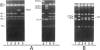Abstract
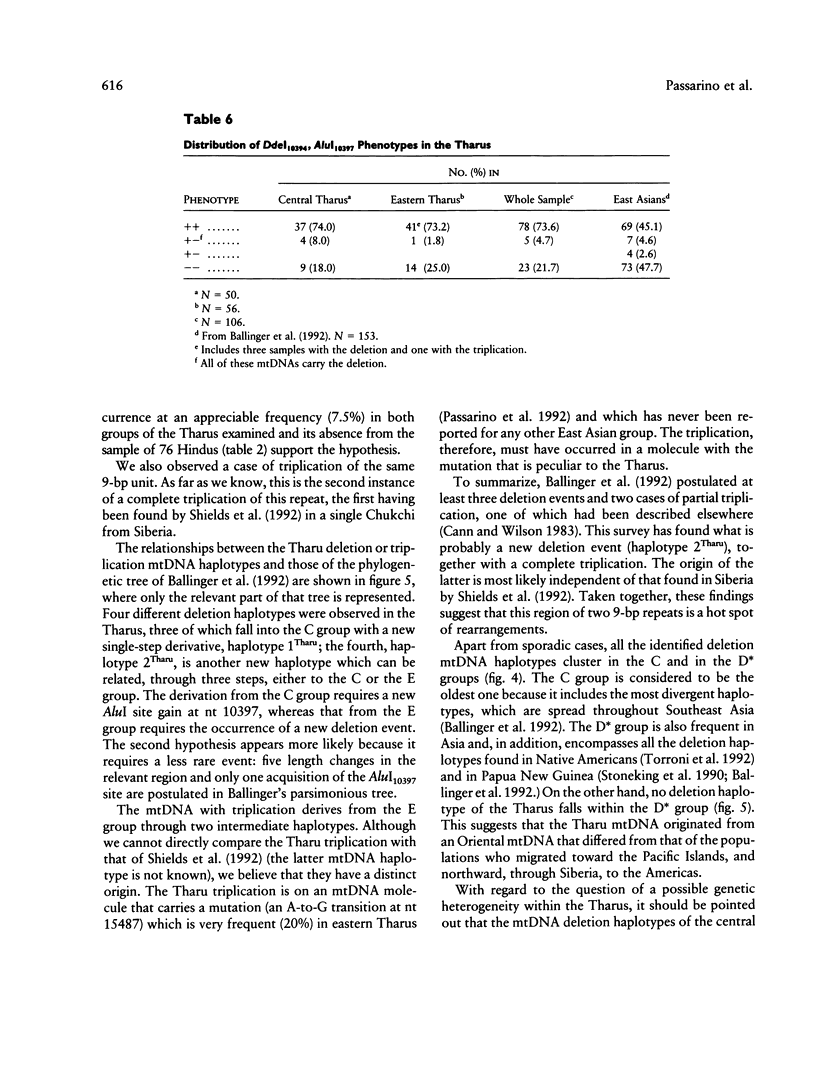
We searched for the East Asian mtDNA 9-bp deletion in the intergenic COII/tRNA(Lys) region in a sample of 107 Tharus (50 from central Terai and 57 from eastern Terai), a population whose anthropological origin has yet to be completely clarified. The deletion, detected by electrophoresis of the PCR-amplified nt 7392-8628 mtDNA fragment after digestion with HaeIII, was found in about 8% of both Tharu groups but was found in none of the 76 Hindus who were examined as a non-Oriental neighboring control population. A complete triplication of the 9-bp unit, the second case so far reported, was also observed in one eastern Tharu. All the mtDNAs with the deletion, and that with the triplication, were further characterized (by PCR amplification of the relevant mtDNA fragments and their digestion with the appropriate enzymes) to locate them in the Ballinger et al. phylogeny of East Asian mtDNA haplotypes. The deletion was found to be associated with four different haplotypes, two of which are reported for the first time. One of the deletions and especially the triplication could be best explained by the assumption of novel length-change events. Ballinger's classification of East Asian mtDNA haplotypes is mainly based on the phenotypes for the DdeI site at nt 10394 and the AluI site at nt 10397. Analysis of the entire Tharu sample revealed that more than 70% of the Tharus have both sites, the association of which has been suggested as an ancient East Asian peculiarity. These results conclusively indicate that the Tharus have a predominantly maternal Oriental ancestry. Moreover, they show at least one and perhaps two further distinct length mutations, and this suggests that the examined region is a hot spot of rearrangements.
