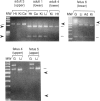
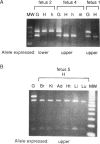
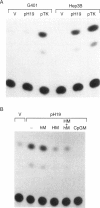
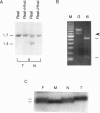
Abstract
Genomic imprinting and monoallelic gene expression appear to play a role in human genetic disease and tumorigenesis. The human H19 gene, at chromosome 11p15, has previously been shown to be monoallelically expressed. Since CpG methylation has been implicated in imprinting, we analyzed methylation of H19 DNA. In fetal and adult organs the transcriptionally silent H19 allele was extensively hypermethylated through the entire gene and its promoter, and, consistent with a functional role for DNA methylation, expression of an H19 promoter-reporter construct was inhibited by in vitro methylation. Gynogenetic ovarian teratomas were found to contain only hypomethylated H19 DNA, suggesting that the expressed H19 allele might be maternal. This was confirmed by analysis of 11p15 polymorphisms in a patient with Wilms tumor. The tumor had lost the maternal 11p15, and H19 expression in the normal kidney was exclusively from this allele. Imprinting of human H19 appears to be susceptible to tissue-specific modulation in somatic development; in one individual, cerebellar cells were found to express only the otherwise silent allele. Implications of these findings for the role of DNA methylation in imprinting and for H19 as a candidate imprinted tumor-suppressor gene are discussed.
Full text
PDF


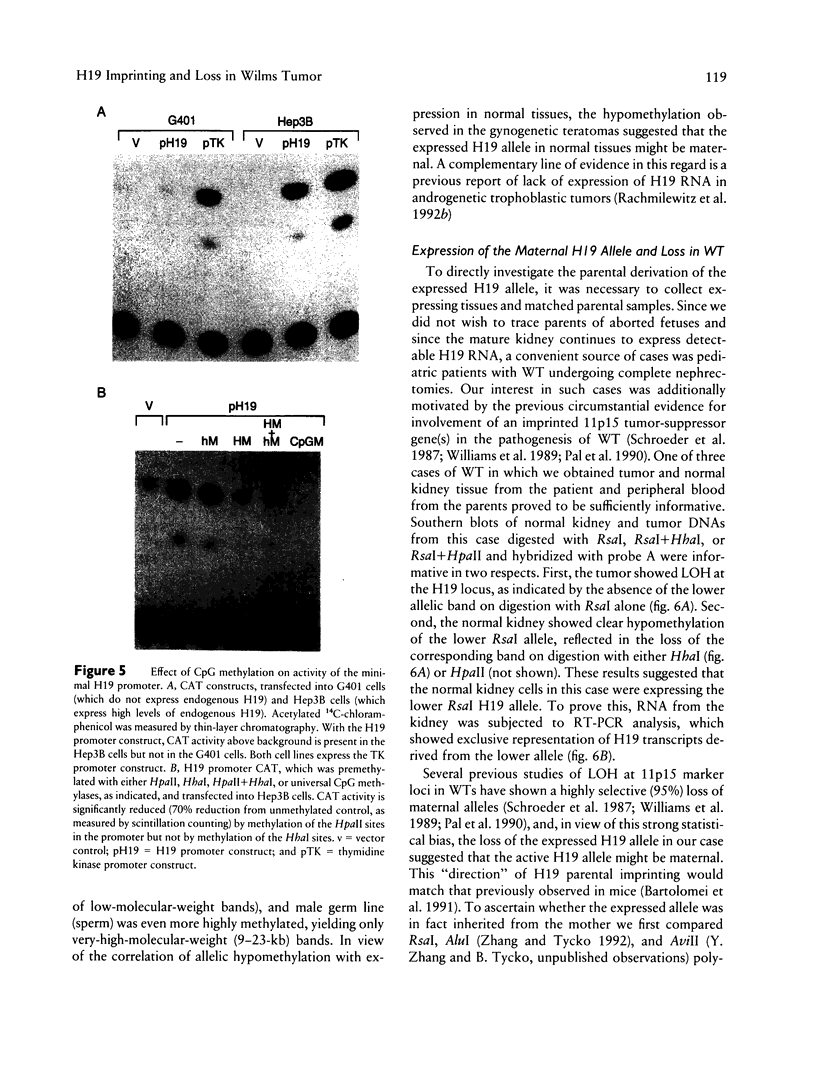
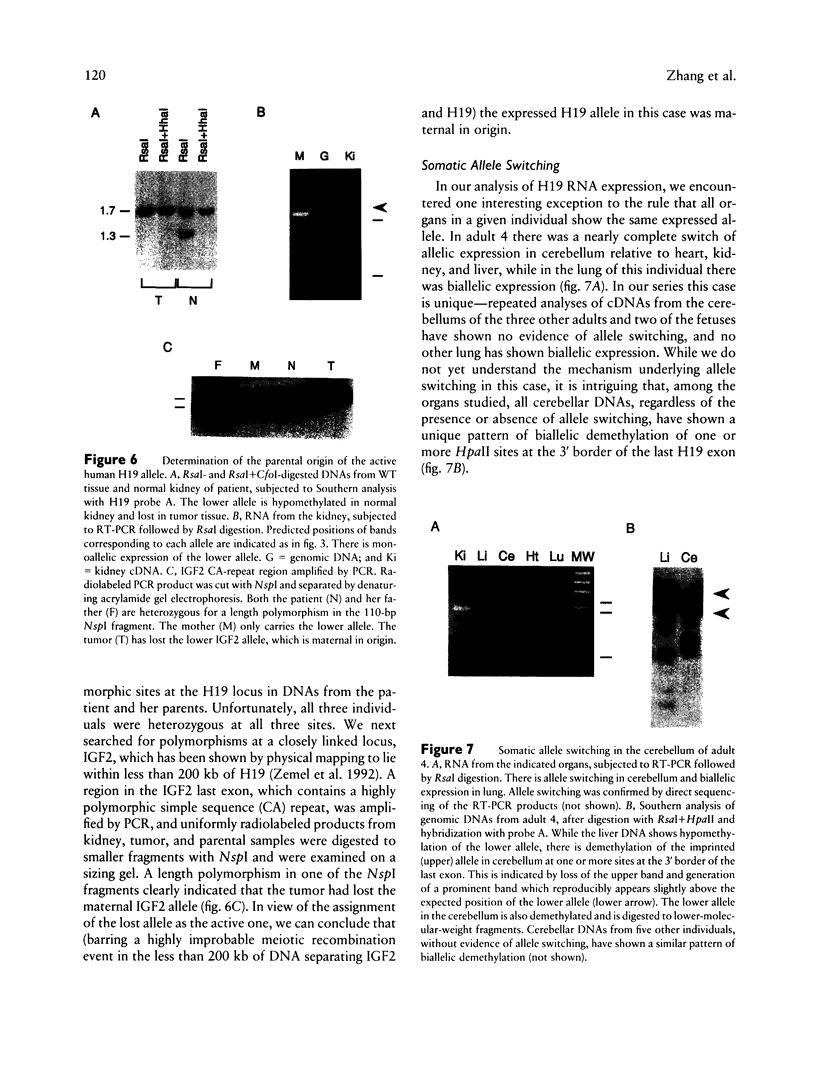




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barlow D. P., Stöger R., Herrmann B. G., Saito K., Schweifer N. The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus. Nature. 1991 Jan 3;349(6304):84–87. doi: 10.1038/349084a0. [DOI] [PubMed] [Google Scholar]
- Bartolomei M. S., Zemel S., Tilghman S. M. Parental imprinting of the mouse H19 gene. Nature. 1991 May 9;351(6322):153–155. doi: 10.1038/351153a0. [DOI] [PubMed] [Google Scholar]
- Bell M. V., Hirst M. C., Nakahori Y., MacKinnon R. N., Roche A., Flint T. J., Jacobs P. A., Tommerup N., Tranebjaerg L., Froster-Iskenius U. Physical mapping across the fragile X: hypermethylation and clinical expression of the fragile X syndrome. Cell. 1991 Feb 22;64(4):861–866. doi: 10.1016/0092-8674(91)90514-y. [DOI] [PubMed] [Google Scholar]
- Bird A. The essentials of DNA methylation. Cell. 1992 Jul 10;70(1):5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- Boyes J., Bird A. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell. 1991 Mar 22;64(6):1123–1134. doi: 10.1016/0092-8674(91)90267-3. [DOI] [PubMed] [Google Scholar]
