irradiation dose and the average number of p53 pulses. More recently, Geva-Zatorsky et al used this system to monitor p53 and Mdm2 dynamics in individual cells for longer time frames (several days) and they observed sustained undamped oscillations in a large fraction of cells. The results of the longer experimental runs showed that the irradiation dose determines the probability that an irradiated cell will oscillate permanently or not, rather than the number of pulses (see Figure 1). Another key finding of this study was that the behavior of isogenic cells showed significant cell–cell differences: some cells showed either no response or a slowly fluctuating signal, whereas other cells showed sustained oscillations with highly variable amplitudes. Mathematical models of the system suggest that low-frequency fluctuations in protein production rates are a source of the noise in the oscillations (Geva-Zatorsky et al, 2006).
Figure 1.
p53–Mdm2 pulses in individual cells. (A) p53-CFP expression levels in two individual cells in response to DNA damage (Lahav et al, 2004). (B) At the population level, the response (p53-CFP levels) appears to increase with increasing DNA damage (Lahav et al, 2004). (C) Observations of p53-CFP levels at the single-cell level for limited experimental durations (up to 16 h) suggest that the number of pulses increases with the increase in DNA damage (Lahav et al, 2004). (D) Longer observations (several days) of p53-CFP and Mdm2-YFP expression levels in individual cells in response to DNA damage (Geva-Zatorsky et al, 2006). (E) The results of long observations on cells at four different radiation doses show that the fraction of cells that oscillate increases with gamma dosage (Geva-Zatorsky et al, 2006).
The NF-κB signaling system is another example of a natural network that has been widely analyzed, usually using experiments performed at the population level. The NF-κB signaling pathway contains a negative feedback loop that drives oscillations in NF-κB nuclear–cytoplasmic localization. Population-level experiments have been used to monitor average NF-κB activity, and have found that damped oscillations in NF-κB localization occur following stimulation (Hoffmann et al, 2002). A more recent study has used time-lapse fluorescence microscopy to observe NF-κB oscillations in individual cells (Nelson et al, 2004). With single-cell resolution, cell-to-cell variation in frequency and amplitude was observed, and this study revealed that asynchronous NF-κB oscillations occur following stimulation.
Tracking the dynamic levels of fluorescent reporter proteins in individual cells can also help to elucidate some of the changes in gene expression that occur during the complex events associated with cell division. For instance, Bean et al (2006) recently investigated the issue of variability in cell-cycle Start by using time-lapse fluorescence microscopy to monitor the expression of a GFP reporter under the control of the G1 cyclin CLN2 promoter in budding yeast. This study found that peaks in fluorescence occur at the time of budding, and monitoring cell-to-cell variability in peak height and peak timing in both wild-type and mutant cells revealed that Start regulators affect variability in intra-Start coherence and variability in Start timing. For instance, deletion of the G1 transcription factor SWI4 resulted in an increase in peak height variability and a decrease in the coherence between bud emergence and CLN2pr-GFP peak levels, suggesting that SWI4 is not essential for Start but necessary for maintaining Start coherence.
Circadian rhythms are one of the most widely studied periodic biological processes. The circadian clock, which consists of a molecular feedback loop, governs the oscillatory expression of clock genes. The oscillatory feedback mechanism, which generates circadian rhythms, resides in individual cells. Recently, several studies have tried to gain insight into how biological clocks control gene expression by monitoring rhythmic behavior in individual cells. Real-time analysis of circadian gene expression has been performed at the single-cell level for cell types ranging from bacterial cells (Mihalcescu et al, 2004) to individual neurons within the mammalian suprachiasmatic nucleus (the circadian center) (Yamaguchi et al, 2003). For instance, a recent study monitored circadian oscillations in fibroblasts and found that individual fibroblasts contain self-sustained circadian oscillations, thus revealing that the damped oscillations observed at the population level result from a loss of synchrony among cells (Welsh et al, 2004; Carr and Whitmore, 2005).
Techniques for single-cell gene expression experiments
A wide variety of new tools have allowed investigators to monitor gene expression with single-cell resolution. Owing to differences in characteristics such as cellular throughput, number of gene products analyzed, sensitivity, and temporal resolution, the gene expression analysis method utilized depends on the objective of the study. For example, flow cytometry is a technique that monitors gene expression at the protein level and is very high throughput in terms of the number of cells analyzed. A flow cytometer can simultaneously measure several parameters at a rate of up to 10 000 cells per second with high precision. Flow cytometry has proven to be useful for monitoring a large number of gene products. A recent study used flow cytometry to monitor the expression of over 2500 proteins in S. cerevisiae, in response to environmental perturbations (Newman et al, 2006). This study demonstrated that there are substantial protein-specific differences in noise, and that protein noise levels are highly correlated with their function and their mode of transcriptional regulation.
‘Static' single-cell measurements, such as those obtained using flow cytometry, are useful for obtaining snapshots of gene expression patterns in individual cells. However, techniques with high temporal resolution are necessary for monitoring the dynamics of gene expression. For example, temporal phenomena such as oscillations in gene expression in individual cells can be observed by using fluorescence-based imaging assays to quantify protein production in individual cells at several time points. Because a flow cytometer can only measure protein levels in each cell at a single time point, flow cytometry cannot be used to observe dynamic gene expression behavior such as oscillations in individual cells. Below, we discuss the tools that not only enable the quantification of protein levels in individual live cells, but also allow for repeated measurements from the same cell over extended time periods and provide information about protein localizations.