Full text
PDF



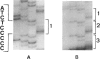
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F. Sequence and organization of the human mitochondrial genome. Nature. 1981 Apr 9;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
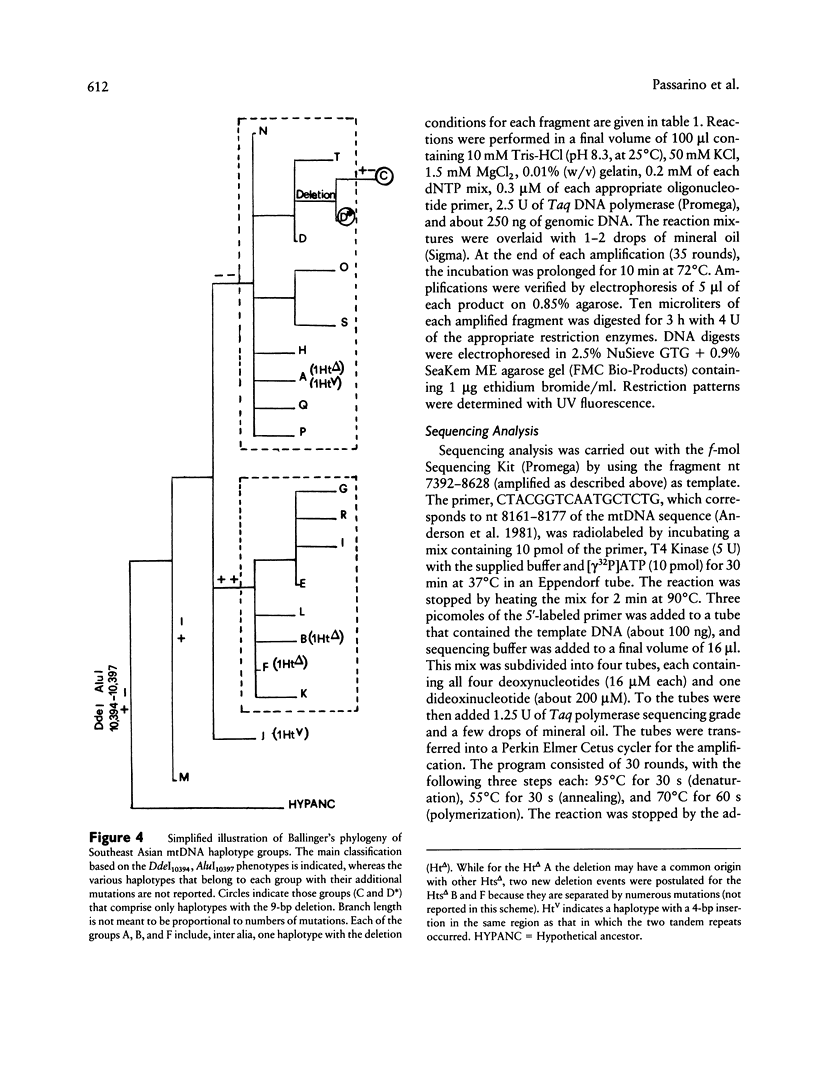
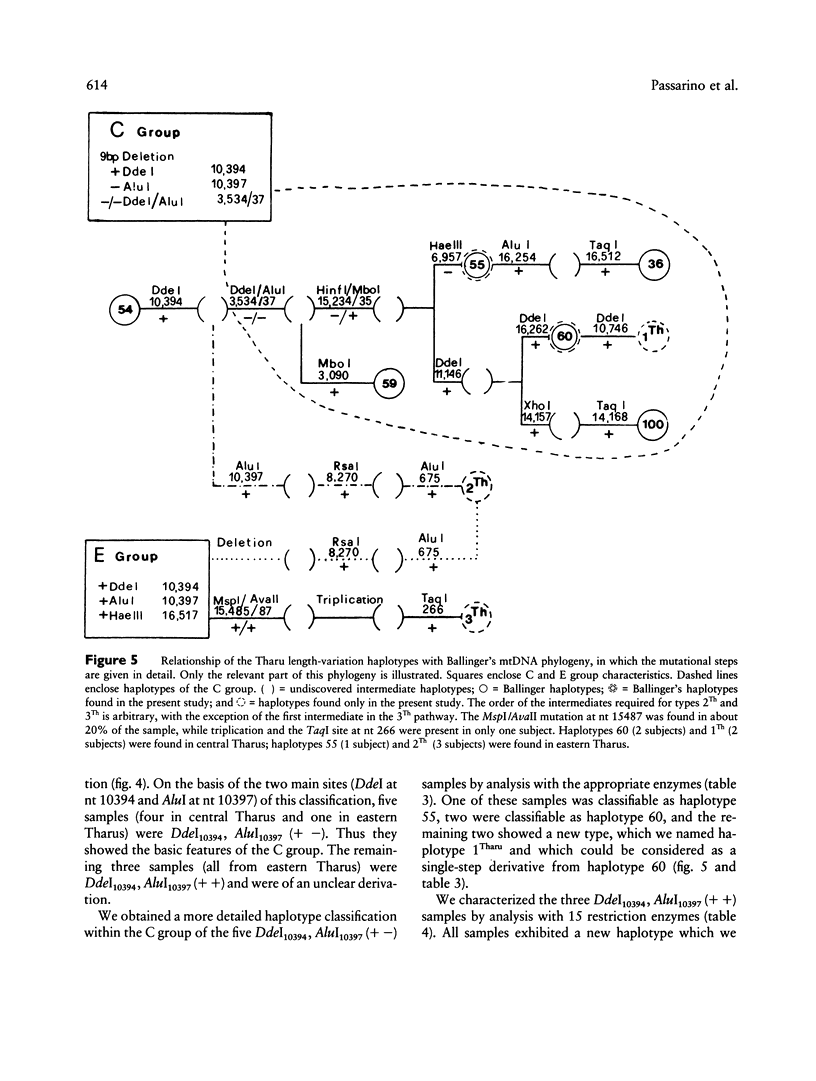
- Ballinger S. W., Schurr T. G., Torroni A., Gan Y. Y., Hodge J. A., Hassan K., Chen K. H., Wallace D. C. Southeast Asian mitochondrial DNA analysis reveals genetic continuity of ancient mongoloid migrations. Genetics. 1992 Jan;130(1):139–152. doi: 10.1093/genetics/130.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc H., Chen K. H., D'Amore M. A., Wallace D. C. Amino acid change associated with the major polymorphic Hinc II site of Oriental and Caucasian mitochondrial DNAs. Am J Hum Genet. 1983 Mar;35(2):167–176. [PMC free article] [PubMed] [Google Scholar]
- Brega A., Gardella R., Semino O., Morpurgo G., Astaldi Ricotti G. B., Wallace D. C., Santachiara Benerecetti A. S. Genetic studies on the Tharu population of Nepal: restriction endonuclease polymorphisms of mitochondrial DNA. Am J Hum Genet. 1986 Oct;39(4):502–512. [PMC free article] [PubMed] [Google Scholar]
- Brown M. D., Voljavec A. S., Lott M. T., Torroni A., Yang C. C., Wallace D. C. Mitochondrial DNA complex I and III mutations associated with Leber's hereditary optic neuropathy. Genetics. 1992 Jan;130(1):163–173. doi: 10.1093/genetics/130.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann R. L., Stoneking M., Wilson A. C. Mitochondrial DNA and human evolution. Nature. 1987 Jan 1;325(6099):31–36. doi: 10.1038/325031a0. [DOI] [PubMed] [Google Scholar]
