Abstract
The ability to sense and respond to changes in oxygen availability is critical for many developmental, physiological, and pathological processes, including angiogenesis, control of blood pressure, and cerebral and myocardial ischemia. Hypoxia-inducible factor-1α (HIF-1α) is a basic-helix–loop–helix (bHLH)containing member of the PER–ARNT–SIM (PAS) family of transcription factors that plays a central role in the response to hypoxia. HIF-1α, and its relatives HIF-2α/endothelial PAS domain protein (EPAS) and HIF-3α, are induced in response to hypoxia and serve to coordinately activate the expression of target genes whose products facilitate cell survival under conditions of oxygen deprivation. When cells are exposed to chronic hypoxia, the protective response can fail, resulting in apoptosis. This study shows that transcription of the gene encoding Nip3, a proapoptotic member of the Bcl-2 family of cell death factors, is strongly induced in response to hypoxia. The Nip3 promoter contains a functional HIF-1-responsive element (HRE) and is potently activated by both hypoxia and forced expression of HIF-1α. Exposure of cultured cells to chronic hypoxia results in the accumulation of a protein recognized by antibodies raised against Nip3. This study demonstrates a direct link between HIF-1α and a proapoptotic member of the Bcl-2 family and offers a reasonable physiological function for members of the Bcl-2 subfamily, including Nip3 and its close relative Nix. These observations indicate that Nip3 may play a dedicated role in the pathological progression of hypoxia-mediated apoptosis, as observed after ischemic injury.
The ability to access and adapt to changes in O2 levels is essential for the viability of all living organisms. Because oxygen is the terminal electron acceptor in oxidative phosphorylation, O2 levels are closely monitored in all cells. If the cellular oxygen concentration fails to match the requirements of energy metabolism or other biochemical reactions dependent on O2, a tightly controlled regulatory pathway is used to respond to the hypoxic environment. A well-characterized mediator of the hypoxic response in mammalian cells is the transcription factor hypoxia-inducible factor-1 (HIF-1) and its close relatives HIF-2/endothelial PER–ARNT–SIM (EPAS) and HIF-3. HIF-1 is a heterodimeric transcription factor composed of HIF-1α and the aryl hydrocarbon receptor nuclear translocator (ARNT) (1). Under normoxic conditions, HIF-1α becomes ubiquitinated and is rapidly degraded by the proteasome (2–4). However, under hypoxic conditions, HIF-1α is both stabilized and activated by posttranslational mechanisms that remain poorly understood (for a review see ref. 5). Upon dimerization with ARNT, HIF-1α is able to induce the expression of target genes containing HREs in their respective transcriptional promoters. A large number of hypoxia-inducible target genes have been identified, most of which have been shown to be regulated by HIF-1 or its close relatives HIF-2/EPAS or HIF-3 (5). Many of these target genes promote cell survival by expediting O2 delivery to oxygen-deprived tissues (e.g., erythropoietin), by promoting the formation of new vasculature (e.g., vascular endothelial growth factor), by increasing glucose transport [e.g., glucose transporter-1 (Glut-1)], and by raising the levels of glycolytic enzymes (e.g., lactate dehydrogenase A).
These adaptive responses serve a critical function in physiological and developmental processes such as angiogenesis and erythropoiesis, as well as pathophysiological states such as ischemia. Persistent oxygen deprivation results in apoptotic cell death. Numerous studies have shown that damage to ischemic tissue resulting from stroke and myocardial infarction is a consequence of programmed cell death (reviewed in ref. 6). HIF-1α has been rigorously proved to play a role in hypoxia-mediated apoptosis. Embryonic stem cells in which the gene encoding HIF-1α has been disrupted do not undergo apoptosis in response to hypoxia, as is the case for their parental counterpart (7). Chinese hamster ovary (CHO) cells lacking proper HIF-1α expression are also resistant to hypoxia-inducible apoptosis (7).
Hallmarks of apoptosis include caspase activation and mitochondrial membrane perturbation affected by members of the Bcl-2 family of cell death factors, some of which are protective (e.g., Bcl-2) and others causative (e.g., Bax). The ratio between proapoptotic and antiapoptotic Bcl-2 factors has been shown to modulate the sensitivity of cells to mitochondrial integrity in response to apoptotic signals (reviewed in ref. 8). This study demonstrates that both the mRNA encoding the proapoptotic Nip3 protein and the Nip3 protein itself accumulate dramatically in response to hypoxia in many different cell lines. Nip3, and its close relative Nix, are the only members of the Bcl-2 family induced at the transcriptional level by hypoxia in CHO-K1 cells. The Nip3 promoter is responsive to both hypoxia and overexpression of HIF-1α via an HRE located upstream from the transcriptional start site. We propose that hypoxia-induced HIF-1 activates expression of the gene encoding Nip3, which in turn primes cells for apoptosis under conditions of persistent oxygen deprivation. This pathway may play an important role in cell death resulting from cerebral and myocardial ischemia.
