Abstract
The AF-6 protein is a multidomain protein that contains two potential Ras-binding domains within its N terminus. Because of this feature, AF-6 has been isolated in both two-hybrid and biochemical approaches and is postulated to be a potential Ras-effector protein. Herein, we show that it is specifically the first Ras-binding domain of AF-6 that mediates this interaction and that the Ras-related Rap1A protein can associate with this motif even more efficiently than the oncogenic Ha-, K-, and N-Ras GTPases. We further demonstrate that both Ras and Rap1 interact with full-length AF-6 in vivo in mammalian cells and that a fraction of Rap1 colocalizes with AF-6 at the membrane. Dominant active Rap1A, in contrast to Ras, when introduced into epithelial MDCK and MCF-7 cells, does not perturb AF-6-specific residency in cell–cell adhesion complexes. In a pursuit to gain further understanding of the role of AF-6 in junctions, we identified profilin as an AF-6-binding protein. Profilin activates monomeric actin units for subsequent polymerization steps at barbed ends of actin filaments and has been shown to participate in cortical actin assembly. To our knowledge, AF-6 is the only integral component in cell–cell junctions discovered thus far that interacts with profilin and thus could modulate actin modeling proximal to adhesion complexes.
The Ras proteins are signal-transducing GTPases that cycle between GDP-bound inactive and GTP-bound active states, relaying signals from various membrane receptors to the nucleus to mediate cellular activities such as cell growth control (1). The importance of Ras genes in the etiology of human cancers is made evident by the frequent findings of activated ras oncogenes in a wide variety of cancers. These cancer cells, as well as oncogenic Ras-transformed cell lines, apart from their deregulated proliferation, are characterized by changes in morphology and cell–cell adhesion (2, 3). Several downstream effector molecules mediating Ras' effect on proliferation have been identified, including Raf, RalGDS, and PI3-kinase proteins (1). Some of these effector molecules are shared by a closely related Ras GTPase, Rap1, which contains a virtually identical effector loop region (4). The first Rap GTPase was identified originally in a screen for revertants of the morphology exerted by K-Ras-transformed cells (5), which suggested a Ras-antagonizing effect. However, more recent studies support fundamental differences between Ras- and Rap1-controlled signaling pathways (4, 6). First, Rap1A, in contrast to Ras, is found largely in mid-Golgi, endocytic vesicles, and lysosomal vesicles, indicating different cellular functions of the proteins (6). On a molecular level, Rap's activation seems to be dictated by a growing number of exchange factors that do not act on the prototypic oncogenic Ras GTPases (6–8). Finally, in developmental systems, the functions of Rap proteins seem to be related primarily to morphological and differentiative events, rather than to those that govern proliferation and cell-fate specification, phenomena that often require signaling via conventional Ras proteins (9, 10).
In addition to the Ras-binding proteins described above, the human AF-6 protein has been identified as a potential Ras-interacting molecule by two different approaches: one based on a two-hybrid interaction assay in yeast (11); the other one based on an affinity purification protocol (12). The AF-6 gene was also found to be fused to the ALL-1 gene in a subset of acute lymphoblastic leukemias caused by chromosomal (t 6;11) translocation events—hence the name ALL-1 fused gene on chromosome 6 (13). Subsequent database analysis led to the prediction of an array of motifs, such as two N-terminal Ras-binding domains (RBDs; ref. 14), U104 and DIL motifs that were described initially in microtubule and actin-based motor proteins, respectively (15), and a PDZ domain followed by an extended C-terminal tail interspersed with proline-enriched patches. The corresponding molecule in rat, namely afadin, is present in two splice variants, the larger of which, l-afadin, was purified as a protein from rat brain extracts associated with filamentous actin. This affinity for F actin resides in a sequence that is unique to l-afadin's C terminus, but is absent in the shorter splice variant, s-afadin, which represents the actual AF-6 homologue (16).
The issue of whether members of the Ras-family of GTPases use AF-6 as a bona fide effector in specific cellular events, however, remains unresolved. Various subcellular localization experiments performed in polarized epithelial cells and tissue sections of intestinal epithelia suggest its distinct residency in cell–cell junctional complexes (16–18). In the former system, AF-6 was assigned to tight junctions, because it was found to bind Zona Occludens (ZO-1), an integral component of tight junctional complexes (17, 19). In the latter, the rat AF-6 homologue seemed to be tethered to adherens-based adhesion complexes (16, 18). Both studies, however, suggest that AF-6, along with other molecules, constitutes a physical link between membrane-located adhesion molecules and the cortical actin cytoskeleton. Consistent with this suggestion are more recent findings demonstrating that the Drosophila homologue of AF-6, Canoe, is targeted to junctional complexes in embryonic epithelia (20). Canoe was first identified by virtue of its severe rough eye phenotype and has been shown to interact genetically with the Notch signaling pathway, a pathway that determines various cell fates in a multitude of developmental processes (21).
Herein, evidence is presented that the Rap1A GTPase, in addition to the classical oncogenic Ras proteins, can interact with AF-6 in two-hybrid assays, as well as in vivo, and that it is the RBD1 domain that confers this binding capacity. We further show that a fraction of dominant active Rap1A colocalizes with AF-6 at the membrane. These data suggest that AF-6 may function as a Rap1 effector molecule. In contrast to oncogenic Ras, cells stably expressing constitutively active Rap1A retain AF-6 at the cellular junctions. It is likely that specific Ras proteins target AF-6 in different developmental and cellular situations to elicit distinct effects. Moreover, in an endeavor to elucidate the role of AF-6 in junctional complexes further, we isolated profilin as an associated protein. Profilin plays a critical role in actin polymerization events, and apart from the F actin association of l-afadin mentioned above, this interaction might provide another more dynamic link between junctional complexes and the actin cytoskeleton.