Technological advances in fluorescent reporter technology
The GFP from the jelly fish Aequorea victoria has become a well-established reporter protein for studying the dynamics of gene expression. Unlike several other commonly used reporters, GFP does not depend on additional cofactors or substrates. The GFP protein has been modified to produce fluorescent proteins with a variety of colors (including blue, cyan, yellow, and red fluorescent proteins), making it possible to study the expression of multiple genes in the same cell. Engineering GFP mutants with specific properties has led to the availability of a wide variety of fluorescent reporter proteins. Several protein properties such as maturation speed and efficiency, extinction coefficient, quantum yield, and photostability need to be taken into consideration when selecting the best fluorescent protein for a particular experiment (Shaner et al, 2005).
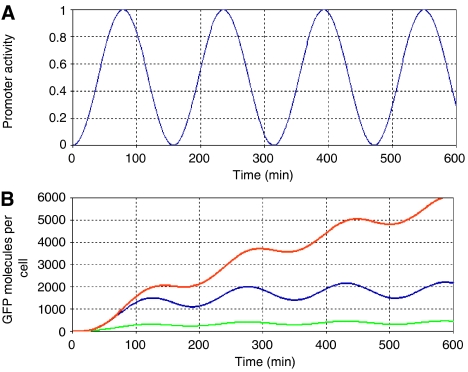
Because GFP is very stable, it is not optimal for monitoring dynamic changes in gene expression. GFP variants with half-lives as short as 30 min in bacteria and yeast (Andersen et al, 1998; Mateus and Avery, 2000) and CFP proteins with half-lives as short as 5 min in yeast (Hackett et al, 2006) have been developed to enable gene expression studies with greater temporal resolution. Fast-degrading fluorescent proteins help to rapidly extinguish the fluorescent signal when the promoter becomes inactive. However, when the promoter is active, fluorescent protein levels must be high enough to produce a fluorescent signal sufficiently higher than background fluorescence (see Figure 2). Thus, the ideal system for monitoring gene expression changes with fast dynamics will utilize a rapidly degrading fluorescent reporter in conjunction with a promoter that yields maximal reporter protein levels when active. The fluorescent reporters employed by the studies mentioned in the first section of this review are listed in Table I.
Figure 2.
Tracking oscillations in gene expression with GFP reporter proteins. (A) Oscillations in promoter activity. (B) Simulated GFP levels for three different systems: The blue curve represents GFP levels for a system that utilizes a destabilized GFP variant using the following parameter values (Elowitz and Leibler, 2000): transcription rate of 0.5 transcripts per second, translation rate of 20 proteins per transcript, mRNA half-life of 2 min, GFP half-life of 90 min (destabilized GFP variant). The orange curve represents GFP levels for a system that utilizes a stable GFP protein with a half-life of several hours (480 min). This system will quickly approach fluorescent saturation levels. The green curve represents GFP levels for a system that utilizes a destabilized GFP variant and has a lower translation rate (5 proteins per transcript). This system will produce a fluorescent signal that is harder to distinguish from background fluorescence.
Table 1.
Fluorescent reporters employed by single-cell gene expression studies
| Reference | Fluorescent reporters used | Analytical method | Organism |
|---|---|---|---|
| Atkinson et al (2003) | CFP | Fluorescence microscopy | E. coli |
| Austin et al (2006) | GFPasv-destabilized GFP variant with half-life around 110 min, GFPaav-destabilized GFP variant (Andersen 98) with half-life around 60 min | Time-lapse microscopy | E. coli |
| Bar-Even et al (2006) | GFP fusion proteins (from Invitrogen's yeast GFP clone collection) containing the coding sequence of Aequorea victoria GFP (S65T). CFP and YFP fusions used for dual-reporter assay | Flow cytometry, fluorescence microscopy | S. cerevisiae |
| Bean et al (2006) | Yeast- and FACS-optimized, destabilized GFP variant with half-life around 30 min (Mateus and Avery, 2000) | Time-lapse microscopy | S. cerevisiae |
| Becskei and Serrano (2000) | EGFP | Fluorescence microscopy | E. coli |
| Becskei et al (2001) | yEGFP (yeast-enhanced green fluorescent protein) | Fluorescence microscopy | S. cerevisiae |
| Colman-Lerner et al (2005) | CFP, YFP | Time-lapse microscopy | S. cerevisiae |
| Elowitz and Leibler (2000) | GFPaav-destabilized GFP variant (Andersen 98) with half-life around 90 min | Time-lapse microscopy | E. coli |
| Elowitz et al (2002) | CFP, YFP (wild-type codons developed by the University of Washington Yeast Resource Center) | Time-lapse microscopy | E. coli |
| Fung et al (2005) | gfpmut3.1AAV-destabilized GFP variant | Time-lapse microscopy | E. coli |
| Gardner et al (2000) | gfpmut3 (FACS-optimized GFP mutant; see Cormack et al, 1996) | Flow cytometry | E. coli |
| Geva-Zatorsky et al (2006) | ECFP from pECFP-C1 (Clontech), EYFP from pEYFP-1 (Clontech) | Time-lapse microscopy | MCF-7 (human breast cancer) |
| Guido et al (2006) | gfpmut3 (FACS-optimized GFP mutant; see Cormack et al, 1996)) | Flow cytometry | E. coli |
| Lahav et al (2004) | ECFP from pECFP-C1 (Clontech), EYFP from pEYFP-1 (Clontech) | Time-lapse microscopy | H1299 & MCF-7 (human lung and breast cancer) |
| Nelson et al (2004) | DsRed and EGFP from pEGFP-N1 and pdsRed-N1 (Clontech) | Time-lapse microscopy | HeLa and SK-N-AS |