Experimental Procedures
Cell Culture.
CV-1, Rat-1, and PAM212 cell lines were maintained in DMEM (HyQ DME; HyClone) containing high glucose and supplemented with 10% FBS (Gemini Biological Products, Woodland, CA) and penicillin/streptomycin (Mediatech, Inc., Herndon, VA; 100 units/ml and 100 μg/ml, respectively). CHO-K1 cells were maintained in HyQ DME/F-12 1:1 medium (HyClone) supplemented with 5% FBS and penicillin/streptomycin. HepG2 and ECV-304 cells were maintained in HyQ DME/F-12 1:1 medium supplemented with 10% FBS and penicillin/streptomycin. All cell lines were grown in the presence of 6.0% CO2 at 37°C. Cells were maintained under hypoxic conditions at 37°C within a modular incubator chamber (Billups-Rothenberg) filled with 5.0% CO2 and either 0.50% or 1.0% O2 (balanced with N2).
Subtractive Hybridization.
Subtractive hybridization was performed with a PCR-Select cDNA Subtraction Kit (CLONTECH) to compare poly(A)+ mRNA populations derived from CHO-K1 cells incubated under normoxic (20% O2) or hypoxic (1% O2) conditions for 16 h. Poly(A)+ mRNA was prepared with an mRNA Purification Kit (Amersham Pharmacia). Individual cDNAs were cloned with the pGEM-T Easy Vector System I (Promega) and used as DNA probes for Northern blot analysis.
Northern Blot Analysis.
Total RNA was prepared from cells with RNA STAT-60 (Tel-Test, Friendswood, TX). Poly(A)+ mRNA was prepared from total RNA with an mRNA Purification Kit (Amersham Pharmacia). RNAs were resolved by electrophoresis in a 1.2% agarose gel in the presence of 1.8% formaldehyde, transferred to nitrocellulose filters, and hybridized with 32P-labeled DNA probes. Gene-specific probes were generated by PCR from CHO-K1 cDNA, using the following oligonucleotide pairs (listed 5′ to 3′): Nip3, GAACATGTCGCAGAGCGGGGA and CCCTCGAGTTACTTCAGAAAGTCTGCTGAG; Glut-1, TGAATTCGAGGAGCTCTTCCACCCTCT and GCGTCGACATATACTATAACTTAGTGTCT; Nix, GGCCTGTGAAGTGGTGTATTGT and ATTATAATCTGCTGGTGGCATT; Bcl-2, CCAAGAATGCAAAGCACATCC and CCCAGCCTCCGTTATCCTG; MCL, CAGCGCAACCACGAGAC and GCAAAAGCCAGCAGCACATT; Bid, AGCCAGATTCTGAAAGTCA and AAGACATCACGGAGCAA; and 28S rRNA, GATCCTTCGATGTCGGCTC and ACTAACCTGTCTCACGACG. A Bcl-W probe was generated from mouse cDNA by using the oligonucleotide pair CAGGCTCAGCCCAGCAA and CTGTCCTCACTGATGCCCA.
A gene-specific probe for actin was obtained by digesting the pTRI-β-actin-mouse antisense control template (Ambion, Austin, TX) with EcoRI and XbaI. Probes for Bcl-XL, Bim, Bax, Bak, and Bik were prepared from human cDNAs (a gift from the laboratory of Xiaodong Wang, Department of Biochemistry, University of Texas Southwestern Medical Center).
Western Blot Analysis.
DNA encoding the first 155 amino acids of the hamster Nip3 polypeptide (replacing the carboxyl-terminal transmembrane domain with a His6 peptide) was cloned into the maltose-binding protein parallel vector (9). Nip3 protein was expressed in Escherichia coli as a fusion with maltose-binding protein in the presence pOFXT7-SL-1, a plasmid that expresses the chaperone system encoded by the groES and groEL genes (10). The Nip3 protein fusion was purified with Ni-NTA agarose (Qiagen, Valencia, CA) and amylose resin (New England Biolabs) and used to immunize rabbits. CHO-K1 cells approaching confluency were maintained in medium supplemented with 0.5% FBS under normoxic or hypoxic (0.5% O2) conditions. Cells were resuspended in sample buffer, and proteins were resolved by SDS/PAGE before Western blot analysis. Immune complexes with the anti-Nip3 polyclonal antibodies were detected by enhanced chemiluminescence, using peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch).
Promoter Isolation.