Methods
Plasmids and Abs.
For the construction of pGAD AF-6 N-terminal domains, AF-6N (amino acids 1–368), AF-6-RBD1 (amino acids 1–140), and AF-6-RBD2 (amino acids 181–368) were PCR amplified from pBSK-AF-6 (a gift from E. Canaani, Weizmann Institute, Rehovot, Israel) and subcloned into pGAD1318 (22). PI3Kδ-RBD was PCR amplified from pBSK-PI3Kδ (a gift from B. Vanhaesebroeck, Ludwig Institute, London) and subcloned into pGAD1318. LexA DNA-binding domain (LBD) Rap1E63 and Rap1N17 constructs were generated by inserting PCR-generated Rap1E63 and Rap1N17 cDNAs into a pLexVJL10 (22). For the construction of the pGAD-profilin constructs, profilin I and profilin II were PCR amplified from a profilin two-hybrid clone and from expressed sequence tag 663,927, respectively, and subcloned into pGAD1318. The plasmids pGADGH c-RafN, pGAD1318 RalGDS-RBD, LBD Ha-RasV12, and LBD RasN17 have been described (23, 24). Full-length Ha-RasV12 and Rap1E63 were subcloned into the pBabe-puro and pDCR vectors (23, 25). pcDNA AF-6-myc was generated by cloning AF-6 lacking the stop codon into pcDNA3.1/Myc-His3 (Invitrogen). To construct glutathione S-transferase (GST)–AF-6 fusion proteins, AF-6 fragments obtained by PCR were subcloned into pGEX-4T1. mAbs against hemagglutinin (HA; clone 12CA5) and Myc (clone 9E10) were purchased from Roche Molecular Biochemicals and Oncogene Science, respectively. Mouse anti-Ras, Rap1, and β-catenin Abs were obtained from Transduction Laboratories (Lexington, KY), and mAb against ZO-1 was purchased from Zymed. Polyclonal Ab against AF-6 (amino acids 1,130–1,612) and profilin were kindly provided by K. Kaibuchi (Nara Institute, Nara, Japan) and T. Pollard (Salk Institute, La Jolla, CA), respectively. Polyclonal Ab against an AF-6-specific peptide (CGRVEQQPDYRRQESRTQD, amino acids 561–576) was generated as described by Harlow and Lane (41).
Retroviral-Mediated Gene Transfer.
For the generation of MDCK-RasV12 and MDCK-Rap1E63 stable cell lines, retroviral-mediated gene transfer was performed as described (26). LinX-A cells (a gift from Greg Hannon, Cold Spring Harbor Laboratory) were transfected with 6 μg of pBabe Ha-RasV12 or pBabe Rap1E63 retroviral plasmids, and the resulting virus was used to infect MDCK cells. Infected cell populations were subjected to puromycin selection (10 μg/ml) for 3 days. Expression of the GTPases was confirmed by Western blot analysis.
Yeast Two-Hybrid Protein–Protein Interactions.
The yeast reporter strain L40 (22) was used for all two-hybrid analyses. The assays to detect protein–protein interactions were performed as described (22). For the yeast two-hybrid screen, a C-terminal fragment (amino acids 910–1,612) of AF-6 fused to LexA to generate pLexA-AF-6C was cotransformed into L40 with a Jurkat cDNA library fused to Gal4-activation domain (GAD) in pGAD1318 (11).
GST Pull-Down, Immunoprecipitation Assays.
GST AF-6 fusion proteins were expressed in BL21 Escherichia coli cells, extracted in bacterial lysis buffer, and purified on glutathione-Sepharose resin (Amersham Pharmacia). Whole-cell lysates of MDCK cells were incubated with 250 μg of fusion protein, and bound proteins were eluted with 50 μl of SDS sample buffer. Proteins were separated by SDS/PAGE and transferred to nitrocellulose membrane. Profilin protein was visualized with a polyclonal anti-profilin Ab and enhanced chemiluminescence (Amersham Pharmacia). Cos1 cells were cotransfected with pcDNA-AF-6-myc and pDCR Ha-RasV12 or pDCR Rap1E63 by using Fugene (Roche Molecular Biochemicals). At 48 h after transfection, the cells were washed twice with PBS and lysed in lysis buffer (27). The supernatants were precleared by incubation with protein G-agarose beads, nutated with 7 μg of anti-HA Ab overnight at 4°C, and then coupled with 30 μl of protein G-agarose for 1 h. The beads were washed five times with lysis buffer, and the proteins were separated by SDS/PAGE. After transfer to nitrocellulose membrane, coimmunoprecipitated AF-6 was detected with anti-myc mAb and enhanced chemiluminescence.
Fluorescence Microscopy.
Cells were plated on coverslips and treated for immunofluorescence as described (28). x,y plane images were collected at 0.5-μm steps through the entire z dimension of labeled cells with a Noran Instruments (Middleton, WI) XL laser scanning confocal attachment mounted on an upright Nikon Optiphot (×40).
Results
Interaction Between Rap1 and AF-6.
