Abstract
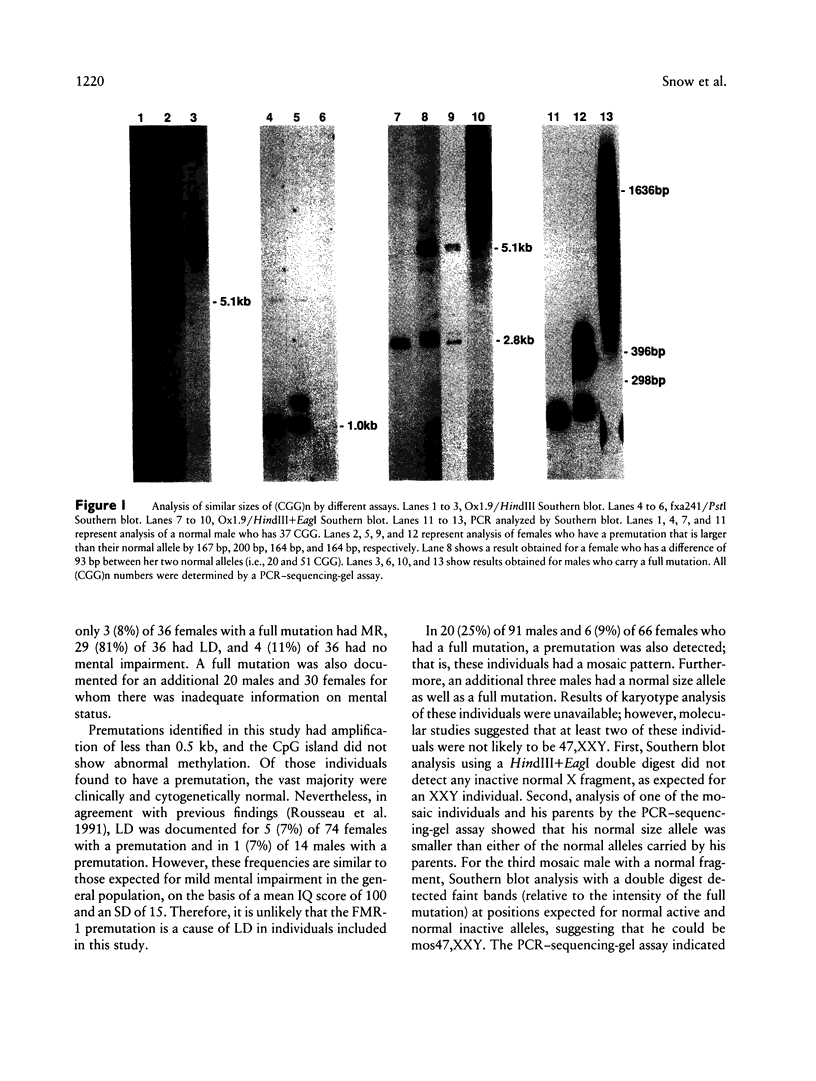
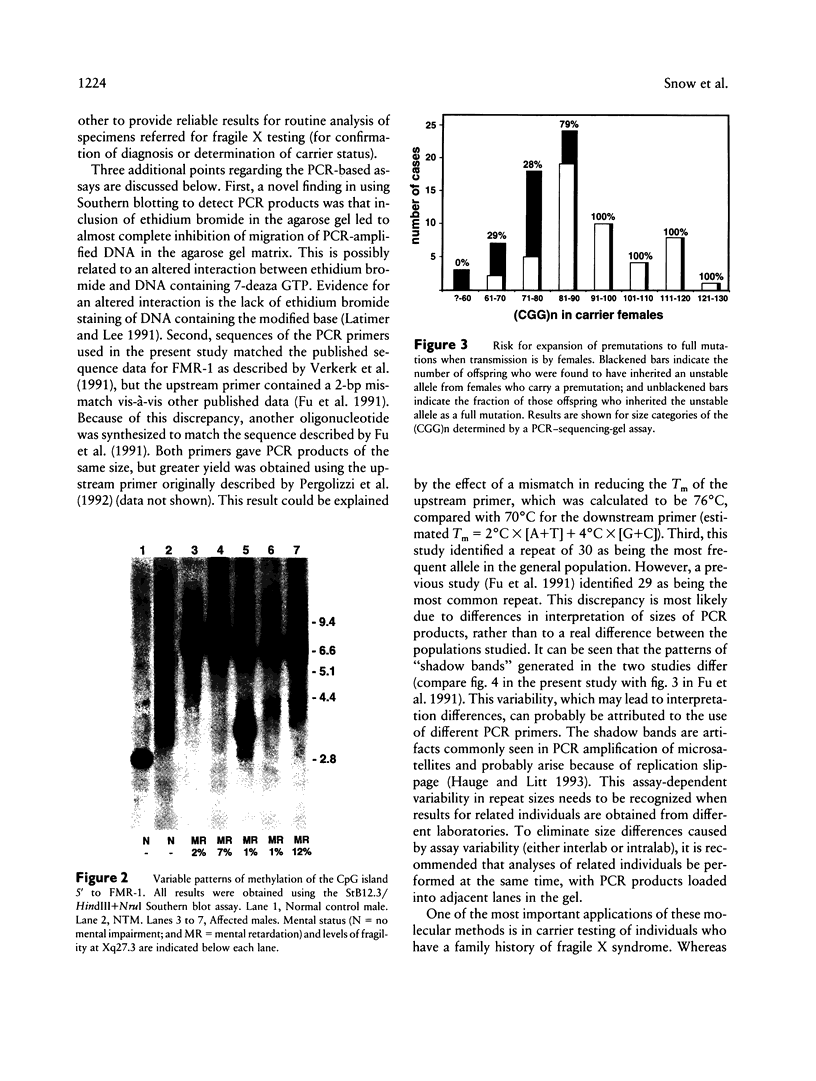
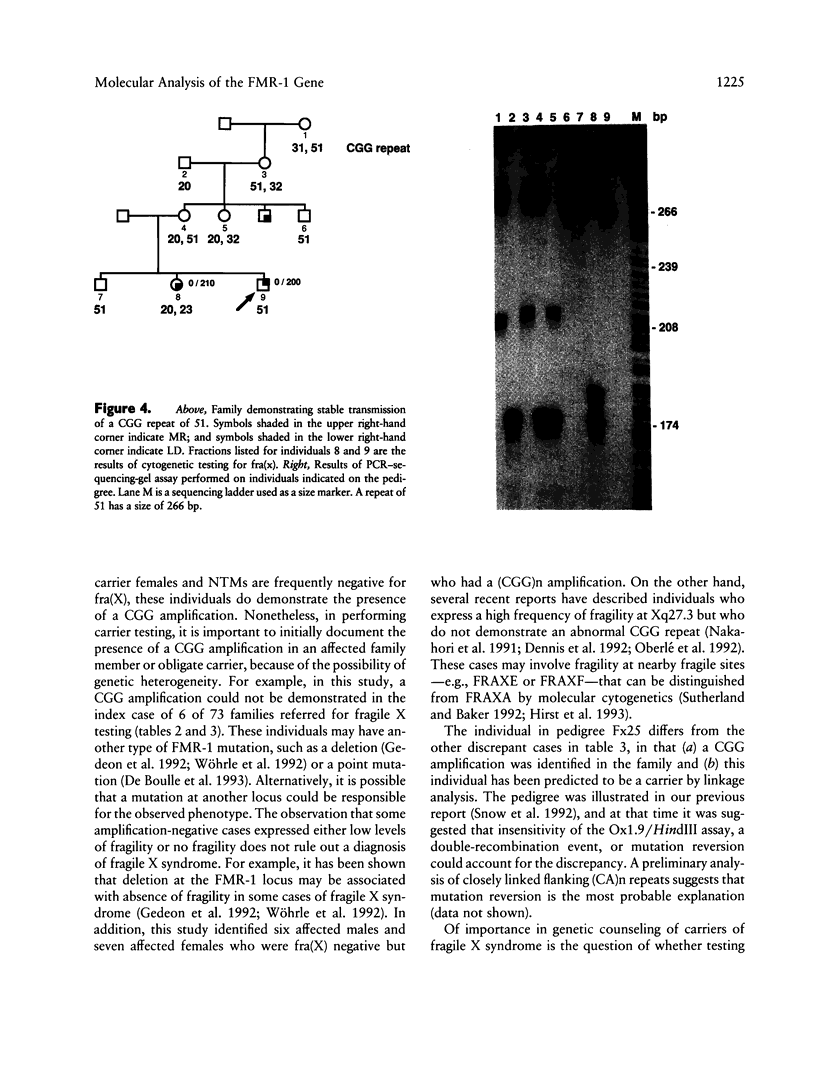
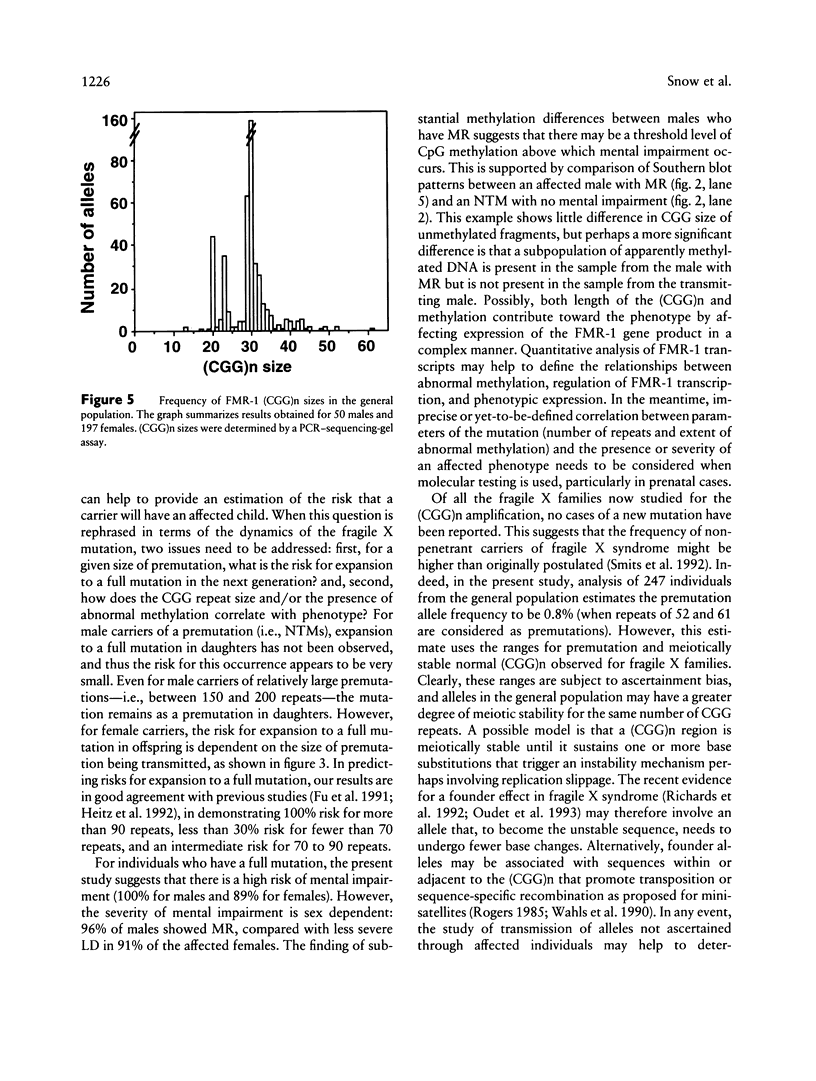
In this study, we have characterized a CGG repeat at the FMR-1 locus in more than 100 families (more than 500 individuals) presenting for fragile X testing and in 247 individuals from the general population. Both Southern blot and PCR-based assays were evaluated for their ability to detect premutations, full mutations, and variability in normal allele sizes. Among the Southern blot assays, the probes Ox1.9 or StB12.3 with a double restriction-enzyme digest were the most sensitive in detecting both small and large amplifications and, in addition, provided information on methylation of an adjacent CpG island. In the PCR-based assays, analysis of PCR products on denaturing DNA sequencing gels allowed the most accurate determination of CGG repeat number up to approximately 130 repeats. A combination of a Southern blot assay with a double digest and the PCR-sequencing-gel assay detected the spectrum of amplification-type mutations at the FMR-1 locus. In the patient population, a CGG repeat of 51 was the largest to be stably inherited, and a repeat of 57 was the smallest size of premutation to be unstably inherited. When premutations were transmitted by females, the size of repeat correlated with risk of expansion to a full mutation in the next generation. Full mutations (large repeats typically associated with an abnormal methylation pattern and mitotic instability) were associated with clinical and cytogenetic manifestations in males but not necessarily in females. In the control population, the CGG repeat ranged from 13 to 61, but 94% of alleles had fewer than 40 repeats. The most frequent allele (34%) was a repeat of 30. One female had an allele (61 repeats) within a range consistent with fragile X premutations, while two other individuals each had a repeat of 52. This suggests that the frequency of unstable alleles in the general population may be approximately 1%.