The 5′ and 3′ termini of the CHO-K1 Nip3 cDNA were obtained by 5′ and 3′ rapid amplification of cDNA ends, with a Marathon cDNA Amplification Kit (CLONTECH). A CHO-K1 λ genomic library (Stratagene) was screened with a 32P-labeled DNA specific for the 5′ terminus of the Nip3 transcript (generated by PCR, using the primers 5′-TTCGGGCTCCTTTGCCG-3′ and 5′-CCATGTCACCATTATAAATAG-3′). A 769-bp SacII clone containing 588 bp of sequence immediately upstream of the Nip3 translation start codon was isolated. This sequence was amplified by PCR, using an oligonucleotide that introduced a NcoI site at the translation start codon (5′-CACACCATGGTAGCTCGGCAGAAGAGCC-3′). This fragment was cloned in-frame with the translation start codon of the luciferase reporter gene in pGL3-Basic (Promega) to generate the Nip3-Luc reporter construct. The HRE1-Mut was constructed by PCR amplification of Nip3-Luc with the oligonucleotides 5′-CACACCATGGTAGCTCGGCAGAAGAGCC-3′ and 5′-CACCCGCACGCGCCCCGCGTTCCTC-3′. After digestion with NcoI, this fragment was ligated into the Nip3-Luc vector digested with PmlI and NcoI. The HRE2-Mut was constructed by PCR amplification of Nip3-Luc with the oligonucleotides 5′-CACACCATGGTAGCTCGGCAGAAGAGCC-3′ and 5′-CCTCCCAGCCAATGGGCGCCGCCGCCGCCGACTGCGTCCTG-3′. After digestion with PflMI and NcoI, this fragment was ligated into the Nip3-Luc vector digested with PflMI and NcoI. The E1b-Luc reporter was constructed by replacing the SalI/BglII fragment of pGL3-TK with the sequence AGATCTCATTATATACCCTGTCGAC.
Transfections.
Human embryonic kidney 293 cells were plated onto 24-well plates (1 × 105 cells per well) in 0.5 ml of HyQ DME/F-12 1:1 medium supplemented with 10% FBS and penicillin/streptomycin 24 h before transfection. Before DNA addition, the medium was changed to HyQ DME (high glucose) medium supplemented with 10% FBS and penicillin/streptomycin. DNAs were prepared by precipitation with 125 μM CaCl2 in the presence of N,N-bis(2-hydroxyethyl)-2-aminoethanesulfonic acid (BES) buffer (140 mM NaCl/25 mM BES/0.75 mM sodium phosphate, pH 6.95). Fifty microliters of precipitation mixture containing 30 ng of reporter DNA and 30 ng of total vector DNA (pcDNA3; Invitrogen) or 15 ng each of HIF-1α and vector DNA was added to each well. Cells were incubated for 19 h under normoxic conditions, or for 5 h under normoxic conditions followed by 14 h under hypoxic conditions (0.5% O2). Cells were resuspended in 100 μl of lysis buffer [30 mM Tricine, pH 7.8/8 mM Mg(OAc)2/0.2 mM EDTA/1% Triton X-100/100 mM 2-mercaptoethanol], and 10 μl was added to 40 μl of lysis buffer + substrate (1.5 mM ATP/0.5 mM CoA/0.5 mM luciferin). Luciferase activity was measured with a microtiter plate luminometer (Torcon Instruments, Torrance, CA). A full-length cDNA encoding human HIF-1α was previously subcloned into pcDNA3 (11).
Results
Nip3 mRNA Levels Increase in Response to Hypoxia.
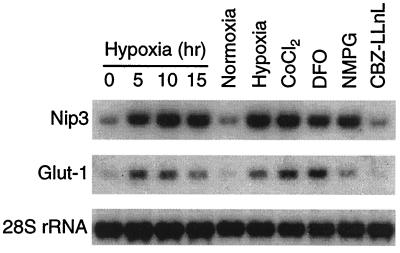
Subtractive hybridization was performed to identify genes induced in the CHO-K1 cell line in response to hypoxia. In addition to several known hypoxia-regulated genes, the subtractive hybridization screen indicated that Nip3 mRNA levels also increase in response to hypoxia. Nip3 is a proapoptotic member of the Bcl-2 family of cell death factors (12–15). As shown in Fig. 1, Nip3 mRNA levels in the CHO-K1 cell line rapidly increase more than 5-fold in response to hypoxia. Among the 16 genes cloned by subtractive hybridization and shown to be responsive to hypoxia (1.0% O2; 15 h) by Northern blot analysis, the Nip3 gene showed the single greatest magnitude of induction (data not shown). Furthermore, the time course of Nip3 induction was observed to parallel that of Glut-1, a well-characterized hypoxia-responsive gene known to depend on the presence of HIF-1α (16). Treatment of cells with “hypoxia mimics” such as divalent cations (CoCl2), iron chelators (deferoxamine mesylate), or reducing agents [5 mM N-(2-mercaptopropionyl)glycine] has been shown to induce HIF-1-responsive genes under normoxic conditions (4, 17). Fig. 1 demonstrates that both Nip3 and Glut-1 mRNAs accumulate substantially in response to these agents. However, treatment of CHO-K1 cells with a proteasome inhibitor (Cbz-Leu-Leu-Leu-norvalinal) did not result in increased levels of either Nip3 or Glut-1 mRNA. Although inhibition of the proteasome is known to stabilize HIF-1α, this form of the transcription factor may not be properly activated by posttranslational regulatory mechanisms such as phosphorylation or redox and does not lead to the induction of HIF-1 target genes (2, 4). These data demonstrate that Nip3 is induced in response to hypoxia in a manner similar to that of previously validated HIF-1 target genes.