| Ozbudak et al (2002) | gfpmut2 (FACS-optimized GFP mutant; see Cormack et al, 1996)) | Flow cytometry | B. subtilis |
| Pedraza and van Oudenaarden (2005) | CFP, YFP, RFP from pECFP, pEYFP, and pDsRed-Express (Clontech) | Fluorescence microscopy | E. coli |
| Raser and O'Shea (2004) | yECFP (yeast-optimized enhanced cyan fluorescent protein) and yVYFP (yeast-optimized Venus yellow fluorescent protein) | Fluorescence microscopy | S. cerevisiae |
| Rosenfeld et al (2002) | gfpmut3 (FACS-optimized GFP mutant; Cormack et al, 1996)) | Fluorimeter | E. coli |
| Rosenfeld et al (2005) | CFP, YFP (yfp gene from pDH5 plasmid, University of Washington Yeast Resource Center) | Time-lapse microscopy | E. coli |
| Sigal et al (2006) | YFP | Time-lapse microscopy | H1299 (human lung cancer) |
| Suel et al (2006) | CFP, YFP | Time-lapse microscopy | B. subtilis |
| Volfson et al (2005) | yEGFP (yeast-enhanced green fluorescent protein) | Flow cytometry | S. cerevisiae |
| Weinberger et al (2005) | Enhanced GFP (Clontech, Palo Alto, CA) | Flow cytometry | Jurkat cells |
Fluorescence-based gene expression assays often involve monitoring the expression of a fluorescent reporter gene from a particular promoter. An alternative method for measuring expression levels involves the use of fluorescent fusion proteins. Fluorescent fusion proteins are created by fusing the coding sequence of a protein of interest to the coding sequence of a fluorescent protein. For the fluorescent protein to be functional, both the fluorescent protein and the host protein must fold correctly (Miyawaki et al, 2003). Fluorescent reporter genes and fluorescent fusion proteins can be expressed in the same cell simultaneously. For example, Rosenfeld et al (2005) recently developed cell strains in which a yellow fluorescent repressor fusion protein represses the expression of a cyan fluorescent reporter gene. This system enabled the simultaneous measurement of transcription factor concentrations (by quantifying YFP levels) and the rate of protein production from a downstream gene (by quantifying CFP levels) (Rosenfeld et al, 2005).
Multiple fluorescent reporter genes can also be expressed in the same cell. For example, Elowitz et al (2002) engineered cell strains in which two fluorescent reporter genes, cyan fluorescent protein (cfp) and yellow fluorescent protein (yfp), were controlled by identical promoters. Correlations between CFP and YFP levels in individual cells were measured to determine the contribution of intrinsic and extrinsic noise to total cell-to-cell variability in gene expression (Elowitz et al, 2002). A more recent study utilized three fluorescent reporter genes (cfp, yfp, and rfp) to investigate how noise is transmitted in a gene network (Pedraza and van Oudenaarden, 2005). In the engineered gene network, the tetracycline repressor (tetR), which is bicistronically transcribed with cfp, downregulates the transcription of yfp. CFP and YFP levels in individual cells were compared to measure how fluctuations in the upstream gene (tetR, cfp) were transmitted to the downstream gene (yfp). A third fluorescent reporter gene, rfp, was expressed from a strong constitutive promoter to determine the contribution of global fluctuations to overall gene expression noise.
Gene expression studies measure fluorescence levels produced by reporter proteins, and these data are used to report expression levels that are roughly proportional to the actual number of fluorescent protein molecules. Recently, a technique has been developed for converting observed fluorescent intensities into numbers of molecules (Rosenfeld et al, 2006). The conversion method follows the dilution of a transiently expressed fluorescent protein as cells grow and divide. At each cell division, asymmetric partitioning of fluorescence is observed. The calibration algorithm uses observed partitioning errors to infer the apparent number of fluorescent proteins per cell. The predicted number of proteins per cell ranged from around 840 molecules in the initial cell to around 10 molecules in the eighth generation of cells.
Typically, fluorescent reporters are used to monitor gene expression dynamics at the translational level. However, a recent study has shown that fluorescent labeling of mRNA molecules can be used to analyze gene expression dynamics at the transcriptional level (Golding et al, 2005). In this study, single mRNA molecules were detected and tracked in individual live E. coli cells by using MS2-GFP fluorescent fusion proteins to tag transcripts containing a tandem array of 96 MS2 binding sites. In addition, the target transcript contained the coding region for a red fluorescent protein, mRFP1, thus allowing the level of mRNA transcript and the level of the encoded protein to be measured simultaneously. The technique for fluorescently labeling mRNA molecules has recently been utilized in eukaryotic cells: transcriptional bursts of a developmental gene were observed in individual cells of the slime mold Dictyostelium discoideum (Chubb et al, 2006).