- Brannan C. I., Dees E. C., Ingram R. S., Tilghman S. M. The product of the H19 gene may function as an RNA. Mol Cell Biol. 1990 Jan;10(1):28–36. doi: 10.1128/mcb.10.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunkow M. E., Tilghman S. M. Ectopic expression of the H19 gene in mice causes prenatal lethality. Genes Dev. 1991 Jun;5(6):1092–1101. doi: 10.1101/gad.5.6.1092. [DOI] [PubMed] [Google Scholar]
- Cattanach B. M., Kirk M. Differential activity of maternally and paternally derived chromosome regions in mice. Nature. 1985 Jun 6;315(6019):496–498. doi: 10.1038/315496a0. [DOI] [PubMed] [Google Scholar]
- Cedar H. DNA methylation and gene activity. Cell. 1988 Apr 8;53(1):3–4. doi: 10.1016/0092-8674(88)90479-5. [DOI] [PubMed] [Google Scholar]
- Davis R. L., Weintraub H., Lassar A. B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987 Dec 24;51(6):987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- DeChiara T. M., Robertson E. J., Efstratiadis A. Parental imprinting of the mouse insulin-like growth factor II gene. Cell. 1991 Feb 22;64(4):849–859. doi: 10.1016/0092-8674(91)90513-x. [DOI] [PubMed] [Google Scholar]
- Dynan W. S. Understanding the molecular mechanism by which methylation influences gene expression. Trends Genet. 1989 Feb;5(2):35–36. doi: 10.1016/0168-9525(89)90016-4. [DOI] [PubMed] [Google Scholar]
- Forejt J., Gregorová S. Genetic analysis of genomic imprinting: an Imprintor-1 gene controls inactivation of the paternal copy of the mouse Tme locus. Cell. 1992 Aug 7;70(3):443–450. doi: 10.1016/0092-8674(92)90168-c. [DOI] [PubMed] [Google Scholar]
- Frank D., Keshet I., Shani M., Levine A., Razin A., Cedar H. Demethylation of CpG islands in embryonic cells. Nature. 1991 May 16;351(6323):239–241. doi: 10.1038/351239a0. [DOI] [PubMed] [Google Scholar]
- Hadchouel M., Farza H., Simon D., Tiollais P., Pourcel C. Maternal inhibition of hepatitis B surface antigen gene expression in transgenic mice correlates with de novo methylation. Nature. 1987 Oct 1;329(6138):454–456. doi: 10.1038/329454a0. [DOI] [PubMed] [Google Scholar]
- Hall J. G. Genomic imprinting: review and relevance to human diseases. Am J Hum Genet. 1990 May;46(5):857–873. [PMC free article] [PubMed] [Google Scholar]
- Han D. K., Liau G. Identification and characterization of developmentally regulated genes in vascular smooth muscle cells. Circ Res. 1992 Sep;71(3):711–719. doi: 10.1161/01.res.71.3.711. [DOI] [PubMed] [Google Scholar]
- Howlett S. K., Reik W. Methylation levels of maternal and paternal genomes during preimplantation development. Development. 1991 Sep;113(1):119–127. doi: 10.1242/dev.113.1.119. [DOI] [PubMed] [Google Scholar]
- Julier C., Hyer R. N., Davies J., Merlin F., Soularue P., Briant L., Cathelineau G., Deschamps I., Rotter J. I., Froguel P. Insulin-IGF2 region on chromosome 11p encodes a gene implicated in HLA-DR4-dependent diabetes susceptibility. Nature. 1991 Nov 14;354(6349):155–159. doi: 10.1038/354155a0. [DOI] [PubMed] [Google Scholar]