Figure 1.
Nip3 mRNA levels increase in CHO-K1 cells in response to hypoxia. CHO-K1 cells were incubated under normoxic (20% O2) or hypoxic (1% O2) conditions or in the presence of 100 μM CoCl2, 100 μM deferoxamine mesylate (DFO), 5 mM N-(2-mercaptopropionyl)glycine (NMPG), or 10 μM Cbz-Leu-Leu-Leu-norvalinal (CBZ-LLnL) under normoxic conditions. Radiolabeled probes specific for Nip3, Glut-1, or 28S rRNA were used to visualize mRNAs from total RNA by Northern blot analysis.
Nip3 Protein Levels Accumulate After Exposure to Prolonged Hypoxia.
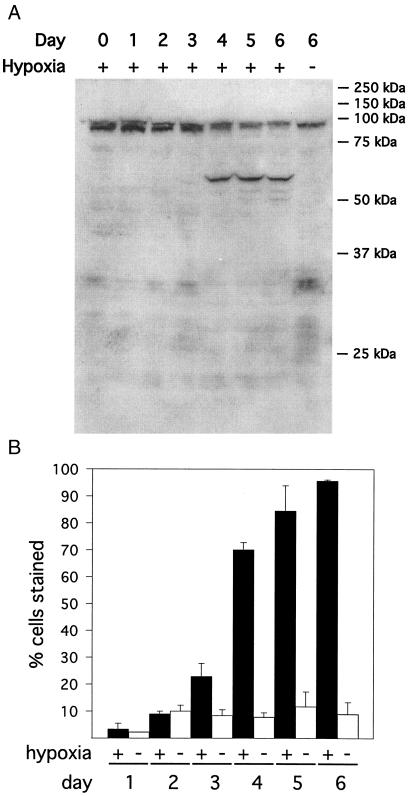
Recombinant Nip3 protein was expressed in E. coli and used to elicit antibodies in rabbits. Rabbit antiserum was used in Western blot assays to visualize Nip3 protein accumulation in CHO-K1 cells after exposure to chronic hypoxia. Because CHO-K1 cells are more resistant to hypoxia than are many cell types, the cells were also incubated in the presence of low serum (7). Hypoxia has previously been shown to increase the number of oligonucleosomes in CHO cells (7), indicating that these cells undergo apoptosis after chronic exposure to hypoxia. As shown in Fig. 2A, a protein product corresponding to an experimental molecular mass of approximately 60 kDa accumulated after several days of exposure to a hypoxic environment. This protein was not observed to accumulate in cells incubated under normoxic conditions. Antiserum from a second rabbit exposed to recombinant Nip3 protein recognized this same 60-kDa protein (data not shown). Although Nip3 protein is calculated to have a molecular mass of 21 kDa, transiently expressed Nip3 protein has been shown to migrate primarily as a 60-kDa species in SDS/PAGE (13, 14). Nip3 protein accumulation in cultured CHO-K1 cells coincides closely with the onset of cell death under physiological conditions, as most cells die between 3 and 6 days of hypoxic exposure (Fig. 2B).
Figure 2.
Nip3 protein levels accumulate after prolonged exposure to hypoxia. (A) Total cell lysates from CHO-K1 cells incubated under normoxic (20% O2) or hypoxic (0.5% O2) conditions for 0–6 days were examined by Western blot analysis, using antiserum raised against recombinant Nip3 protein. The endogenous Nip3 protein has an observed molecular mass of approximately 60 kDa (13, 14). (B) CHO-K1 cells were maintained under normoxic (20% O2) or hypoxic (0.5% O2) conditions for 1–6 days. Cells were harvested and incubated in the presence of 0.2% trypan blue in PBS for 30 min at room temperature. Viable cells exclude trypan blue and are not stained. Values represent the average ± SD of three samples.
Nip3 mRNA Levels Are Induced by Hypoxia in a Variety of Cell Lines.