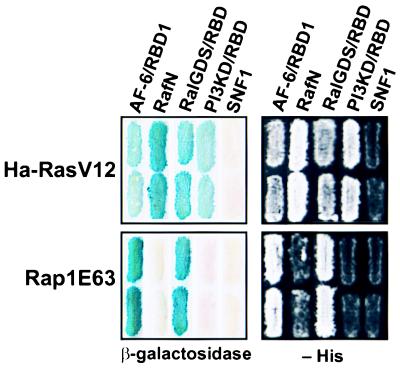
We previously identified peptides, comprising amino acids 1–180 and 1–205 of rat and human AF-6, respectively, in yeast two-hybrid screens looking for oncogenic Ras (Ha-RasV12)-interacting proteins (11). Database searches revealed that the complete AF-6 protein contains two predicted RBDs: the first comprising amino acids 39–133; the second containing amino acids 256–348 (14). To assess whether both domains are able to interact with AF-6, we constructed LexA DNA-binding fusion constructs expressing the first (RBD1), second (RBD2), or both RBDs (AF-6N) of human AF-6. As shown in Table 1, AF-6N and the first domain (RBD1), with even higher affinity, associated with Ha-RasV12. The second domain (RBD2) failed to bind to Ha-RasV12. Similar results were obtained with K-Ras and N-Ras; however, N-Ras showed a weaker interaction (data not shown). We also investigated a possible association between the Ras-related protein Rap1 and AF-6 and noticed that the strength of interaction between Rap1 and AF-6 exceeded that exerted by Ras/AF-6 complexes (Table 1). Both wild-type and constitutively activated (CA) mutant forms of Ha-Ras and Rap1 interact with AF-6; however, none of the dominant-negative mutant forms showed binding activity toward AF-6. A similar binding profile has been demonstrated between Ras and its well established effector, Raf (24). Furthermore, we made use of the two-hybrid system to see how AF-6 compares to c-Raf, RalGDS, and δ-PI3-kinase in terms of its ability to bind to Ras and Rap1. As shown in Fig. 1, RasV12 is able to interact with all targets, the strongest interaction being with c-Raf. CA Rap1E63 shows strong interaction with AF-6 and RalGDS; however, no association was observed with δ-PI3-kinase, and only very weak interaction was seen with c-Raf. These results indicate that both GTPases, Ras and Rap1, use effector molecules that are only partially identical and that they exhibit differential binding profiles toward the targets listed above.
Table 1.
Interaction between Ras/Rap1 and AF-6
| LBD fusion | β-Galactosidase activity of GAD-fused AF-6 domain
|
||
|---|---|---|---|
| AF-6N | AF-6-RBD1 | AF-6-RBD2 | |
| RasV12 | 99.0 ± 1.6 | 210.0 ± 1.8 | 0.7 ± 1.4 |
| RasN17 | 0.6 ± 1.4 | 0.9 ± 1.4 | 0.6 ± 1.1 |
| RapE63 | 120.0 ± 1.7 | 350.0 ± 1.9 | 1.2 ± 1.1 |
| RapN17 | 0.7 ± 1.7 | 0.8 ± 1.9 | 0.9 ± 1.1 |
| Lamin | 0.7 ± 1.3 | 0.8 ± 1.1 | 0.7 ± 1.5 |
Interaction between Ras/Rap1 and AF-6. The AF-6N, AF-6-RBD1, and AF-6-RBD2 GAD fusion products were transformed individually into the yeast reporter strain L40 with LBD fusions containing constitutively active and negative mutant forms of Ha-Ras and Rap1, as well as lamin as a negative control. Transformants were grown in selective synthetic medium, and βgalactosidase activity was assayed with O-nitrophenyl-β-galactosidase; values (miller units) are the means ± SD of triplicate determinations.
Figure 1.
Interaction between Ras/Rap1 and their potential targets. The LexA two-hybrid tester strain L40 was transformed with plasmids expressing CA Ras and Rap1 fused to LBD, and their potential targets AF-6, c-Raf, RalGDS, and δ-PI3-kinase (PI3KD) fused to GAD. Transformants were assayed for β-galactosidase expression (Left) and for their ability to grow on medium lacking histidine (−His; Right).
Although an in vivo interaction between oncogenic Ras and full-length AF-6 has been reported (27, 29), this interaction remained to be defined for Rap1 and AF-6. To assess whether Rap1 interacts with AF-6 in mammalian cells, myc-tagged full-length AF-6 protein was coexpressed with HA-tagged Rap1E63 in Cos1 cells. HA-tagged Ha-RasV12 was included as a positive control. Cos cells were used, because the introduction of wild-type AF-6 cDNA into MDCK and MCF7 cells did not allow high levels of expression. As shown in Fig. 2A, GTPase-bound AF-6 could be detected by blotting with an anti-myc Ab after immunoprecipitation of HA-tagged Rap1. To corroborate this finding further, we sought to compare the immunofluorescence patterns of Rap1A and AF-6. To this end, a green fluorescent protein-tagged Rap1AE63 expression construct was introduced into epithelial MCF7 cells. We selected low Rap1E63-expressing cells and assessed the distribution of Rap1E63 as well as its colocalization with endogenous AF-6. As observed previously by other investigators (6), we noticed that Rap1A staining is highly reminiscent of a typical Golgi and vesicular distribution. However, a fraction of the protein can also be detected clearly at the plasma membrane (see Fig. 2B). Staining the same cells with an AF-6-directed Ab revealed colocalization between the two proteins at the plasma membrane, opening the possibility of an interplay between Rap1A and AF-6 at the plasma membrane. Taken together, these results suggest that AF-6 is a potential effector, not only of oncogenic Ras, but also of activated Rap1.
Figure 2.