- Kafri T., Ariel M., Brandeis M., Shemer R., Urven L., McCarrey J., Cedar H., Razin A. Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line. Genes Dev. 1992 May;6(5):705–714. doi: 10.1101/gad.6.5.705. [DOI] [PubMed] [Google Scholar]
- Laird C. D. Proposed mechanism of inheritance and expression of the human fragile-X syndrome of mental retardation. Genetics. 1987 Nov;117(3):587–599. doi: 10.1093/genetics/117.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leff S. E., Brannan C. I., Reed M. L., Ozçelik T., Francke U., Copeland N. G., Jenkins N. A. Maternal imprinting of the mouse Snrpn gene and conserved linkage homology with the human Prader-Willi syndrome region. Nat Genet. 1992 Dec;2(4):259–264. doi: 10.1038/ng1292-259. [DOI] [PubMed] [Google Scholar]
- Leibovitch M. P., Nguyen V. C., Gross M. S., Solhonne B., Leibovitch S. A., Bernheim A. The human ASM (adult skeletal muscle) gene: expression and chromosomal assignment to 11p15. Biochem Biophys Res Commun. 1991 Nov 14;180(3):1241–1250. doi: 10.1016/s0006-291x(05)81329-4. [DOI] [PubMed] [Google Scholar]
- Linder D., McCaw B. K., Hecht F. Parthenogenic origin of benign ovarian teratomas. N Engl J Med. 1975 Jan 9;292(2):63–66. doi: 10.1056/NEJM197501092920202. [DOI] [PubMed] [Google Scholar]
- Luckow B., Schütz G. CAT constructions with multiple unique restriction sites for the functional analysis of eukaryotic promoters and regulatory elements. Nucleic Acids Res. 1987 Jul 10;15(13):5490–5490. doi: 10.1093/nar/15.13.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath J., Solter D. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell. 1984 May;37(1):179–183. doi: 10.1016/0092-8674(84)90313-1. [DOI] [PubMed] [Google Scholar]
- Meehan R. R., Lewis J. D., McKay S., Kleiner E. L., Bird A. P. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell. 1989 Aug 11;58(3):499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- Monk M., Boubelik M., Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development. 1987 Mar;99(3):371–382. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- Oberlé I., Rousseau F., Heitz D., Kretz C., Devys D., Hanauer A., Boué J., Bertheas M. F., Mandel J. L. Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science. 1991 May 24;252(5009):1097–1102. doi: 10.1126/science.252.5009.1097. [DOI] [PubMed] [Google Scholar]
- Pachnis V., Brannan C. I., Tilghman S. M. The structure and expression of a novel gene activated in early mouse embryogenesis. EMBO J. 1988 Mar;7(3):673–681. doi: 10.1002/j.1460-2075.1988.tb02862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal N., Wadey R. B., Buckle B., Yeomans E., Pritchard J., Cowell J. K. Preferential loss of maternal alleles in sporadic Wilms' tumour. Oncogene. 1990 Nov;5(11):1665–1668. [PubMed] [Google Scholar]