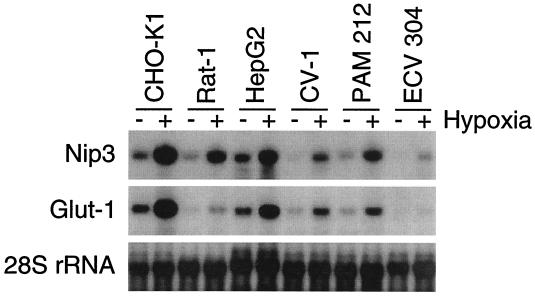
Nip3 has been shown to be expressed in a wide variety of cell/tissue types (13). Because HIF-1 is a ubiquitously expressed transcription factor that serves as a central component of a conserved oxygen-sensing and response pathway in mammalian cells, genes containing HREs within their promoters are often induced in response to hypoxia in different cell lines. In addition to CHO-K1 cells, both Nip3 and Glut-1 mRNAs were observed to accumulate in response to hypoxia in the CV-1 (monkey kidney), Rat-1 (rat fibroblast), PAM212 (human epithelial), HepG2 (human hepatocellular carcinoma), and ECV-304 (human bladder carcinoma) cell lines, as demonstrated by Northern blot analysis (Fig. 3).
Figure 3.
Nip3 mRNA levels are induced in response to hypoxia in several different cell lines. Cells were incubated under normoxic (20% O2) or hypoxic (0.5% O2) conditions for 15 h. Radiolabeled probes specific for Nip3, Glut-1, or 28S rRNA were used to visualize mRNAs from total RNA by Northern analysis.
Nip3 and Nix mRNAs Are the Only Members of the Bcl-2 Family Induced by Hypoxia.
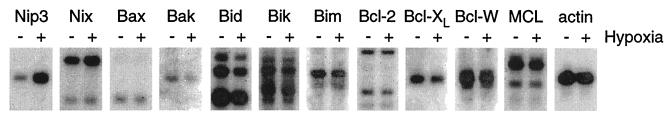
Radiolabeled probes specific for almost all mammalian members of the Bcl-2 family of apoptosis factors were used to evaluate changes in mRNA expression in response to hypoxia by Northern blot analysis. As shown in Fig. 4, Nip3 is induced more than 5-fold in CHO-K1 cells after a 10-h incubation in an atmosphere containing 0.5% O2. The only other member of the Bcl-2 family whose gene was observed to be induced by hypoxia is Nix (2-fold). The proapoptotic Nix polypeptide is the closest relative to Nip3 among all Bcl-2 family members, sharing 56% amino acid identity with Nip3 (18, 19). No other member of the Bcl-2 family was induced by hypoxia as assayed at the level of mRNA accumulation. mRNA levels for Bax, Bik, and Bim remained essentially unchanged after hypoxia, whereas mRNA levels actually decreased slightly for Bak and Bid. No expression was observed for genes encoding the proapoptotic factors Blk, Bok, or Bad in the CHO-K1 cell line (data not shown). In addition, almost no change was observed in mRNA levels for the antiapoptotic members of the Bcl-2 family, including Bcl-2, Bcl-XL, Bcl-W, and MCL (Fig. 4). No expression was detected for A1/Bfl-1 or Boo/Diva in this cell line (data not shown). These data indicate that among known Bcl-2 family members in CHO-K1 cells, only the mRNAs encoding Nip3 and the closely related Nix protein are induced by hypoxia.
Figure 4.
Nip3 and Nix are the only members of the Bcl-2 family of apoptotic factors induced in response to hypoxia. CHO-K1 cells were incubated under normoxic (20% O2) or hypoxic (0.5% O2) conditions for 10 h. Radiolabeled probes specific for the indicated genes were used to visualize mRNAs from poly(A)+ mRNA by Northern analysis.
The Nip3 Promoter Confers Hypoxia Inducibility to a Reporter Gene and Can Be Activated by HIF-1α.