Single-cell assays
Fluorescent reporter levels in individual cells can be monitored by using fluorescence microscopy to visualize cells grown on a glass slide or dish, and images can be captured using digital cameras. Although this approach requires more time and has a much lower throughput than methods such as flow cytometry, in vivo imaging enables the real-time quantification of gene expression in single live cells. Fluorescence-based imaging assays can also be used to observe protein localizations (Nelson et al, 2004; Sigal et al, 2006), whereas methods such as flow cytometry cannot provide spatial resolution.
Recent technical advances in live-cell imaging, such as the development of extremely sensitive cameras, high-precision automated stages, and faster computers with greater storage capacity, have enabled the rapid acquisition and storage of large-format microscopic images (Shav-Tal et al, 2004). Such improvements allow the tracking of gene expression dynamics with increased precision and higher temporal resolution. Typical time-lapse microscopy experiments provide useful and very detailed information on the dynamic behavior of a small number of cells, but typically run only for a short time period and often do not lead to good statistics over a population. The duration of a typical time-lapse experiment is often limited by cell stacking effects: the experimental run is either ended when cells begin to grow out of the focal plane, or if the run is continued the cells that are out of focus must be discarded from the analysis.
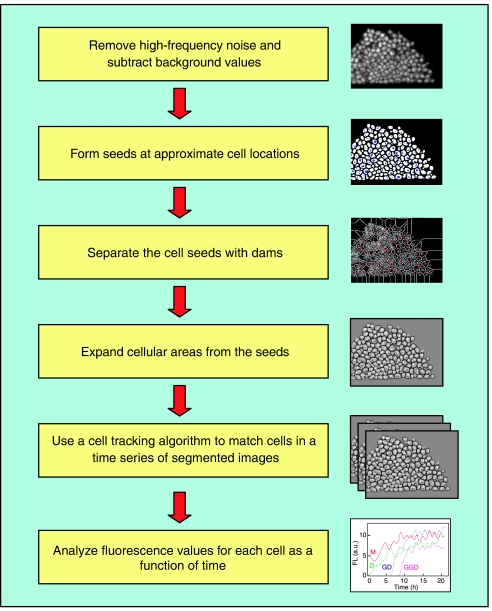
Image processing software allows gene expression data to be extracted from fluorescence images. The extraction of single-cell expression levels from a fluorescence image requires segmenting the images into individual cells and quantifying the fluorescence within the segment boundaries. Software packages such as Metamorph software (Universal Imaging, Westchester, PA), Openlab software (Improvision, Lexington, MA), and AQM Advance 6.0 software (Kinetic Imaging, UK) are available for image analysis. However, many studies utilize custom-written software that is often developed using MATLAB (The Mathworks Inc.) or IDL (The Interactive Data Language, Research Systems Inc., Boulder, CO). Gene expression dynamics can be tracked at the single-cell level by assembling cellular fluorescence levels for a set of time-lapse images. Extracting time-series gene expression data from a sequence of fluorescence images involves segmenting each image into individual cells and then matching cells in a sequence of images (see Figure 3). Image segmentation can be achieved with the following main steps: flat-field correcting and removing high-frequency noise, determining approximate cell locations using a seeding technique, producing a set of dams that separate cell seeds, and expanding the cellular area from the seed to a specified intensity level. Matching cells in a sequence of images can be accomplished using cluster analysis with the center of mass coordinates and total fluorescence integrated over the cellular area used as cluster variables (Cookson et al, 2005).
Figure 3.
Steps involved in processing time-lapse images (Cookson et al, 2005). The first step in the image analysis process involves filtering high-frequency noise and subtracting background and cellular autofluorescence values. Next, each image is segmented into individual cells by using a seeding technique to determine approximate cell locations, producing a set of dams that separate the cell seeds, and expanding the cellular area from the seed to a specified intensity level. A cell tracking algorithm is applied to a time series of segmented images to obtain a time course for each cell.
Image analysis tools can also be used to detect cell division events based on the abrupt decrease in cellular fluorescence values that occurs after cell division. Recently, Sigal et al investigated cell-cycle-dependent changes in protein dynamics by synchronizing cells a posteriori based on the detection of cell division events in individual cells in time-lapse images. An alternative method for examining the effects of cell-cycle position involves arresting cells at certain stages in the cell cycle (Colman-Lerner et al, 2005). Retroactive in silico synchronization allows cell-cycle effects to be examined without perturbing cellular behavior. Using the in silico synchronization method, cell-cycle dependence of nuclear protein levels was observed in a subset of the nuclear proteins that were examined. This study also utilized custom software to generate displays (termed ‘synchrograms') to visualize protein localization dynamics which are easier to discern by eye than changes in protein amounts. Using synchrograms, diverse cell-cycle-dependent localization patterns were observed in a subset of the examined proteins (Sigal et al, 2006).