(A) Association of Rap1 and full-length AF-6 in vivo in mammalian cells. Cos1 cells were transfected with myc-tagged full-length AF-6 together with empty vector, HA-tagged Rap1E63, or HA-tagged RasV12. Protein expression was confirmed by immunoblotting cell lysates with anti-HA Ab for detection of Ras and Rap1 and anti-myc Ab for detection of AF-6. Ha-tagged GTPases were immunoprecipitated with anti-HA Ab, and associated AF-6 was probed with anti-myc Ab on a Western blot (WB). (B) Comparative localization of overexpressed Rap1AE63 and endogenous AF-6 in epithelial cells. A cDNA encoding green fluorescent protein (GFP)-Rap1AE63 was introduced into MCF7 cells, and the distribution of the GTPase was assessed in vivo by confocal microscopy. The distribution of endogenous AF-6 was visualized with an AF-6-specific Ab and rhodamine-conjugated anti-rabbit IgGs.
Role of AF-6 in Junctional Complex Organization.
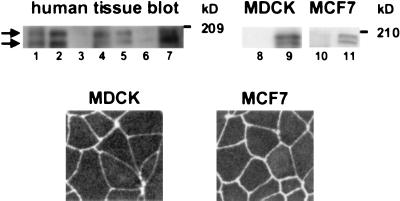
As an initial step toward understanding the role of AF-6, its tissue distribution and subcellular localization were examined. Takai and coworkers (16) reported that the rat homologue of AF-6, afadin, exists as two isoforms: the longer isoform being ubiquitously expressed, and the shorter being restricted to brain. However, we observed two bands in dog MDCK and human breast MCF7 epithelial cells when using Abs raised against human AF-6. Furthermore, when using the same Abs on a multiple human tissue blot from lung, kidney, spleen, testis, ovary, heart, and pancreas, we also noted two bands with molecular masses of approximately 195 and 180 kDa (see Fig. 3 Upper). These data suggest a wider distribution for the shorter isoform of human AF-6.
Figure 3.
(Upper) Distribution of AF-6 in multiple tissues and cell lines. (Lanes 1–7) A multiple human tissue blot from lung, kidney, spleen, testis, ovary, heart, and pancreas (Geno Technology, St. Louis) was immunoblotted with anti-AF-6 Ab. (Lanes 8–11) Immunoblot analysis of MDCK and MCF7 cell lysates with preimmune serum (lanes 8 and 10) and anti-AF-6 Ab (lanes 9 and 11), respectively. Two bands with molecular masses of approximately 195 and 180 kDa (kD) can be seen (arrows). (Lower) Localization of AF-6 in MDCK and MCF7 cells. Confluent MDCK and MCF7 cells were stained with rabbit polyclonal Ab against AF-6, followed by Texas red-conjugated anti-rabbit IgGs.
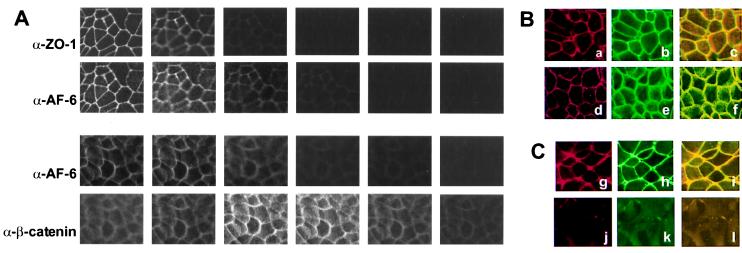
As shown in Fig. 3 Lower, AF-6 resides at cell–cell contact sites in both MDCK and MCF7 cell lines. Because two other laboratories reported tight versus adherens junctional localizations of human and rat AF-6 proteins, we reassessed its distribution in MCF7 and MDCK cells. Toward this end, we performed colocalization studies by laser scanning confocal microscopy by using ZO-1 and β-catenin as markers for tight junctions and adherens junctions, respectively. Cells were costained with either monoclonal anti-ZO-1 and polyclonal anti-AF-6 or monoclonal anti-β-catenin and polyclonal anti-AF-6, and multiple serial optical sections were taken from the apical to the basolateral side. Six serial optical sections (one section every 2 μm) are shown for each staining (Fig. 4A). Both ZO-1 and AF-6 staining was concentrated at the apical side of MCF7 cells but with AF-6 having a slightly broader lateral distribution. No AF-6 staining was observed in more basal sections containing strong β-catenin staining. Similar observations were made when MDCK cells were used (data not shown). En face views from stacked serial sections showed colocalization between AF-6 and ZO-1, which was not as apparent for AF-6 and β-catenin (Fig. 4B). These data suggest that AF-6 in MCF7 and MDCK cells colocalizes with ZO-1 rather than with β-catenin. We want to point out that AF-6 distribution was observed to vary dependent on the adherens state of the cells. Cells showed increasing intensity of AF-6 staining and membrane localization as they establish full adherens potential with neighboring cells. In cells that are embedded in a completely confluent cell layer, the presence of AF-6 in the cytoplasm appears to increase.
Figure 4.
(A) Confocal images of MCF7 cells comparing the localization of AF-6, ZO-1, and β-catenin. Confluent MCF7 cells were doubly stained with a rabbit polyclonal Ab against AF-6 and a mouse mAb against ZO-1 or a mouse mAb against β-catenin. As secondary Abs, Texas red-conjugated anti-rabbit IgGs and FITC-conjugated anti-mouse IgGs were used. Six serial optical sections (one section every 2 μm) are shown for each staining. From left to right, apical to basolateral side. (B) Localization of AF-6, ZO-1, and β-catenin in confluent control MCF7 cells. As described above, MCF7 cells were costained with a rabbit polyclonal Ab against AF-6 (a and d) and a mouse mAb against ZO-1 (b) or a mouse mAb against β-catenin (e). AF-6 is shown in red; ZO-1 and β-catenin are shown in green; and overlay is shown in yellow (c and f). (C) Localization of AF-6 and ZO-1 in Rap1E63-expressing (g–i) and Ha-RasV12-expressing (j–l) MDCK cells. The indicated cells were costained with a rabbit polyclonal Ab against AF-6 (g and j) and a mouse mAb against ZO-1 (h and k).