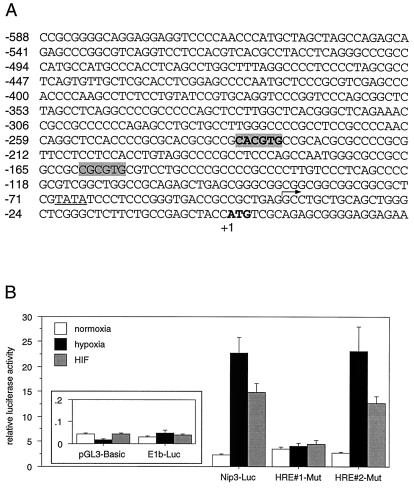
The 5′ and 3′ ends of the CHO-K1 Nip3 cDNA were identified by rapid amplification of cDNA ends, and a full-length cDNA was obtained by PCR amplification. A CHO-K1 genomic library was screened with a DNA probe consisting of the 5′ portion of the Nip3 cDNA to obtain the sequence upstream of the putative transcriptional start site (Fig. 5A). A 588-bp fragment just upstream of the Nip3 translation start codon was found to contain a canonical TATA box located 25 bp upstream of the 5′ end of the cDNA, as well as two possible HREs (HRE1: 5′-CACGTG-3′; HRE2: 5′-CGCGTG-3′) fitting the consensus 5′-RCGTG-3′ (20). This 588-bp segment of the Nip3 gene was cloned upstream from a luciferase reporter gene and used to monitor promoter activity in 293 cells. Reporter constructs analyzed in this study included ones containing no promoter (pGL3-Basic), a minimal promoter containing a TATA box (E1b-Luc), the Nip3 promoter (Nip3-Luc), and the Nip3 promoter containing site-directed mutations in each of the two putative HREs. As shown in Fig. 5B, luciferase expression was induced 10-fold by the Nip3 promoter under hypoxic conditions. Cotransfection of HIF-1α also resulted in a substantial induction of luciferase expression from the Nip3-Luc reporter under normoxic conditions. Mutation of the HRE located 234 bp upstream of the Nip3 translation start codon (HRE1-Mut) led to the complete loss of both hypoxia- and HIF-1α-mediated responsiveness. However, mutation of the consensus HRE located 160 bp upstream of the Nip3 translation start codon (HRE2-Mut) did not affect promoter activity. Neither hypoxia nor HIF-1α transfection led to an induction of luciferase enzyme activity specified by pGL3-Basic or by the minimal promoter within E1b-Luc. These data demonstrate that the DNA sequences located immediately upstream of the Nip3 gene confer transcriptional induction in response to both hypoxia and forced expression of HIF-1α. Furthermore, the ability of the Nip3 promoter to respond to hypoxia- or HIF-1α-mediated transcriptional activation depends on the presence of an HRE consensus sequence located 234 bp upstream of the Nip3 translation start codon.
Figure 5.
The Nip3 promoter is responsive to hypoxia and to HIF-1α. (A) The promoter sequence of Nip3 from CHO-K1 cells contains two consensus HREs (highlighted in gray). HRE1 is shown in boldface. The translation start codon of the Nip3 polypeptide at position +1 is in boldface. The 5′ end of the mRNA as determined by 5′ rapid amplification of cDNA ends is indicated by an arrow and is located 25 nucleotides downstream of a putative TATA box (underlined). HRE1-Mut changed the putative HRE beginning at position −234 from 5′-CACGTG-3′ to 5′-CACCAC-3′. HRE2-Mut changed the putative HRE beginning at position −160 from 5′-CGCGTG-3′ to 5′-CGACTG-3′. (B) Luciferase reporter constructs containing no promoter (pGL3-Basic), a minimal TATA box containing promoter (E1B-Luc), the Nip3 promoter, or the Nip3 promoter containing a mutation in the first (HRE#1-Mut) or second (HRE#2-Mut) putative HRE was transfected into 293 cells along with pcDNA3 (vector) or HIF-1α. Cells transfected with vector were incubated for 19 h in an atmosphere containing 20% O2 (normoxia) or for 5 h in an atmosphere containing 20% O2 followed by 14 h in an atmosphere containing 0.5% O2 (hypoxia). Cells transfected with HIF-1α were incubated for 19 h under normoxic (20% O2) conditions. The values represent the average luciferase activity of six samples; bars indicate standard error.
Discussion
Hypoxia-induced apoptosis plays an important role in the pathology of many diseases, including ischemic damage to the heart and brain. Neuronal cell death after both transient global ischemia and focal ischemia demonstrates features of apoptosis, including DNA laddering and caspase activation 24–72 h after the hypoxic insult (reviewed in ref. 21). Cardiomyocytes similarly undergo apoptosis after ischemia (22–25). HIF-1α activity is known to be induced in the brain as a consequence of ischemia (26) and appears to be required for delayed neuronal cell death (26). The present study shows that transcription of the gene encoding Nip3 is significantly induced in response to hypoxia. Nip3 is likely to be a direct target gene for HIF-1α, based on the presence of a functional HRE located within its hypoxia-responsive promoter. These findings suggest that the levels of Nip3 mRNA and its protein product are substantially elevated in hypoxic cells. No member of the Bcl-2 family of cell death factors other than Nip3 has been shown to be directly regulated at the level of transcription by hypoxia.
Like other members of the Bcl-2 family, Nip3 (and its close relative Nix) contains a BH3 domain and a hydrophobic domain at its carboxyl terminus that likely mediates membrane targeting and/or dimerization (13–15). Nip3 has been shown to form heterodimers with antiapoptotic Bcl-2 family members such as Bcl-2 and Bcl-XL (12–15) and may promote apoptosis by sequestering these factors. Nip3 overexpression during transient transfection has been shown to induce apoptosis. However, the onset of apoptosis induced by Nip3 is generally slower than that observed upon transfection of more potent apoptotic factors such as Bid or Bax (13–15). Nip3 appears to be ubiquitously expressed (13) and is shown in this study to be strongly induced by hypoxia in numerous cell lines.