Microfluidic ‘lab on a chip' technologies
Owing to the stochastic nature of gene expression, the optimal experimental setup for analyzing gene expression dynamics will be capable of both monitoring the behavior of a large population of cells and of tracking individual cells. Flow cytometry can be used to obtain gene expression data for thousands of cells, but only provides a snapshot of gene expression at single time points. Traditional microscopy experiments can track gene expression dynamics in individual cells, but can only monitor a relatively small population of cells. Microfluidic ‘lab on a chip' technologies can be used to track gene expression changes in individual cells, enable large populations of cells to be monitored, and allow the precise control of the cellular microenvironment.
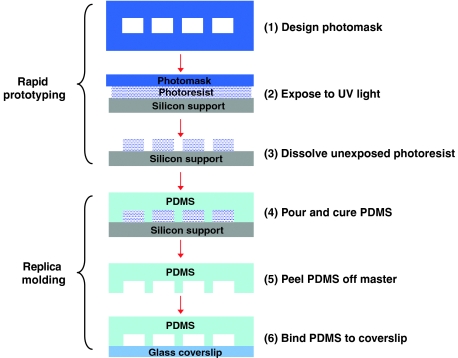
Microfluidics involves the manipulation of very small fluid volumes, enabling the creation and control of nanoliter volume reactors. Microfluidic devices can be fabricated using soft lithography and rapid prototyping techniques (McDonald et al, 2000). In rapid prototyping, a computer-aided design program is used to create a design for channels, which are printed at high resolution onto transparency film. The transparency film then serves as the photomask. Master molds are generated by using the photomask in contact lithography to produce a positive relief of photoresist on a silicon wafer. The master mold is durable and can be used indefinitely. In replica molding, poly(dimethylsiloxane) (PDMS) is poured over the master and heat cured to generate a negative replica of the master. The PDMS is then removed from the mold and sealed against a glass coverslip to form the device features and channels (see Figure 4). The advantages of using soft lithography to fabricate microfluidic devices include the low cost of production, the ease and speed of fabrication, and the reduction in the amount of reagents consumed.
Figure 4.
Diagram of the main steps involved in fabricating a microfluidic device. The first step in the fabrication process is to develop a photomask design using a computer drawing package and to print the design onto transparency film at high resolution. The second step is to coat a silicon support with photoresist and expose the photoresist to ultraviolet light through the photomask. The third step is to dissolve the unexposed photoresist to obtain a reusable master. The master consists of cured photoresist on the silicon support with a pattern defined by the photomask. The fourth step is to pour PDMS over the master and cure the PDMS by baking at 80°C for 1 h. The fifth step is to release the hardened PDMS from the master. The sixth step is to bind the PDMS to the glass coverslip.
Microfluidic systems can accurately control the biochemical composition of the cellular microenvironment. Precise control of temperature, nutrient supply, waste removal, and fluidic stress are important for recreating the in vivo cellular microenvironment. In addition, microfluidic devices can generate arbitrarily defined stable or dynamic concentration gradients (Dertinger et al, 2001). Temperature gradients can also be established using microfluidic devices, allowing the acquisition of data as a function of temperature (Mao et al, 2002). Furthermore, microfluidic devices can be used to introduce specific perturbations of the cellular microenvironment. For instance, Takayama et al (2001) demonstrated that a microfluidic device with multiple laminar inlet flows can be used to deliver small molecules to selected domains inside single mammalian cells.
Recently, Balaban et al (2004) employed microfluidic devices to observe phenotypic switching of growth rates in individual E. coli cells. Microfluidic devices containing narrow PDMS grooves were developed to force the formation of linear microcolonies during cell growth, thus allowing growth rates of the progeny of individual cells to be determined from the lengths of the linear microcolonies. The study found that persister cells, cells that survive ampicillin treatment, had slower growth rates than normal cells before exposure to antibiotics. The authors conclude that phenotypic switching between fast growth states and slow growth states enables a fraction of a genetically homogenous population to survive exposure to antibiotic treatment.
Microfluidic devices have also been used to extend the duration of gene expression experiments. The duration of a typical time-lapse experiment is limited by cell stacking effects. This limitation can be overcome by utilizing a microfluidic device in which the geometry of the device confines cells to a monolayer, thus preventing cells from growing out of the focal plane and allowing longer experimental runs. Recently, Cookson et al (2005) designed such a microfluidic device to acquire long-term single-cell dynamical measurements over a large population. This device was used to acquire single-cell fluorescence data from S. cerevesiae over many cellular generations.
Microfluidic systems combine the advantages of flow cytometry and conventional microscopy by providing a method that is capable of tracking gene expression dynamics in individual cells without sacrificing the ability to generate good statistics over a population. Furthermore, microfluidic technology allows for highly specific control of the cellular microenvironment. Microfluidic devices can be scaled, allowing high-throughput experiments to be performed. For example, microfluidic networks containing arrays of individual cell culture chambers have been developed to enable monitoring of gene expression dynamics for multiple experimental conditions. Thompson et al (2004) used this approach to monitor changes in activation of a transcription factor in response to eight different concentrations of a cytokine.