We next wanted to determine whether activated mutant forms of Ras or Rap1 could influence the localization of AF-6 at the cell–cell junctions. Toward this end, we created stable MDCK cell lines expressing either Ha-RasV12 or Rap1E63. We observed that the MDCK Ha-RasV12-expressing cells never gained a polygonal cell shape, instead maintaining a morphology more characteristic of fibroblasts and mesenchymal cells. Most of the cells lost their cell–cell contacts completely or were associated only loosely with their neighboring cells. In these cells, AF-6 and ZO-1 staining was absent from the cell borders, and a diffuse staining was apparent in the cytoplasm (Fig. 4C). In contrast, Rap1E63-expressing stable MDCK cells displayed a typical epithelial morphology and the localizations of AF-6 and ZO-1 in these cells were not perturbed (Fig. 4C). It is noteworthy that transiently transfected MCF7 cells with high levels of Rap1E63 expression displayed aberrations from the normally observed juxtaposed adhering cells.
Identification of Profilin as an AF-6-Binding Protein.
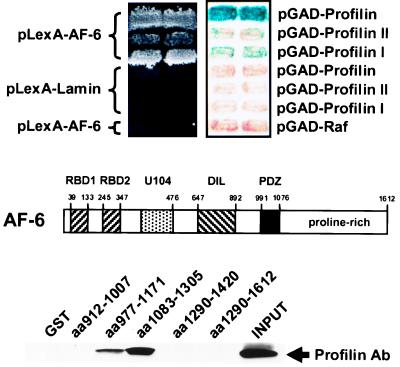
The specific subcellular localization and domain structure of the AF-6 protein suggest that AF-6 may be a component of a more complex protein network. To gain further information about unknown binding partners, we performed a two-hybrid interaction trap assay by using the C-terminal portion of the protein (amino acids 912–1,612). Independent clones (n = 2.3 × 106) of a human Jurkat T cell cDNA library were screened, yielding 530 colonies that showed prototrophy on medium lacking histidine. Further analyses of the candidates yielded a major cDNA species that encoded the small G actin-binding protein profilin. Because all of the cDNAs carried additional noncoding sequences upstream of the protein-coding region, we PCR amplified the ORF of profilin I and fused it to GAD. In addition, the ORF of profilin II, an isoform that is predominantly expressed in brain tissues, was amplified from an expressed sequence tag and treated similarly. Fig. 5 Top summarizes the results of the two-hybrid assay and indicates that interactions were strongest with molecules containing additional sequences 5′ of the profilin-specific ATG. One reason for this observation may be stabilization of the overall fold, or alternatively, the epitope interacting with AF-6 could be more favorably positioned. However, both profilin I and II, the former more pronounced than the latter, bind to the original bait.
Figure 5.
Interaction of the C terminus of AF-6 and profilin. (Top) Two-hybrid interaction. An isolated profilin I cDNA containing an additional leader sequence and PCR-amplified profilin I and profilin II cDNAs fused to GAD were cotransformed with LBD AF-6N into L40 and were assessed for β-galactosidase expression and for their ability to grow on medium lacking histidine. pLex-Lamin and pGADGH were used as negative controls. (Middle) Structure of AF-6. (Bottom) AF-6–profilin association examined in GST pull-down assays. Overlapping fragments of the AF-6 C-terminal 700 amino acids were expressed as GST fusions and used to bind profilin from a whole-cell MDCK lysate. Bound profilin was visualized with an α-profilin Ab in a subsequent Western blot analysis.
To reevaluate this two-hybrid interaction on a biochemical level and to map the profilin-binding region within the AF-6 protein, we performed GST pull-down assays. The complete bait sequence previously used in the two-hybrid screen was fused in overlapping fragments to GST and expressed in E. coli. Purified fusion proteins were tested subsequently for their ability to bind profilin out of an MDCK whole-cell lysate. Two overlapping fusions retained profilin, as shown by Western blot analysis with profilin-directed antiserum (Fig. 5 Bottom). This study indicated that the AF-6-binding site for profilin is located within the sequence spanned by overlapping fusions (amino acids 977–1,171 and amino acids 1,083–1,305). Surprisingly, this region does not contain proline-rich stretches characteristic of profilin-interacting proteins. However, it does have one copy of a ZPPX motif at amino acids 1,163–1,166 (Z is a proline, glycine, alanine, or occasionally a charged residue, and X is preferentially a hydrophobic residue). This motif was proposed by Witke et al. (30) to be a feature in a number of profilin-associated proteins. Furthermore, we were also able to detect an in vivo interaction between profilin and AF-6. AF-6-bound profilin could be detected with a profilin-directed Ab in an immunoprecipitate obtained with a myc Ab from Cos cells transfected with pcDNA-AF-6-myc (data not shown). The fact that profilin plays an important role in actin polymerization raises the question as to whether AF-6, by recruiting profilin, may regulate actin modeling proximal to junctional complexes.
Discussion
AF-6 was isolated by virtue of its propensity to interact with oncogenic Ha-Ras proteins (11, 12). The characterization of the complete cDNA of human and rodent AF-6 homologues and subsequent prediction of the motifs arranged therein revealed two N-terminal RBDs. These motifs, as structural and functional entities, are also present in other Ras-binding proteins such as Raf, RalGDS, and Rin-1 (14) and seem to interact specifically with the activated GTP-bound forms of Ras-GTPases. Because the effector specificity of a given GTPase is dictated predominately by the amino acid composition of its effector loop, we thought it likely that Rap1A, another GTPase, which shares identical amino acids 32–40 (4), might also associate with AF-6.