A critical consequence of cerebral and myocardial ischemia is the prolonged exposure of these tissues to hypoxia. The HIF-1-mediated hypoxic response is reasonably predicted to promote survival of these tissues by means of the promotion of erythropoiesis, neovascularization, and adaptive glycolytic growth (27). Failure to adequately adapt to a persistent hypoxic environment leads to apoptotic cell death. Based on the findings reported in this study, the hypoxic induction of Nip3 is hypothesized to play a direct role in the progression of hypoxia-mediated apoptosis in these disease states. As a likely HIF-1 target gene, Nip3 is induced shortly after the onset of hypoxia. The accumulation of Nip3 protein is consistent with the onset of cell death in cultured CHO-K1 cells resulting from chronic exposure to a hypoxic environment. There is a substantial delay between Nip3 mRNA accumulation and Nip3 protein accumulation in the CHO-K1 cell line. This delay may a be consequence of intrinsic Nip3 instability (13) or may indicate a requirement for additional apoptotic factors/events to promote high levels of Nip3 protein accumulation. It has not yet been determined whether this delay is observed in other cells exposed to hypoxia and whether Nip3 expression is necessary or sufficient for the progression of cell death.
Nip3-induced apoptosis appears to be relatively slow, perhaps because of its modest apoptotic activity relative to the very potent members of the Bcl-2 family such as Bid or Bax (15) or its instability (13). Ischemic tissues also exhibit a delay in the execution of the cell death pathway after hypoxic insult. This delay in apoptosis may provide a critical opportunity for hypoxic tissues to adapt to oxygen deprivation by means of activation of an initial, HIF-1-dependent, protective response. However, sustained O2 deprivation might specify the sufficient accumulation of Nip3 to promote cell death. If so, this model hypothesizes that HIF-1 may play a direct role in the balance between cell survival and eventual cell death during hypoxia. It is not yet known whether Nip3 expression is induced in neurons and cardiomyocytes during ischemic injury, and whether hypoxia-induced expression of Nip3 indeed helps to specify the delayed progression of oxygen-starved cells toward apoptosis. Finally, should this hypothesis prove to be correct, it will be of interest to consider the adaptive benefits of a system capable of cleansing and eliminating hypoxia-damaged tissue.
Acknowledgments
I thank Dr. Steven L. McKnight, in whose laboratory I have worked as a postdoctoral fellow; the members of the McKnight laboratory for advice and support; Carolyn Michnoff for injecting the rabbits; Olivier Fayet for providing the pOFXT7-SL-1 plasmid; and Deepak Nijhawan and Dr. Xiadong Wang from the Department of Biochemistry at the University of Texas Southwestern Medical Center for their helpful advice and provision of reagents. This work was funded by a National Research Service Award from the National Institutes of Health (R.K.B.), grants provided to Steven L. McKnight from the National Institutes of Health (DK52031 and MH59388), and unrestricted endowment funds from an anonymous donor.
Abbreviations
- HIF-1
hypoxia-inducible factor-1
- HRE
HIF-1-responsive element
- Glut-1
glucose transporter-1
- CHO
Chinese hamster ovary
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF283504).
References
- 1.Wang G L, Jiang B H, Rue E A, Semenza G L. Proc Natl Acad Sci USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kallio P J, Wilson W J, O'Brien S, Makino Y, Poellinger L. J Biol Chem. 1999;274:6519–6525. doi: 10.1074/jbc.274.10.6519. [DOI] [PubMed] [Google Scholar]
- 3.Huang L E, Gu J, Schau M, Bunn H F. Proc Natl Acad Sci USA. 1998;95:7987–7992. doi: 10.1073/pnas.95.14.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salceda S, Caro J. J Biol Chem. 1997;272:22642–22647. doi: 10.1074/jbc.272.36.22642. [DOI] [PubMed] [Google Scholar]