- Cann R. L., Wilson A. C. Length mutations in human mitochondrial DNA. Genetics. 1983 Aug;104(4):699–711. doi: 10.1093/genetics/104.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra V. P. Studies on serum groups in the Kumaon region, India. Humangenetik. 1970 Aug 17;10(1):35–43. doi: 10.1007/BF00297638. [DOI] [PubMed] [Google Scholar]
- Denaro M., Blanc H., Johnson M. J., Chen K. H., Wilmsen E., Cavalli-Sforza L. L., Wallace D. C. Ethnic variation in Hpa 1 endonuclease cleavage patterns of human mitochondrial DNA. Proc Natl Acad Sci U S A. 1981 Sep;78(9):5768–5772. doi: 10.1073/pnas.78.9.5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
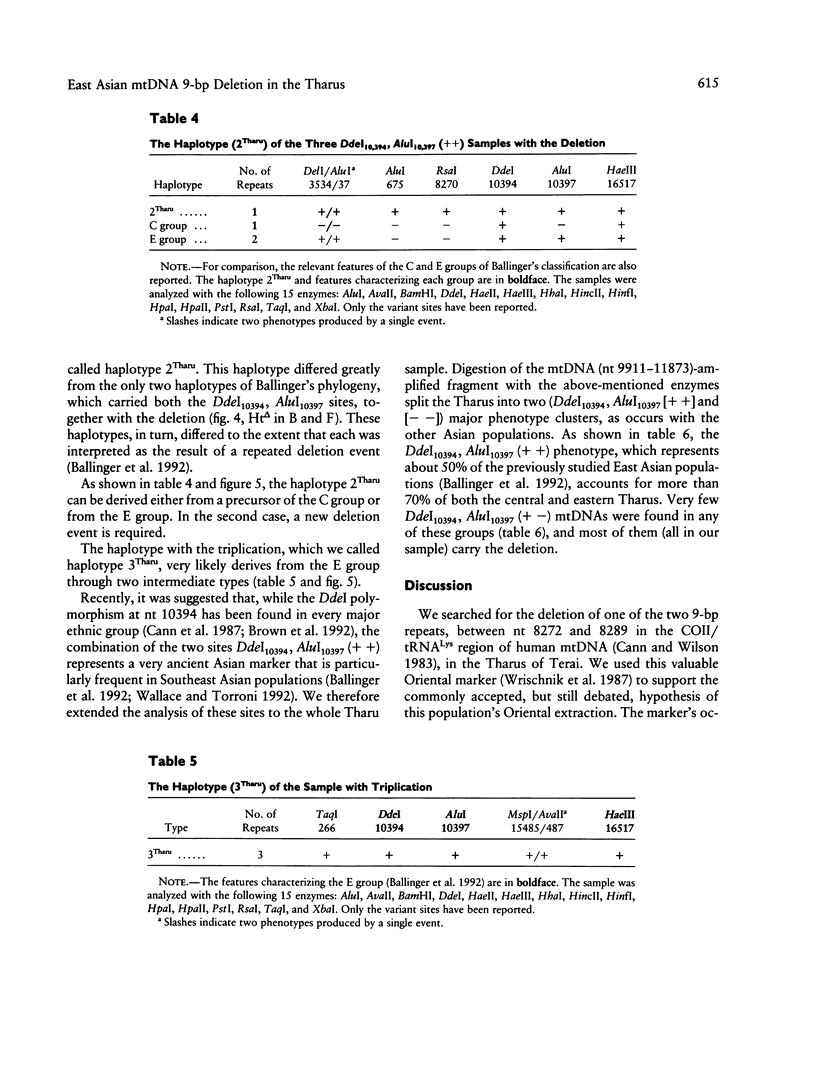
- Harihara S., Hirai M., Suutou Y., Shimizu K., Omoto K. Frequency of a 9-bp deletion in the mitochondrial DNA among Asian populations. Hum Biol. 1992 Apr;64(2):161–166. [PubMed] [Google Scholar]
- Hertzberg M., Mickleson K. N., Serjeantson S. W., Prior J. F., Trent R. J. An Asian-specific 9-bp deletion of mitochondrial DNA is frequently found in Polynesians. Am J Hum Genet. 1989 Apr;44(4):504–510. [PMC free article] [PubMed] [Google Scholar]
- Horai S., Matsunaga E. Mitochondrial DNA polymorphism in Japanese. II. Analysis with restriction enzymes of four or five base pair recognition. Hum Genet. 1986 Feb;72(2):105–117. doi: 10.1007/BF00283927. [DOI] [PubMed] [Google Scholar]
- Modiano G., Morpurgo G., Terrenato L., Novelletto A., Di Rienzo A., Colombo B., Purpura M., Mariani M., Santachiara-Benerecetti S., Brega A. Protection against malaria morbidity: near-fixation of the alpha-thalassemia gene in a Nepalese population. Am J Hum Genet. 1991 Feb;48(2):390–397. [PMC free article] [PubMed] [Google Scholar]