In this study, we further dissected the AF-6 N-terminal portion by separating the predicted RBDs from each other to investigate their binding potentials for Ras and Rap GTPases. The complete N terminus, harboring both Ras-binding pockets, was able to interact with CA Ras and Rap1 GTPases, with the binding to Rap1A significantly exceeding that to Ha-Ras. This profile was mimicked and even magnified by RBD1 of AF-6 alone, whereas RBD2 was virtually incompetent to bind Ras and Rap1A. These observations stand in agreement with recent in vitro studies published by Linnemann et al. (31). Based on kinetic and thermodynamic observations, the authors concluded that the first 141 amino acids of AF-6 form a complex with CA Rap1A at higher affinity than with CA Ha-Ras. Whereas it has been demonstrated that the full-length AF-6 protein can undergo complex formation with Ha-Ras (27, 29) and with the more recently identified M-Ras (27) in a cell-based context, this complex formation remained to be shown for Rap1A. Herein, we show by coimmunoprecipitation that Rap1A can also tether full-length AF-6 into a physical complex when coexpressed in Cos1 cells. Furthermore, we find a certain portion of Rap1A, when expressed at low levels in epithelial MCF7 cells, to be present at the plasma membrane where it colocalizes with endogenous AF-6 protein. These findings, together with the binding profile described above, is consistent with the concept of a functional interaction between AF-6 and Rap1A.
Localization studies in MDCK and MCF7 cells revealed that AF-6 resides in cell–cell junctional complexes. In particular, we found that, in these cell lines, AF-6 colocalizes with ZO-1 to tight junctions, consistent with previous studies in MDCK cells (17), where a direct interaction between AF-6 and ZO-1 was also reported. Other studies examining mouse small intestinal sections showed a residency in E cadherin-based adherens junctions of the rodent homologue afadin (16, 18). This difference in specific residency between the different systems remains unclear. As mentioned above, AF-6 in rats seems to be present in two isoforms, and the smaller variant, s-afadin, was shown to be expressed specifically in rat brain (16). However, we observed that AF-6 and the larger variant are both present in a number of human tissues and cell lines other than those derived from brain. Thus, it is likely that a larger F actin-associated isoform of AF-6 in human tissues will serve as a linker molecule between the membrane and the actin cytoskeleton. With regards to the shorter isoform, profilin (as discussed below) may mediate a link to the actin cytoskeleton. Given the localization of AF-6 at cell–cell junctions and its multidomain structure, it is tempting to speculate that AF-6 functions as a scaffolding protein involved in the maintenance, establishment, or function of junctional complexes. This idea is in favor with the mouse AF-6 loss-of-function phenotype. Herein, the structural organization of cell–cell junctions, both of adherens and tight junctions, is abolished, and the proper progression of embryogenesis is stalled early in development (32, 33). On a cellular level, the concept of tight junctions originating from adherens junctions during the process of polarization is an interesting one (19) and could explain the disruption of both types of complexes by loss of a single protein.
The fact that AF-6 binds to Ras and even more strongly to Rap1 raises the question of whether AF-6 is a mediator of Ras, Rap1, or both signaling pathways. The primary function attributed to Ras is its role in transformation and differentiation. In contrast to the well established Raf, RalGDS, and PI3-kinase molecules, we could not detect any effect of AF-6 on transformation in a classical foci formation assay (data not shown). However, a contribution of AF-6 in epithelial transformation can not be excluded per se. Oncogenic transformation by Ras is also usually accompanied by a loss of cell–cell adhesion properties and morphological changes (2); hence, we sought to investigate whether oncogenic Ras could perturb AF-6 localization. We observed that the introduction of CA Ha-Ras into MDCK cells causes disruption of intercellular adhesion architecture. Under these circumstances, AF-6 and ZO-1 do not accumulate at the membrane. The precise mechanism by which Ha-Ras elicits this morphological phenotype remains elusive, and thus far, a possible model of Ras targeting AF-6 to interfere with cell–cell adhesion awaits further investigation. A central question in this context is whether activation of Ras dissociates AF-6 directly from junctional complexes or whether such a dissociation may be a secondary effect provoked by events downstream of other signaling pathways. Nonetheless, hints for a role of AF-6 in Ras signaling come from Drosophila, where the AF-6 homologue Canoe has been genetically linked to Ras in the development of the compound eye (34).
As mentioned earlier, recent studies underline fundamental differences between Ras and Rap signaling pathways (4, 6). In Drosophila, Rap1 has been implicated in several aspects of morphogenesis (9). Its role in mammalian cells, on the other hand, remains unclear. Rap1A does exhibit some effect on adhesion, although, thus far, this effect has been shown only for cell–extracellular matrix interactions (4, 6). CA Rap1A stably expressed in MDCK cells, in contrast to RasV12, does not give rise to a disruption of cell–cell contacts, and the infected cells maintain their epithelial morphology. ZO-1 and β-catenin, as tight and adherens junctional markers, respectively, remain associated with their membrane-based complexes, as does AF-6. Studies assessing the adhesive properties of cells lacking Rap1 function will be required to establish a clear role for Rap1 in cell–cell adhesion. It is noteworthy, however, that in gastrulating Drosophila embryos mutant for Rap1, the cell morphology is greatly impaired. The cells display aberrant shapes, and cell migration processes are also significantly perturbed (9). Thus, it is an intriguing possibility that AF-6 may mediate some of Rap1's effects on morphogenesis. Whether AF-6 indeed serves as a bona fide effector molecule for Ras and/or Rap GTPases in vivo and which developmental or physiological situation might favor which of the GTPases remain open issues that need more profound experimentation. Moreover, we can also not exclude the possibility that AF-6 as a scaffolding molecule may serve to recruit the GTPases to cell–cell junctions.