- 5.Semenza G L. Annu Rev Cell Dev Biol. 1999;15:551–578. doi: 10.1146/annurev.cellbio.15.1.551. [DOI] [PubMed] [Google Scholar]
- 6.Lipton P. Physiol Rev. 1999;79:1431–1568. doi: 10.1152/physrev.1999.79.4.1431. [DOI] [PubMed] [Google Scholar]
- 7.Carmeliet P, Dor Y, Herbert J M, Fukumura D, Brusselmans K, Dewerchin M, Neeman M, Bono F, Abramovitch R, Maxwell P, et al. Nature (London) 1998;394:485–490. doi: 10.1038/28867. [DOI] [PubMed] [Google Scholar]
- 8.Gross A, McDonnell J M, Korsmeyer S J. Genes Dev. 1999;13:1899–1911. doi: 10.1101/gad.13.15.1899. [DOI] [PubMed] [Google Scholar]
- 9.Sheffield P, Garrard S, Derewenda Z. Protein Expression Purif. 1999;15:34–39. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- 10.Castanié M-P, Bergès H, Oreglia J, Prère M-F, Fayet O. Anal Biochem. 1997;254:150–152. doi: 10.1006/abio.1997.2423. [DOI] [PubMed] [Google Scholar]
- 11.Tian H, McKnight S L, Russell D W. Genes Dev. 1997;11:72–82. doi: 10.1101/gad.11.1.72. [DOI] [PubMed] [Google Scholar]
- 12.Boyd J M, Malstrom S, Subramanian T, Venkatesh L K, Schaeper U, Elangovan B, D'Sa-Eipper C, Chinnadurai G. Cell. 1994;79:341–351. doi: 10.1016/0092-8674(94)90202-x. [DOI] [PubMed] [Google Scholar]
- 13.Chen G, Ray R, Dubik D, Shi L, Cizeau J, Bleackley R C, Saxena S, Gietz R D, Greenberg A H. J Exp Med. 1997;186:1975–1983. doi: 10.1084/jem.186.12.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ray R, Chen G, Vande Velde C, Cizeau J, Park J H, Reed J C, Gietz R D, Greenberg A H. J Biol Chem. 2000;275:1439–1448. doi: 10.1074/jbc.275.2.1439. [DOI] [PubMed] [Google Scholar]
- 15.Yasuda M, Theodorakis P, Subramanian T, Chinnadurai G. J Biol Chem. 1998;273:12415–12421. doi: 10.1074/jbc.273.20.12415. [DOI] [PubMed] [Google Scholar]
- 16.Wood S M, Wiesener M S, Yeates K M, Okada N, Pugh C W, Maxwell P H, Ratcliffe P J. J Biol Chem. 1998;273:8360–8368. doi: 10.1074/jbc.273.14.8360. [DOI] [PubMed] [Google Scholar]
- 17.Wang G L, Semenza G L. Blood. 1993;82:3610–3615. [PubMed] [Google Scholar]
- 18.Chen G, Cizeau J, Vande Velde C, Park J H, Bozek G, Bolton J, Shi L, Dubik D, Greenberg A. J Biol Chem. 1999;274:7–10. doi: 10.1074/jbc.274.1.7. [DOI] [PubMed] [Google Scholar]
- 19.Matsushima M, Fujiwara T, Takahashi E, Minaguchi T, Eguchi Y, Tsujimoto Y, Suzumori K, Nakamura Y. Genes Chromosomes Cancer. 1998;21:230–235. [PubMed] [Google Scholar]
- 20.Semenza G L, Jiang B H, Leung S W, Passantino R, Concordet J P, Maire P, Giallongo A. J Biol Chem. 1996;271:32529–32537. doi: 10.1074/jbc.271.51.32529. [DOI] [PubMed] [Google Scholar]
- 21.Halterman M W, Federoff H J. Exp Neurol. 1999;159:65–72. doi: 10.1006/exnr.1999.7160. [DOI] [PubMed] [Google Scholar]
- 22.Tanaka M, Ito H, Adachi S, Akimoto H, Nishikawa T, Kasajima T, Marumo F, Hiroe M. Circ Res. 1994;75:426–433. doi: 10.1161/01.res.75.3.426. [DOI] [PubMed] [Google Scholar]
- 23.Black S C, Huang J Q, Rezaiefar P, Radinovic S, Eberhart A, Nicholson D W, Rodger I W. J Mol Cell Cardiol. 1998;30:733–742. doi: 10.1006/jmcc.1998.0660. [DOI] [PubMed] [Google Scholar]
- 24.Cheng W, Kajstura J, Nitahara J A, Li B, Reiss K, Liu Y, Clark W A, Krajewski S, Reed J C, Olivetti G, Anversa P. Exp Cell Res. 1996;226:316–327. doi: 10.1006/excr.1996.0232. [DOI] [PubMed] [Google Scholar]
- 25.Narula J, Haider N, Virmani R, DiSalvo T G, Kolodgie F D, Hajjar R J, Schmidt U, Semigran M J, Dec G W, Khaw B A. N Engl J Med. 1996;335:1182–1189. doi: 10.1056/NEJM199610173351603. [DOI] [PubMed] [Google Scholar]
- 26.Halterman M W, Miller C C, Federoff H J. J Neurosci. 1999;19:6818–6824. doi: 10.1523/JNEUROSCI.19-16-06818.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Semenza G L. J Lab Clin Med. 1998;131:207–214. doi: 10.1016/s0022-2143(98)90091-9. [DOI] [PubMed] [Google Scholar]