Abstract
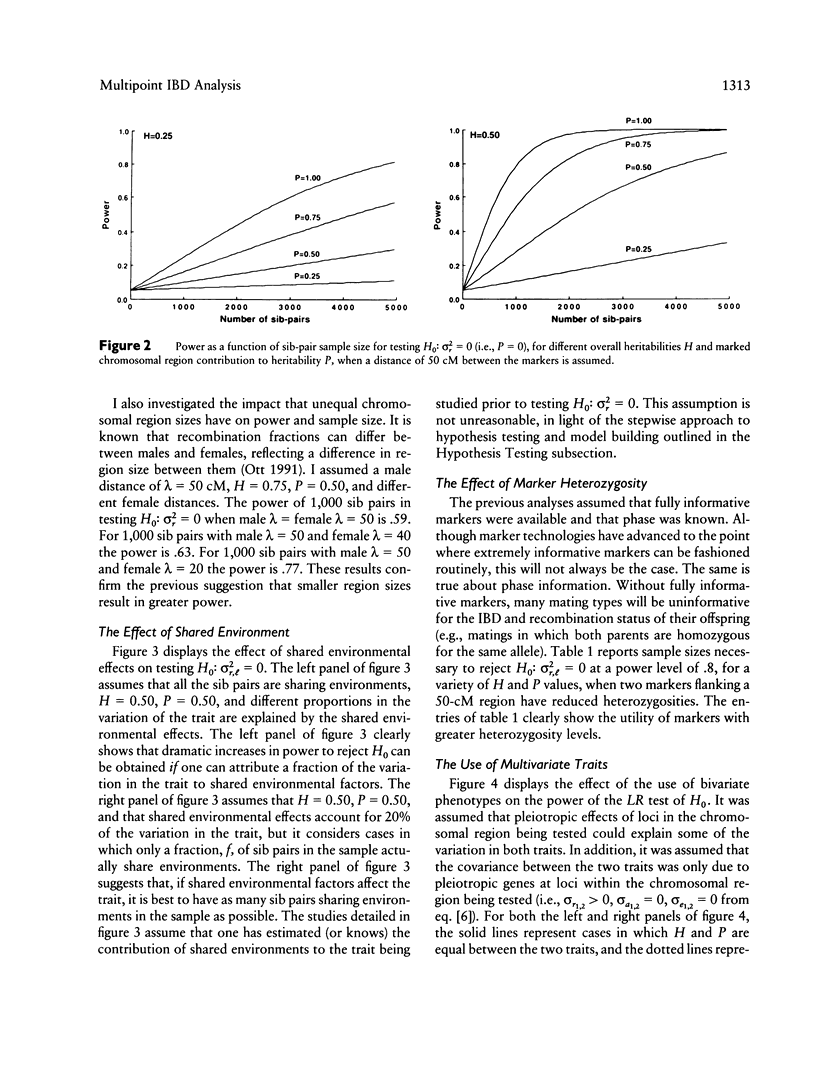
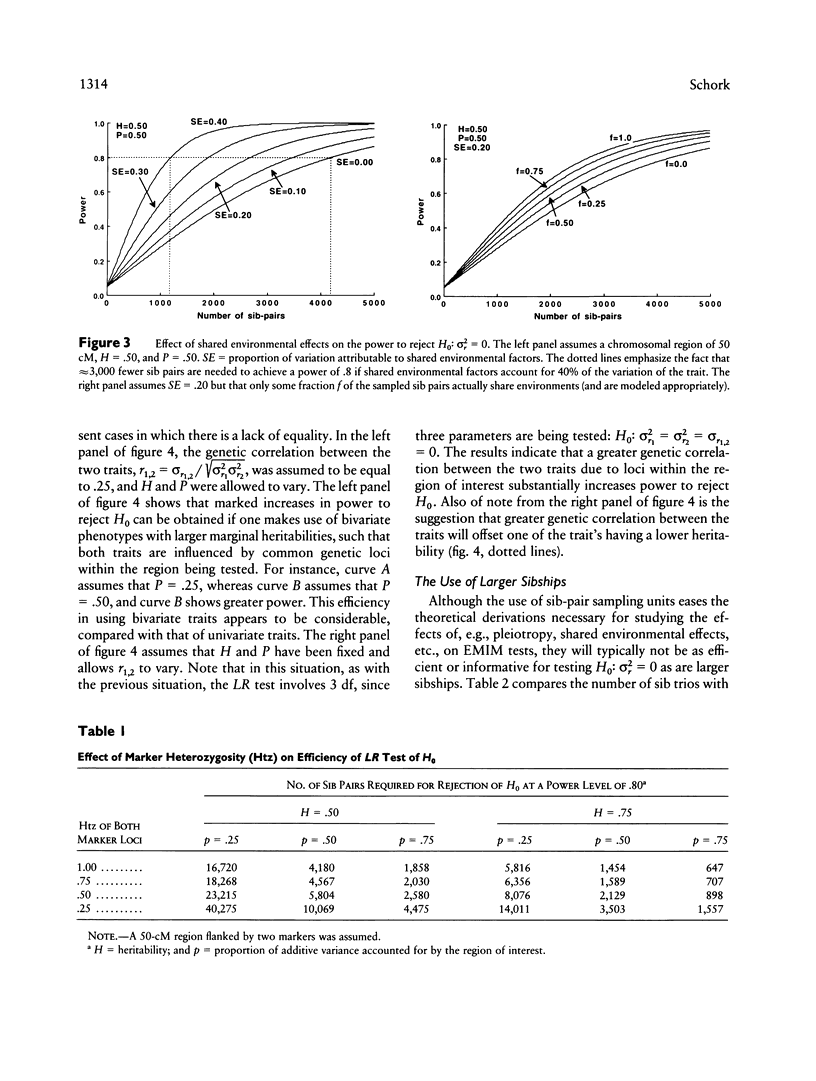
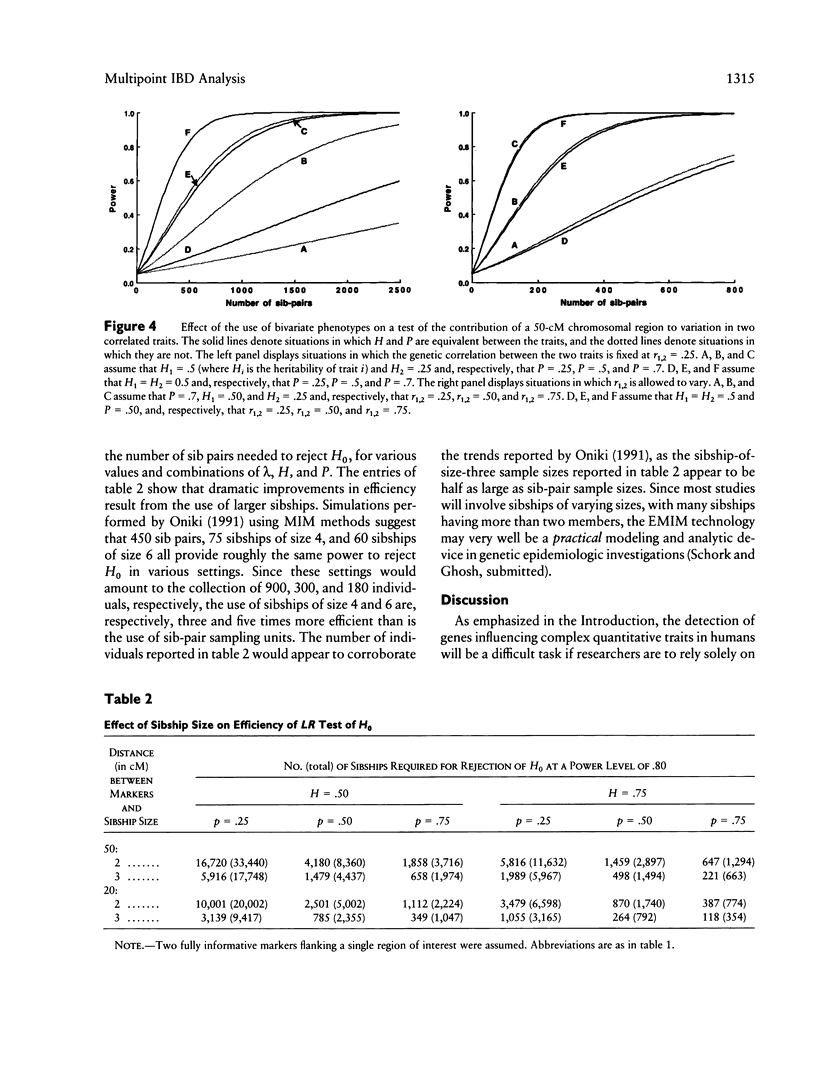
Goldgar introduced a novel marker-based method for partitioning the variation of a quantitative trait into specific chromosomal regions. Unlike traditional linkage mapping methods, Goldgar's method does not require the estimation of statistical quantities characterizing each locus thought to influence the trait under scrutiny (e.g., allele frequencies, penetrances, etc.). Goldgar's method is thus more flexible and less model dependent than many traditional marker-based genetic analysis techniques. Unfortunately, however, many of the properties of Goldgar's method have not been investigated. In this paper, the utility of an extended version of Goldgar's approach is studied in settings in which sibships are taken as the sampling unit of interest. The extensions discussed resolve around the incorporation of a wider variety of effects and factors into Goldgar's basic model. Analytic studies pertaining to power, sample-size requirements, and estimation procedures for the proposed extended version of Goldgar's method are described. Hypothesis-testing strategies are also discussed. The results of the analytic studies indicate that, although an extended sib-pair version of Goldgar's variance-partitioning approach to modeling the chromosomal determinants of a quantitative trait will be useful only for traits with high heritabilities or when fine-scale genetic maps can be employed. Goldgar's technique as a whole has promise, as it can be made relatively robust statistically, refined through some simple and intuitive extensions, and can be easily adapted to work with more complex sampling units. Further extensions of Goldgar's methods are proposed, and areas in need of additional research are discussed.