Future expectations
Single-cell time-lapse gene expression studies are necessary for observing gene expression dynamics in heterogeneous cell populations. Further advances in single-cell time-lapse experimental techniques will likely lead to novel insights relating to our understanding of gene regulation. Future gene expression studies are apt to be capable of monitoring reporter levels with increased sensitivity. For instance, two recent studies have reported the development of single-protein molecule detection techniques, which have been used to examine low-level gene expression in individual cells (Cai et al, 2006; Yu et al, 2006). The method developed by Yu et al utilizes a membrane-targeting fluorescent fusion protein that can be detected with single-molecule sensitivity as a result of the slower diffusion times for membrane-bound protein molecules. The ultrasensitive protein detection technique developed by Cai et al is based on the β-gal reporter enzyme that hydrolyzes a synthetic substrate to produce a fluorescent product. Cai et al modified the classic β-gal assay to achieve high sensitivity by using a microfluidic chamber to contain fluorescent product molecules that are actively pumped out of the cell.
Future studies are likely to extend the length of time-lapse experiments, and to monitor gene expression in a tightly controlled environment, by drawing upon the capabilities of microfluidic technology. Furthermore, the development of parallelized microfluidic systems containing hundreds of chambers will enable a large number of cellular populations to be analyzed simultaneously (Martin et al, 2003).
Acknowledgments
We warmly acknowledge the section editor, Thomas Lemberger, along with our three anonymous reviewers, for insightful and helpful suggestions for improving the manuscript. This work was supported by the National Institutes of Health Grant GM69811-01 and the National Science Foundation Graduate Research Fellowship Program.
References
- Andersen JB, Sternberg C, Poulsen LK, Bjorn SP, Givskov M, Molin S (1998) New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl Environ Microbiol 64: 2240–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson MR, Savageau MA, Myers JT, Ninfa AJ (2003) Development of genetic circuitry exhibiting toggle switch or oscillatory behavior in Escherichia coli. Cell 113: 597–607 [DOI] [PubMed] [Google Scholar]
- Austin DW, Allen MS, McCollum JM, Dar RD, Wilgus JR, Sayler GS, Samatova NF, Cox CD, Simpson ML (2006) Gene network shaping of inherent noise spectra. Nature 439: 608–611 [DOI] [PubMed] [Google Scholar]
- Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S (2004) Bacterial persistence as a phenotypic switch. Science 305: 1622–1625 [DOI] [PubMed] [Google Scholar]
- Bar-Even A, Paulsson J, Maheshri N, Carmi M, O'Shea E, Pilpel Y, Barkai N (2006) Noise in protein expression scales with natural protein abundance. Nat Genet 38: 636–643 [DOI] [PubMed] [Google Scholar]
- Bean JM, Siggia ED, Cross FR (2006) Coherence and timing of cell cycle start examined at single-cell resolution. Mol Cell 21: 3–14 [DOI] [PubMed] [Google Scholar]
- Becskei A, Seraphin B, Serrano L (2001) Positive feedback in eukaryotic gene networks: cell differentiation by graded to binary response conversion. EMBO J 20: 2528–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becskei A, Serrano L (2000) Engineering stability in gene networks by autoregulation. Nature 405: 590–593 [DOI] [PubMed] [Google Scholar]
- Benzer S (1953) Induced synthesis of enzymes in bacteria analyzed at the cellular level. Biochim Biophys Acta 11: 383–395 [DOI] [PubMed] [Google Scholar]
- Blake WJ, Kaern M, Cantor CR, Collins JJ (2003) Noise in eukaryotic gene expression. Nature 422: 633–637 [DOI] [PubMed] [Google Scholar]
- Carr AJ, Whitmore D (2005) Imaging of single light-responsive clock cells reveals fluctuating free-running periods. Nat Cell Biol 7: 319–321 [DOI] [PubMed] [Google Scholar]
- Cai L, Friedman N, Xie XS (2006) Stochastic protein expression in individual cells at the single molecule level. Nature 440: 358–362 [DOI] [PubMed] [Google Scholar]
- Chubb JR, Trcek T, Shenoy SM, Singer RH (2006) Transcriptional pulsing of a developmental gene. Curr Biol 16: 1018–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman-Lerner A, Gordon A, Serra E, Chin T, Resnekov O, Endy D, Pesce CG, Brent R (2005) Regulated cell-to-cell variation in a cell-fate decision system. Nature 437: 699–706 [DOI] [PubMed] [Google Scholar]
- Cookson S, Ostroff N, Pang WL, Volfson D, Hasty J (2005) Monitoring dynamics of single-cell gene expression over multiple cell cycles. Mol Systems Biol 1: 24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack BP, Valdivia RH, Falkow S (1996) FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173: 33–38 [DOI] [PubMed] [Google Scholar]