- Passarino G., Semino O., Pepe G., Shrestha S. L., Modiano G., Santachiara Benerecetti A. S. MtDNA polymorphisms among Tharus of eastern Terai (Nepal). Gene Geogr. 1992 Dec;6(3):139–147. [PubMed] [Google Scholar]
- Shields G. F., Hecker K., Voevoda M. I., Reed J. K. Absence of the Asian-specific region V mitochondrial marker in Native Beringians. Am J Hum Genet. 1992 Apr;50(4):758–765. [PMC free article] [PubMed] [Google Scholar]
- Stoneking M., Jorde L. B., Bhatia K., Wilson A. C. Geographic variation in human mitochondrial DNA from Papua New Guinea. Genetics. 1990 Mar;124(3):717–733. doi: 10.1093/genetics/124.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrenato L., Shrestha S., Dixit K. A., Luzzatto L., Modiano G., Morpurgo G., Arese P. Decreased malaria morbidity in the Tharu people compared to sympatric populations in Nepal. Ann Trop Med Parasitol. 1988 Feb;82(1):1–11. doi: 10.1080/00034983.1988.11812202. [DOI] [PubMed] [Google Scholar]
- Torroni A., Schurr T. G., Yang C. C., Szathmary E. J., Williams R. C., Schanfield M. S., Troup G. A., Knowler W. C., Lawrence D. N., Weiss K. M. Native American mitochondrial DNA analysis indicates that the Amerind and the Nadene populations were founded by two independent migrations. Genetics. 1992 Jan;130(1):153–162. doi: 10.1093/genetics/130.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace D. C., Torroni A. American Indian prehistory as written in the mitochondrial DNA: a review. Hum Biol. 1992 Jun;64(3):403–416. [PubMed] [Google Scholar]
- Wrischnik L. A., Higuchi R. G., Stoneking M., Erlich H. A., Arnheim N., Wilson A. C. Length mutations in human mitochondrial DNA: direct sequencing of enzymatically amplified DNA. Nucleic Acids Res. 1987 Jan 26;15(2):529–542. doi: 10.1093/nar/15.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]