To elucidate further the role of AF-6 in junctional structures and to shed more light on the molecular composition of AF-6-containing complexes, we set out to isolate other AF-6-binding proteins. Using two-hybrid techniques, we discovered profilin to be a binding partner of AF-6 and corroborated this finding in vitro by mapping a fragment within the AF-6 protein's C terminus, C-terminal of the PDZ domain, that retained profilin-binding activity. AF-6 differs from other profilin ligands such as VASP (35), MENA (36), diaphanous (37), and the ARP2/3 complex (38, 39) in that it does not interact with profilin via proline-rich stretches. It does, however, contain repeats of a ZPPX motif that was proposed as a feature in a number of profilin-associated proteins by Witke et al. (30) and is present in the overlap of the two profilin-binding GST fusions amino acids 977–1,171 and amino acids 1,083–1,305 at amino acids 1,163–1,166. Profilin is an essential element in the actin-assembling machinery. Its association with monomeric actin and bringing it to the barbed ends of actin chains seems indispensable for polymerization (40). Cell–cell adhesion complexes are linked to a dense filamentous actin network by a number of crosslinking molecules such as α-catenin, α-actinin, and vinculin in the case of adherens junctions and ZO-1 in tight junctions. Thus far, actin polymerization activity has not been ascribed to junctional complexes, and the possibility that AF-6, by sequestering profilin to these multiprotein complexes, might indirectly provide such a function is surely an intriguing one.
Acknowledgments
We thank Drs. K. Kaibuchi, T. Pollard, E. Canaani, and B. Vanhaesebroeck for providing reagents and A. Schmitz and C. D'Souza-Schorey for helpful discussion. This work is supported by National Institutes of Health and U.S. Army Medical Research Grants (to L.V.A.). B.B. is supported by a Leopoldina postdoctoral fellowship, and E.-E.G. is supported by a National Institutes of Health training grant.
Abbreviations
- RBD
Ras-binding domain
- ZO
Zona Occludens
- GST
glutathione S-transferase
- HA
hemagglutinin
- GAD
Gal4-activation domain
- LBD
LexA DNA-binding domain
- CA
constitutively activated
References
- 1.Katz M E, McCormick F. Curr Opin Genet Dev. 1997;7:75–79. doi: 10.1016/s0959-437x(97)80112-8. [DOI] [PubMed] [Google Scholar]
- 2.Kinch M S, Burridge K. Biochem Soc Trans. 1995;23:446–450. doi: 10.1042/bst0230446. [DOI] [PubMed] [Google Scholar]
- 3.Potempa S, Ridley A J. Mol Biol Cell. 1998;9:2185–2200. doi: 10.1091/mbc.9.8.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bos J L. EMBO J. 1998;23:6776–6782. doi: 10.1093/emboj/17.23.6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kitayama H, Sugimoto Y, Matsuzaki T, Ikawa Y, Noda M. Cell. 1989;56:77–84. doi: 10.1016/0092-8674(89)90985-9. [DOI] [PubMed] [Google Scholar]
- 6.Zwartkruis F J, Bos J L. Exp Cell Res. 1999;253:157–165. doi: 10.1006/excr.1999.4695. [DOI] [PubMed] [Google Scholar]
- 7.de Rooij J, Zwartkruis F J, Verheijen M H, Cool R H, Nijman S M, Wittinghofer A, Bos J L. Nature (London) 1998;396:474–477. doi: 10.1038/24884. [DOI] [PubMed] [Google Scholar]
- 8.Kawasaki H, Springett G M, Mochizuki N, Toki S, Nakaya M, Matsuda M, Housman D E, Graybiel A M. Science. 1998;282:2275–2279. doi: 10.1126/science.282.5397.2275. [DOI] [PubMed] [Google Scholar]
- 9.Asha H, de Ruiter N D, Wang M, Hariharan I K. EMBO J. 1999;18:605–615. doi: 10.1093/emboj/18.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chant J, Corrado K, Pringle J R, Herskowitz I. Cell. 1991;65:1213–1224. doi: 10.1016/0092-8674(91)90016-r. [DOI] [PubMed] [Google Scholar]
- 11.Van Aelst L, White M, Wigler M. Cold Spring Harbor Symp Quant Biol. 1994;59:181–186. doi: 10.1101/sqb.1994.059.01.022. [DOI] [PubMed] [Google Scholar]
- 12.Kuriyama M, Harada N, Kuroda S, Yammamoto T, Nakafuku M, Iwamatsu A, Yamamoto D, Prasad R, Croce C, Canaani E, et al. J Biol Chem. 1996;271:607–610. doi: 10.1074/jbc.271.2.607. [DOI] [PubMed] [Google Scholar]
- 13.Prasad R, Gu Y, Alder H, Nakamura T, Canaani O, Saito H, Huebner K, Gale R P, Nowell P C, Kuriyama K, et al. Cancer Res. 1993;53:5624–5628. [PubMed] [Google Scholar]
- 14.Ponting C P, Benjamin D R. Trends Biochem Sci. 1996;21:422–425. doi: 10.1016/s0968-0004(96)30038-8. [DOI] [PubMed] [Google Scholar]
- 15.Ponting C P. Trends Biochem Sci. 1995;20:265–266. doi: 10.1016/s0968-0004(00)89040-4. [DOI] [PubMed] [Google Scholar]