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bell M. V., Hirst M. C., Nakahori Y., MacKinnon R. N., Roche A., Flint T. J., Jacobs P. A., Tommerup N., Tranebjaerg L., Froster-Iskenius U. Physical mapping across the fragile X: hypermethylation and clinical expression of the fragile X syndrome. Cell. 1991 Feb 22;64(4):861–866. doi: 10.1016/0092-8674(91)90514-y. [DOI] [PubMed] [Google Scholar]
- De Boulle K., Verkerk A. J., Reyniers E., Vits L., Hendrickx J., Van Roy B., Van den Bos F., de Graaff E., Oostra B. A., Willems P. J. A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nat Genet. 1993 Jan;3(1):31–35. doi: 10.1038/ng0193-31. [DOI] [PubMed] [Google Scholar]
- Dennis N. R., Curtis G., Macpherson J. N., Jacobs P. A. Two families with Xq27.3 fragility, no detectable insert in the FMR-1 gene, mild mental impairment, and absence of the Martin-Bell phenotype. 1992 Apr 15-May 1Am J Med Genet. 43(1-2):232–236. doi: 10.1002/ajmg.1320430137. [DOI] [PubMed] [Google Scholar]
- Erster S. H., Brown W. T., Goonewardena P., Dobkin C. S., Jenkins E. C., Pergolizzi R. G. Polymerase chain reaction analysis of fragile X mutations. Hum Genet. 1992 Sep-Oct;90(1-2):55–61. doi: 10.1007/BF00210744. [DOI] [PubMed] [Google Scholar]
- Fu Y. H., Kuhl D. P., Pizzuti A., Pieretti M., Sutcliffe J. S., Richards S., Verkerk A. J., Holden J. J., Fenwick R. G., Jr, Warren S. T. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell. 1991 Dec 20;67(6):1047–1058. doi: 10.1016/0092-8674(91)90283-5. [DOI] [PubMed] [Google Scholar]
- Gedeon A. K., Baker E., Robinson H., Partington M. W., Gross B., Manca A., Korn B., Poustka A., Yu S., Sutherland G. R. Fragile X syndrome without CCG amplification has an FMR1 deletion. Nat Genet. 1992 Aug;1(5):341–344. doi: 10.1038/ng0892-341. [DOI] [PubMed] [Google Scholar]