- Dertinger SKW, Chiu DT, Jeon NL, Whitesides GM (2001) Generation of gradients having complex shapes using microfluidic networks. Anal Chem 73: 1240–1246 [Google Scholar]
- Elowitz MB, Leibler S (2000) A synthetic oscillatory network of transcriptional regulators. Nature 403: 335–338 [DOI] [PubMed] [Google Scholar]
- Elowitz MB, Levine AJ, Siggia ED, Swain PS (2002) Stochastic gene expression in a single cell. Science 297: 1183–1186 [DOI] [PubMed] [Google Scholar]
- Fung E, Wong WW, Suen JK, Bulter T, Lee SG, Liao JC (2005) A synthetic gene-metabolic oscillator. Nature 435: 118–122 [DOI] [PubMed] [Google Scholar]
- Gardner TS, Cantor CR, Collins JJ (2000) Construction of a genetic toggle switch in Escherichia coli. Nature 403: 339–342 [DOI] [PubMed] [Google Scholar]
- Geva-Zatorsky N, Rosenfeld N, Itzkovitz S, Milo R, Sigal A, Dekel E, Yarnitzky T, Liron Y, Polak P, Lahav G, Alon U (2006) Oscillations and variability in the p53 system. Mol Syst Biol 2: 2006.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldbeter A (2002) Computational approaches to cellular rhythms. Nature 420: 238–245 [DOI] [PubMed] [Google Scholar]
- Golding I, Paulsson J, Zawilski SM, Cox EC (2005) Real-time kinetics of gene activity in individual bacteria. Cell 123: 1025–1036 [DOI] [PubMed] [Google Scholar]
- Guido NJ, Wang X, Adalsteinsson D, McMillen D, Hasty J, Cantor CR, Elston TC, Collins JJ (2006) A bottom-up approach to gene regulation. Nature 439: 856–860 [DOI] [PubMed] [Google Scholar]
- Hackett EA, Esch RK, Maleri S, Errede B (2006) A family of destabilized cyan fluorescent proteins as transcriptional reporters in S. cerevisiae. Yeast 23: 333–349 [DOI] [PubMed] [Google Scholar]
- Hasty J, McMillen D, Collins JJ (2002) Engineered gene circuits. Nature 420: 224–230 [DOI] [PubMed] [Google Scholar]
- Hoffmann A, Levchenko A, Scott ML, Baltimore D (2002) The IkappaB–NF-kappaB signaling module: temporal control and selective gene activation. Science 298: 1241–1245 [DOI] [PubMed] [Google Scholar]
- Kaern M, Elston TC, Blake WJ, Collins JJ (2005) Stochasticity in gene expression: from theories to phenotypes. Nat Rev Genet 6: 451–464 [DOI] [PubMed] [Google Scholar]
- Lahav G, Rosenfeld N, Sigal A, Geva-Zatorsky N, Levine AJ, Elowitz MB, Alon U (2004) Dynamics of the p53–Mdm2 feedback loop in individual cells. Nat Genet 36: 147–150 [DOI] [PubMed] [Google Scholar]
- Lev Bar-Or R, Maya R, Segel LA, Alon U, Levine AJ, Oren M (2000) Generation of oscillations by the p53-Mdm2 feedback loop: a theoretical and experimental study. Proc Natl Acad Sci USA 97: 11250–11255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloney PC, Rotman B (1973) Distribution of suboptimally induces-D-galactosidase in Escherichia coli. The enzyme content of individual cells. J Mol Biol 73: 77–91 [DOI] [PubMed] [Google Scholar]
- Mao H, Yang T, Cremer PS (2002) A microfluidic device with a linear temperature gradient for parallel and combinatorial measurements. J Am Chem Soc 124: 4432–4435 [DOI] [PubMed] [Google Scholar]
- Martin K, Henkel T, Baier V, Grodrian A, Schon T, Roth M, Michael Kohler J, Metze J (2003) Generation of larger numbers of separated microbial populations by cultivation in segmented-flow microdevices. Lab Chip 3: 202–207 [DOI] [PubMed] [Google Scholar]
- Mateus C, Avery SV (2000) Destabilized green fluorescent protein for monitoring dynamic changes in yeast gene expression with flow cytometry. Yeast 16: 1313–1323 [DOI] [PubMed] [Google Scholar]
- McAdams HH, Arkin A (1999) It's a noisy business! genetic regulation at the nanomolar scale. Trends Genet 15: 65–69 [DOI] [PubMed] [Google Scholar]
- McDonald JC, Duffy DC, Anderson JR, Chiu DT, Wu H, Schueller OJ, Whitesides GM (2000) Fabrication of microfluidic systems in poly(dimethylsiloxane). Electrophoresis 21: 27–40 [DOI] [PubMed] [Google Scholar]
- Michael D, Oren M (2003) The p53–Mdm2 module and the ubiquitin system. Semin Cancer Biol 13: 49–58 [DOI] [PubMed] [Google Scholar]
- Mihalcescu I, Hsing W, Leibler S (2004) Resilient circadian oscillator revealed in individual cyanobacteria. Nature 430: 81–85 [DOI] [PubMed] [Google Scholar]
- Miyawaki A, Sawano A, Kogure T (2003) Lighting up cells: labelling proteins with fluorophores. Nat Cell Biol 5 (Suppl): S1–S7 [PubMed] [Google Scholar]
- Nelson DE, Ihekwaba AE, Elliott M, Johnson JR, Gibney CA, Foreman BE, Nelson G, See V, Horton CA, Spiller DG, Edwards SW, McDowell HP, Unitt JF, Sullivan E, Grimley R, Benson N, Broomhead D, Kell DB, White MR (2004) Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science 306: 704–708 [DOI] [PubMed] [Google Scholar]
- Newman JR, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, Derisi JL, Weissman JS (2006) Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature 441: 840–846 [DOI] [PubMed] [Google Scholar]