- Poirier F., Chan C. T., Timmons P. M., Robertson E. J., Evans M. J., Rigby P. W. The murine H19 gene is activated during embryonic stem cell differentiation in vitro and at the time of implantation in the developing embryo. Development. 1991 Dec;113(4):1105–1114. doi: 10.1242/dev.113.4.1105. [DOI] [PubMed] [Google Scholar]
- Ponce M. R., Micol J. L. PCR amplification of long DNA fragments. Nucleic Acids Res. 1992 Feb 11;20(3):623–623. doi: 10.1093/nar/20.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachmilewitz J., Gileadi O., Eldar-Geva T., Schneider T., de-Groot N., Hochberg A. Transcription of the H19 gene in differentiating cytotrophoblasts from human placenta. Mol Reprod Dev. 1992 Jul;32(3):196–202. doi: 10.1002/mrd.1080320303. [DOI] [PubMed] [Google Scholar]
- Rachmilewitz J., Goshen R., Ariel I., Schneider T., de Groot N., Hochberg A. Parental imprinting of the human H19 gene. FEBS Lett. 1992 Aug 31;309(1):25–28. doi: 10.1016/0014-5793(92)80731-u. [DOI] [PubMed] [Google Scholar]
- Reik W., Collick A., Norris M. L., Barton S. C., Surani M. A. Genomic imprinting determines methylation of parental alleles in transgenic mice. Nature. 1987 Jul 16;328(6127):248–251. doi: 10.1038/328248a0. [DOI] [PubMed] [Google Scholar]
- Riggs A. D., Pfeifer G. P. X-chromosome inactivation and cell memory. Trends Genet. 1992 May;8(5):169–174. doi: 10.1016/0168-9525(92)90219-t. [DOI] [PubMed] [Google Scholar]
- Sanford J. P., Clark H. J., Chapman V. M., Rossant J. Differences in DNA methylation during oogenesis and spermatogenesis and their persistence during early embryogenesis in the mouse. Genes Dev. 1987 Dec;1(10):1039–1046. doi: 10.1101/gad.1.10.1039. [DOI] [PubMed] [Google Scholar]
- Sapienza C., Paquette J., Tran T. H., Peterson A. Epigenetic and genetic factors affect transgene methylation imprinting. Development. 1989 Sep;107(1):165–168. doi: 10.1242/dev.107.1.165. [DOI] [PubMed] [Google Scholar]
- Sapienza C., Peterson A. C., Rossant J., Balling R. Degree of methylation of transgenes is dependent on gamete of origin. Nature. 1987 Jul 16;328(6127):251–254. doi: 10.1038/328251a0. [DOI] [PubMed] [Google Scholar]
- Sasaki H., Jones P. A., Chaillet J. R., Ferguson-Smith A. C., Barton S. C., Reik W., Surani M. A. Parental imprinting: potentially active chromatin of the repressed maternal allele of the mouse insulin-like growth factor II (Igf2) gene. Genes Dev. 1992 Oct;6(10):1843–1856. doi: 10.1101/gad.6.10.1843. [DOI] [PubMed] [Google Scholar]
- Schroeder W. T., Chao L. Y., Dao D. D., Strong L. C., Pathak S., Riccardi V., Lewis W. H., Saunders G. F. Nonrandom loss of maternal chromosome 11 alleles in Wilms tumors. Am J Hum Genet. 1987 May;40(5):413–420. [PMC free article] [PubMed] [Google Scholar]