- Hauge X. Y., Litt M. A study of the origin of 'shadow bands' seen when typing dinucleotide repeat polymorphisms by the PCR. Hum Mol Genet. 1993 Apr;2(4):411–415. doi: 10.1093/hmg/2.4.411. [DOI] [PubMed] [Google Scholar]
- Heitz D., Devys D., Imbert G., Kretz C., Mandel J. L. Inheritance of the fragile X syndrome: size of the fragile X premutation is a major determinant of the transition to full mutation. J Med Genet. 1992 Nov;29(11):794–801. doi: 10.1136/jmg.29.11.794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst M. C., Barnicoat A., Flynn G., Wang Q., Daker M., Buckle V. J., Davies K. E., Bobrow M. The identification of a third fragile site, FRAXF, in Xq27--q28 distal to both FRAXA and FRAXE. Hum Mol Genet. 1993 Feb;2(2):197–200. doi: 10.1093/hmg/2.2.197. [DOI] [PubMed] [Google Scholar]
- Hirst M. C., Nakahori Y., Knight S. J., Schwartz C., Thibodeau S. N., Roche A., Flint T. J., Connor J. M., Fryns J. P., Davies K. E. Genotype prediction in the fragile X syndrome. J Med Genet. 1991 Dec;28(12):824–829. doi: 10.1136/jmg.28.12.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacky P. B., Ahuja Y. R., Anyane-Yeboa K., Breg W. R., Carpenter N. J., Froster-Iskenius U. G., Fryns J. P., Glover T. W., Gustavson K. H., Hoegerman S. F. Guidelines for the preparation and analysis of the fragile X chromosome in lymphocytes. Am J Med Genet. 1991 Feb-Mar;38(2-3):400–403. doi: 10.1002/ajmg.1320380249. [DOI] [PubMed] [Google Scholar]
- Kremer E. J., Pritchard M., Lynch M., Yu S., Holman K., Baker E., Warren S. T., Schlessinger D., Sutherland G. R., Richards R. I. Mapping of DNA instability at the fragile X to a trinucleotide repeat sequence p(CCG)n. Science. 1991 Jun 21;252(5013):1711–1714. doi: 10.1126/science.1675488. [DOI] [PubMed] [Google Scholar]