- 16.Mandai K, Nakanishi H, Satoh A, Obaishi H, Wada M, Nishioka H, Itoh M, Mizoguchi A, Aoki T, Fujimoto T, et al. J Cell Biol. 1997;139:517–528. doi: 10.1083/jcb.139.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamamoto T, Harada N, Kano K, Taya S, Canaani E, Matsuura Y, Mizoguchi A, Ide C, Kaibuchi K. J Cell Biol. 1997;139:785–795. doi: 10.1083/jcb.139.3.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mandai K, Nakanishi H, Satoh A, Takahashi K, Satoh K, Nishioka H, Mizoguchi A, Takai Y. J Cell Biol. 1999;144:1001–1017. doi: 10.1083/jcb.144.5.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitic L L, Anderson J M. Annu Rev Physiol. 1998;60:121–142. doi: 10.1146/annurev.physiol.60.1.121. [DOI] [PubMed] [Google Scholar]
- 20.Takahashi K, Matsuo T, Katsube T, Ueda R, Yamamoto D. Mech Dev. 1998;78:97–111. doi: 10.1016/s0925-4773(98)00151-8. [DOI] [PubMed] [Google Scholar]
- 21.Miyamoto H, Nihonmatsu I, Kondo S, Ueda R, Togashi S, Hirata K, Kkegami Y, Yamamoto D. Genes Dev. 1995;9:612–625. doi: 10.1101/gad.9.5.612. [DOI] [PubMed] [Google Scholar]
- 22.Van Aelst L. Methods Mol Biol. 1997;84:201–222. doi: 10.1385/0-89603-488-7:201. [DOI] [PubMed] [Google Scholar]
- 23.White M A, Nicolette C, Minden A, Polverino A, Van Aelst L, Karin M, Wigler M H. Cell. 1995;80:533–541. doi: 10.1016/0092-8674(95)90507-3. [DOI] [PubMed] [Google Scholar]
- 24.Van Aelst L, Barr M, Marcus S, Polverino A, Wigler M. Proc Natl Acad Sci USA. 1993;90:6213–6217. doi: 10.1073/pnas.90.13.6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morgenstein J P, Land H. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maestro R, Dei Tos A P, Hamamori Y, Krasnokutsky S, Sarto V, Kedes L, Doglioni C, Beach D H, Hannon G J. Genes Dev. 1999;13:2207–2217. doi: 10.1101/gad.13.17.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quilliam L A, Castro A F, Rogers-Graham K S, Martin C B, Der C J, Bi C. J Biol Chem. 1999;274:23850–23857. doi: 10.1074/jbc.274.34.23850. [DOI] [PubMed] [Google Scholar]
- 28.D'Souza-Schorey C, Boshans R, McDonough M, Stahl P, Van Aelst L. EMBO J. 1997;16:5445–5454. doi: 10.1093/emboj/16.17.5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamamoto T, Harada N, Kawano Y, Taya S, Kaibuchi K. Biochem Biophys Res Commun. 1999;259:103–107. doi: 10.1006/bbrc.1999.0731. [DOI] [PubMed] [Google Scholar]
- 30.Witke W, Podtelejnikov A V, Di Nardo A, Sutherland J D, Gurniak C B, Dotti C, Mann M. EMBO J. 1998;17:967–976. doi: 10.1093/emboj/17.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Linnemann T, Geyer M, Jaitner B K, Block C, Kalbitzer H R, Wittinghofer A, Herrmann C. J Biol Chem. 1999;274:13556–13562. doi: 10.1074/jbc.274.19.13556. [DOI] [PubMed] [Google Scholar]
- 32.Zhadanov A B, Provance D W J, Speer C A, Coffin J D, Goss D, Blixt J A, Reichert C M, Mercer J A. Curr Biol. 1999;9:880–888. doi: 10.1016/s0960-9822(99)80392-3. [DOI] [PubMed] [Google Scholar]
- 33.Ikeda W, Nakanishi H, Miyoshi J, Mandai K, Ishizaki H, Tanaka M, Togawa A, Takahashi K, Nishioka H, Yoshida H, et al. J Cell Biol. 1999;146:1117–1131. doi: 10.1083/jcb.146.5.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsuo T, Takahashi K, Kondo S, Kaibuchi K, Yamamoto D. Development (Cambridge, UK) 1997;124:2671–2680. doi: 10.1242/dev.124.14.2671. [DOI] [PubMed] [Google Scholar]
- 35.Reinhard M, Giehl K, Abel K, Haffner C, Jarchau T, Hoppe V, Jockusch B M, Walter U. EMBO J. 1995;14:1583–1589. doi: 10.1002/j.1460-2075.1995.tb07146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gertler F B, Niebuhr K, Reinhard M, Wehland J, Soriano P. Cell. 1996;18:227–239. doi: 10.1016/s0092-8674(00)81341-0. [DOI] [PubMed] [Google Scholar]
- 37.Watanabe N, Madaule P, Reid T, Ishizaki T, Wantanabe G, Kakizuka A, Saito Y, Nakao K, Jockusch B M, Narumiya S. EMBO J. 1997;16:3044–3056. doi: 10.1093/emboj/16.11.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Machesky L M, Gould K L. Curr Opin Cell Biol. 1999;11:117–121. doi: 10.1016/s0955-0674(99)80014-3. [DOI] [PubMed] [Google Scholar]
- 39.Mullins R D, Kelleher J F, Xu J, Pollard T D. Mol Biol Cell. 1998;9:841–852. doi: 10.1091/mbc.9.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sohn R H, Goldschmidt-Clermont P J. BioEssays. 1994;16:465–472. doi: 10.1002/bies.950160705. [DOI] [PubMed] [Google Scholar]
- 41.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]