- Scrable H., Cavenee W., Ghavimi F., Lovell M., Morgan K., Sapienza C. A model for embryonal rhabdomyosarcoma tumorigenesis that involves genome imprinting. Proc Natl Acad Sci U S A. 1989 Oct;86(19):7480–7484. doi: 10.1073/pnas.86.19.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle A. G., Beechey C. V. Complementation studies with mouse translocations. Cytogenet Cell Genet. 1978;20(1-6):282–303. doi: 10.1159/000130859. [DOI] [PubMed] [Google Scholar]
- Surani M. A., Barton S. C., Norris M. L. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature. 1984 Apr 5;308(5959):548–550. doi: 10.1038/308548a0. [DOI] [PubMed] [Google Scholar]
- Sussenbach J. S. The gene structure of the insulin-like growth factor family. Prog Growth Factor Res. 1989;1(1):33–48. doi: 10.1016/0955-2235(89)90040-9. [DOI] [PubMed] [Google Scholar]
- Swain J. L., Stewart T. A., Leder P. Parental legacy determines methylation and expression of an autosomal transgene: a molecular mechanism for parental imprinting. Cell. 1987 Aug 28;50(5):719–727. doi: 10.1016/0092-8674(87)90330-8. [DOI] [PubMed] [Google Scholar]
- Toth M., Lichtenberg U., Doerfler W. Genomic sequencing reveals a 5-methylcytosine-free domain in active promoters and the spreading of preimposed methylation patterns. Proc Natl Acad Sci U S A. 1989 May;86(10):3728–3732. doi: 10.1073/pnas.86.10.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkerk A. J., Pieretti M., Sutcliffe J. S., Fu Y. H., Kuhl D. P., Pizzuti A., Reiner O., Richards S., Victoria M. F., Zhang F. P. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991 May 31;65(5):905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- Wiles M. V. Isolation of differentially expressed human cDNA clones: similarities between mouse and human embryonal carcinoma cell differentiation. Development. 1988 Nov;104(3):403–413. doi: 10.1242/dev.104.3.403. [DOI] [PubMed] [Google Scholar]
- Williams J. C., Brown K. W., Mott M. G., Maitland N. J. Maternal allele loss in Wilms' tumour. Lancet. 1989 Feb 4;1(8632):283–284. doi: 10.1016/s0140-6736(89)91300-7. [DOI] [PubMed] [Google Scholar]
- Yoo-Warren H., Pachnis V., Ingram R. S., Tilghman S. M. Two regulatory domains flank the mouse H19 gene. Mol Cell Biol. 1988 Nov;8(11):4707–4715. doi: 10.1128/mcb.8.11.4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemel S., Bartolomei M. S., Tilghman S. M. Physical linkage of two mammalian imprinted genes, H19 and insulin-like growth factor 2. Nat Genet. 1992 Sep;2(1):61–65. doi: 10.1038/ng0992-61. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Tycko B. Monoallelic expression of the human H19 gene. Nat Genet. 1992 Apr;1(1):40–44. doi: 10.1038/ng0492-40. [DOI] [PubMed] [Google Scholar]