- Latimer L. J., Lee J. S. Ethidium bromide does not fluoresce when intercalated adjacent to 7-deazaguanine in duplex DNA. J Biol Chem. 1991 Jul 25;266(21):13849–13851. [PubMed] [Google Scholar]
- Macpherson J. N., Nelson D. L., Jacobs P. A. Frequent small amplifications in the FMR-1 gene in fra(X) families: limits to the diagnosis of 'premutations'. J Med Genet. 1992 Nov;29(11):802–806. doi: 10.1136/jmg.29.11.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton N. E., Macpherson J. N. Population genetics of the fragile-X syndrome: multiallelic model for the FMR1 locus. Proc Natl Acad Sci U S A. 1992 May 1;89(9):4215–4217. doi: 10.1073/pnas.89.9.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahori Y., Knight S. J., Holland J., Schwartz C., Roche A., Tarleton J., Wong S., Flint T. J., Froster-Iskenius U., Bentley D. Molecular heterogeneity of the fragile X syndrome. Nucleic Acids Res. 1991 Aug 25;19(16):4355–4359. doi: 10.1093/nar/19.16.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberlé I., Boué J., Croquette M. F., Voelckel M. A., Mattei M. G., Mandel J. L. Three families with high expression of a fragile site at Xq27.3, lack of anomalies at the FMR-1 CpG island, and no clear phenotypic association. 1992 Apr 15-May 1Am J Med Genet. 43(1-2):224–231. doi: 10.1002/ajmg.1320430136. [DOI] [PubMed] [Google Scholar]
- Oberlé I., Rousseau F., Heitz D., Kretz C., Devys D., Hanauer A., Boué J., Bertheas M. F., Mandel J. L. Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science. 1991 May 24;252(5009):1097–1102. doi: 10.1126/science.252.5009.1097. [DOI] [PubMed] [Google Scholar]
- Oudet C., Mornet E., Serre J. L., Thomas F., Lentes-Zengerling S., Kretz C., Deluchat C., Tejada I., Boué J., Boué A. Linkage disequilibrium between the fragile X mutation and two closely linked CA repeats suggests that fragile X chromosomes are derived from a small number of founder chromosomes. Am J Hum Genet. 1993 Feb;52(2):297–304. [PMC free article] [PubMed] [Google Scholar]