Full text
PDF










Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beaty T. H., Self S. G., Liang K. Y., Connolly M. A., Chase G. A., Kwiterovich P. O. Use of robust variance components models to analyse triglyceride data in families. Ann Hum Genet. 1985 Oct;49(Pt 4):315–328. doi: 10.1111/j.1469-1809.1985.tb01707.x. [DOI] [PubMed] [Google Scholar]
- Blangero J., Konigsberg L. W. Multivariate segregation analysis using the mixed model. Genet Epidemiol. 1991;8(5):299–316. doi: 10.1002/gepi.1370080503. [DOI] [PubMed] [Google Scholar]
- Blangero J., MacCluer J. W., Kammerer C. M., Mott G. E., Dyer T. D., McGill H. C., Jr Genetic analysis of apolipoprotein A-I in two dietary environments. Am J Hum Genet. 1990 Sep;47(3):414–428. [PMC free article] [PubMed] [Google Scholar]
- Carey G. Inference about genetic correlations. Behav Genet. 1988 May;18(3):329–338. doi: 10.1007/BF01260933. [DOI] [PubMed] [Google Scholar]
- Donnelly K. P. The probability that related individuals share some section of genome identical by descent. Theor Popul Biol. 1983 Feb;23(1):34–63. doi: 10.1016/0040-5809(83)90004-7. [DOI] [PubMed] [Google Scholar]
- Goldgar D. E. Multipoint analysis of human quantitative genetic variation. Am J Hum Genet. 1990 Dec;47(6):957–967. [PMC free article] [PubMed] [Google Scholar]
- Goldgar D. E., Oniki R. S. Comparison of a multipoint identity-by-descent method with parametric multipoint linkage analysis for mapping quantitative traits. Am J Hum Genet. 1992 Mar;50(3):598–606. [PMC free article] [PubMed] [Google Scholar]
- Haseman J. K., Elston R. C. The investigation of linkage between a quantitative trait and a marker locus. Behav Genet. 1972 Mar;2(1):3–19. doi: 10.1007/BF01066731. [DOI] [PubMed] [Google Scholar]
- Hultén M. Chiasma distribution at diakinesis in the normal human male. Hereditas. 1974;76(1):55–78. doi: 10.1111/j.1601-5223.1974.tb01177.x. [DOI] [PubMed] [Google Scholar]
- Lander E. S., Botstein D. Strategies for studying heterogeneous genetic traits in humans by using a linkage map of restriction fragment length polymorphisms. Proc Natl Acad Sci U S A. 1986 Oct;83(19):7353–7357. doi: 10.1073/pnas.83.19.7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange K., Boehnke M. Extensions to pedigree analysis. IV. Covariance components models for multivariate traits. Am J Med Genet. 1983 Mar;14(3):513–524. doi: 10.1002/ajmg.1320140315. [DOI] [PubMed] [Google Scholar]
- Moll P. P., Powsner R., Sing C. F. Analysis of genetic and environmental sources of variation in serum cholesterol in Tecumseh, Michigan. V. Variance components estimated from pedigrees. Ann Hum Genet. 1979 Jan;42(3):343–354. doi: 10.1111/j.1469-1809.1979.tb00668.x. [DOI] [PubMed] [Google Scholar]
- Nelson S. F., McCusker J. H., Sander M. A., Kee Y., Modrich P., Brown P. O. Genomic mismatch scanning: a new approach to genetic linkage mapping. Nat Genet. 1993 May;4(1):11–18. doi: 10.1038/ng0593-11. [DOI] [PubMed] [Google Scholar]
- Schork N. J., Boehnke M., Terwilliger J. D., Ott J. Two-trait-locus linkage analysis: a powerful strategy for mapping complex genetic traits. Am J Hum Genet. 1993 Nov;53(5):1127–1136. [PMC free article] [PubMed] [Google Scholar]
- Schork N. J., Schork M. A. The relative efficiency and power of small-pedigree studies of the heritability of a quantitative trait. Hum Hered. 1993 Jan-Feb;43(1):1–11. doi: 10.1159/000154106. [DOI] [PubMed] [Google Scholar]
- Sing C. F., Haviland M. B., Templeton A. R., Zerba K. E., Reilly S. L. Biological complexity and strategies for finding DNA variations responsible for inter-individual variation in risk of a common chronic disease, coronary artery disease. Ann Med. 1992 Dec;24(6):539–547. doi: 10.3109/07853899209167008. [DOI] [PubMed] [Google Scholar]
- Sing C. F., Orr J. D. Analysis of genetic and environmental sources of variation in serum cholesterol in Tecumseh, Michigan. III. Identification of genetic effects using 12 polymorphic genetic blood marker systems. Am J Hum Genet. 1976 Sep;28(5):453–464. [PMC free article] [PubMed] [Google Scholar]