- Novick A, Weiner M (1957) Enzyme induction as an all-or-none phenomenon. Proc Natl Acad Sci USA 43: 553–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A (2002) Regulation of noise in the expression of a single gene. Nat Genet 31: 69–73 [DOI] [PubMed] [Google Scholar]
- Pedraza JM, van Oudenaarden A (2005) Noise propagation in gene networks. Science 307: 1965–1969 [DOI] [PubMed] [Google Scholar]
- Ramsey S, Ozinsky A, Clark A, Smith KD, de Atauri P, Thorsson V, Orrell D, Bolouri H (2006) Transcriptional noise and cellular heterogeneity in mammalian macrophages. Philos Trans R Soc London B Biol Sci 361: 495–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao CV, Wolf DM, Arkin AP (2002) Control, exploitation and tolerance of intracellular noise. Nature 420: 231–237 [DOI] [PubMed] [Google Scholar]
- Raser JM, O'Shea EK (2004) Control of stochasticity in eukaryotic gene expression. Science 304: 1811–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raser JM, O'Shea EK (2005) Noise in gene expression: origins, consequences, and control. Science 309: 2010–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigney DR, Schieve WC (1977) Stochastic model of linear, continuous protein synthesis in bacterial populations. J Theor Biol 69: 761–766 [DOI] [PubMed] [Google Scholar]
- Rosenfeld N, Elowitz MB, Alon U (2002) Negative autoregulation speeds the response times of transcription networks. J Mol Biol 323: 785–793 [DOI] [PubMed] [Google Scholar]
- Rosenfeld N, Perkins TJ, Alon U, Elowitz MB, Swain PS (2006) A fluctuation method to quantify in vivo fluorescence data. Biophys J 91: 759–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB (2005) Gene regulation at the single-cell level. Science 307: 1962–1965 [DOI] [PubMed] [Google Scholar]
- Savageau MA (1974) Comparison of classical and autogenous systems of regulation in inducible operons. Nature 252: 546–549 [DOI] [PubMed] [Google Scholar]
- Shaner NC, Steinbach PA, Tsien RY (2005) A guide to choosing fluorescent proteins. Nat Methods 2: 905–909 [DOI] [PubMed] [Google Scholar]
- Shav-Tal Y, Singer RH, Darzacq X (2004) Imaging gene expression in single living cells. Nat Rev Mol Cell Biol 5: 855–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigal A, Milo R, Cohen A, Geva-Zatorsky N, Klein Y, Alaluf I, Swerdlin N, Perzov N, Danon T, Liron Y, Raveh T, Carpenter AE, Lahav G, Alon U (2006) Dynamic proteomics in individual human cells uncovers widespread cell-cycle dependence of nuclear proteins. Nat Methods 3: 525–531 [DOI] [PubMed] [Google Scholar]
- Sprinzak D, Elowitz MB (2005) Reconstruction of genetic circuits. Nature 438: 443–448 [DOI] [PubMed] [Google Scholar]
- Spudich JL, Koshland DE Jr (1976) Non-genetic individuality: chance in the single cell. Nature 262: 467–471 [DOI] [PubMed] [Google Scholar]
- Suel GM, Garcia-Ojalvo J, Liberman LM, Elowitz MB (2006) An excitable gene regulatory circuit induces transient cellular differentiation. Nature 440: 545–550 [DOI] [PubMed] [Google Scholar]
- Swain PS, Elowitz MB, Siggia ED (2002) Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc Natl Acad Sci USA 99: 12795–12800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama S, Ostuni E, LeDuc P, Naruse K, Ingber DE, Whitesides GM (2001) Subcellular positioning of small molecules. Nature 411: 1016. [DOI] [PubMed] [Google Scholar]
- Thattai M, van Oudenaarden A (2001) Intrinsic noise in gene regulatory networks. Proc Natl Acad Sci USA 98: 8614–8619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DM, King KR, Wieder KJ, Toner M, Yarmush ML, Jayaraman A (2004) Dynamic gene expression profiling using a microfabricated living cell array. Anal Chem 76: 4098–4103 [DOI] [PubMed] [Google Scholar]
- Volfson D, Marciniak J, Blake WJ, Ostroff N, Tsimring LS, Hasty J (2005) Origins of extrinsic variability in eukaryotic gene expression. Nature 439: 861–864 [DOI] [PubMed] [Google Scholar]
- Weinberger LS, Burnett JC, Toettcher JE, Arkin AP, Schaffer DV (2005) Stochastic gene expression in a lentiviral positive-feedback loop: HIV-1 Tat fluctuations drive phenotypic diversity. Cell 122: 169–182 [DOI] [PubMed] [Google Scholar]
- Welsh DK, Yoo SH, Liu AC, Takahashi JS, Kay SA (2004) Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr Biol 14: 2289–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Isejima H, Matsuo T, Okura R, Yagita K, Kobayashi M, Okamura H (2003) Synchronization of cellular clocks in the suprachiasmatic nucleus. Science 302: 1408–1412 [DOI] [PubMed] [Google Scholar]
- Yu J, Xiao J, Ren X, Lao K, Xie XS (2006) Probing gene expression in live cells, one protein molecule at a time. Science 311: 1600–1603 [DOI] [PubMed] [Google Scholar]