- Pergolizzi R. G., Erster S. H., Goonewardena P., Brown W. T. Detection of full fragile X mutation. Lancet. 1992 Feb 1;339(8788):271–272. doi: 10.1016/0140-6736(92)91334-5. [DOI] [PubMed] [Google Scholar]
- Pieretti M., Zhang F. P., Fu Y. H., Warren S. T., Oostra B. A., Caskey C. T., Nelson D. L. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell. 1991 Aug 23;66(4):817–822. doi: 10.1016/0092-8674(91)90125-i. [DOI] [PubMed] [Google Scholar]
- Richards R. I., Holman K., Friend K., Kremer E., Hillen D., Staples A., Brown W. T., Goonewardena P., Tarleton J., Schwartz C. Evidence of founder chromosomes in fragile X syndrome. Nat Genet. 1992 Jul;1(4):257–260. doi: 10.1038/ng0792-257. [DOI] [PubMed] [Google Scholar]
- Rogers J. H. The origin and evolution of retroposons. Int Rev Cytol. 1985;93:187–279. doi: 10.1016/s0074-7696(08)61375-3. [DOI] [PubMed] [Google Scholar]
- Rousseau F., Heitz D., Biancalana V., Blumenfeld S., Kretz C., Boué J., Tommerup N., Van Der Hagen C., DeLozier-Blanchet C., Croquette M. F. Direct diagnosis by DNA analysis of the fragile X syndrome of mental retardation. N Engl J Med. 1991 Dec 12;325(24):1673–1681. doi: 10.1056/NEJM199112123252401. [DOI] [PubMed] [Google Scholar]
- Smits A., Smeets D., Hamel B., Dreesen J., van Oost B. High prevalence of the fra(X) syndrome cannot be explained by a high mutation rate. 1992 Apr 15-May 1Am J Med Genet. 43(1-2):345–352. doi: 10.1002/ajmg.1320430153. [DOI] [PubMed] [Google Scholar]
- Snow K., Doud L., Hagerman R., Hull C., Hirst M. C., Davies K. E., Thibodeau S. L. Analysis of mutations at the fragile X locus using the DNA probe Ox1.9. 1992 Apr 15-May 1Am J Med Genet. 43(1-2):244–254. doi: 10.1002/ajmg.1320430139. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. S., Nelson D. L., Zhang F., Pieretti M., Caskey C. T., Saxe D., Warren S. T. DNA methylation represses FMR-1 transcription in fragile X syndrome. Hum Mol Genet. 1992 Sep;1(6):397–400. doi: 10.1093/hmg/1.6.397. [DOI] [PubMed] [Google Scholar]
- Sutherland G. R., Baker E. Characterisation of a new rare fragile site easily confused with the fragile X. Hum Mol Genet. 1992 May;1(2):111–113. doi: 10.1093/hmg/1.2.111. [DOI] [PubMed] [Google Scholar]
- Sutherland G. R., Gedeon A., Kornman L., Donnelly A., Byard R. W., Mulley J. C., Kremer E., Lynch M., Pritchard M., Yu S. Prenatal diagnosis of fragile X syndrome by direct detection of the unstable DNA sequence. N Engl J Med. 1991 Dec 12;325(24):1720–1722. doi: 10.1056/NEJM199112123252407. [DOI] [PubMed] [Google Scholar]
- Verkerk A. J., Pieretti M., Sutcliffe J. S., Fu Y. H., Kuhl D. P., Pizzuti A., Reiner O., Richards S., Victoria M. F., Zhang F. P. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991 May 31;65(5):905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- Wahls W. P., Wallace L. J., Moore P. D. Hypervariable minisatellite DNA is a hotspot for homologous recombination in human cells. Cell. 1990 Jan 12;60(1):95–103. doi: 10.1016/0092-8674(90)90719-u. [DOI] [PubMed] [Google Scholar]
- Wöhrle D., Kotzot D., Hirst M. C., Manca A., Korn B., Schmidt A., Barbi G., Rott H. D., Poustka A., Davies K. E. A microdeletion of less than 250 kb, including the proximal part of the FMR-I gene and the fragile-X site, in a male with the clinical phenotype of fragile-X syndrome. Am J Hum Genet. 1992 Aug;51(2):299–306. [PMC free article] [PubMed] [Google Scholar]
- Yu S., Pritchard M., Kremer E., Lynch M., Nancarrow J., Baker E., Holman K., Mulley J. C., Warren S. T., Schlessinger D. Fragile X genotype characterized by an unstable region of DNA. Science. 1991 May 24;252(5009):1179–1181. doi: 10.1126/science.252.5009.1179. [DOI] [PubMed] [Google Scholar]