Abstract
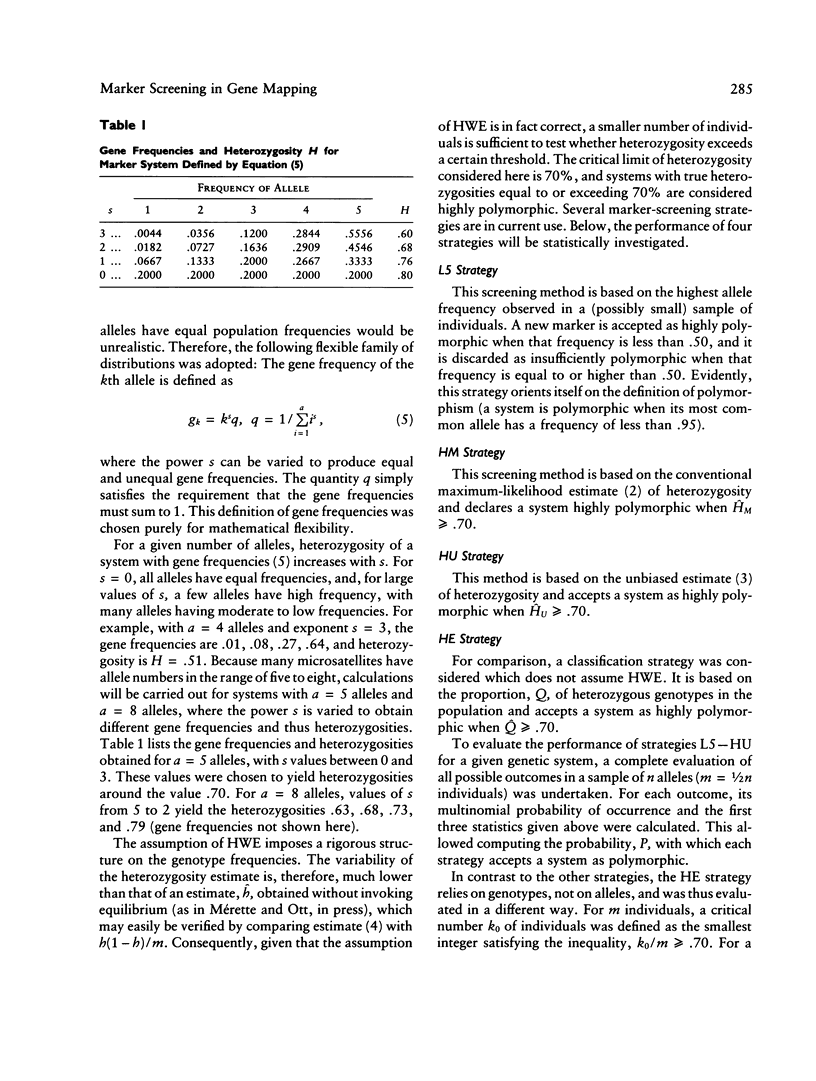
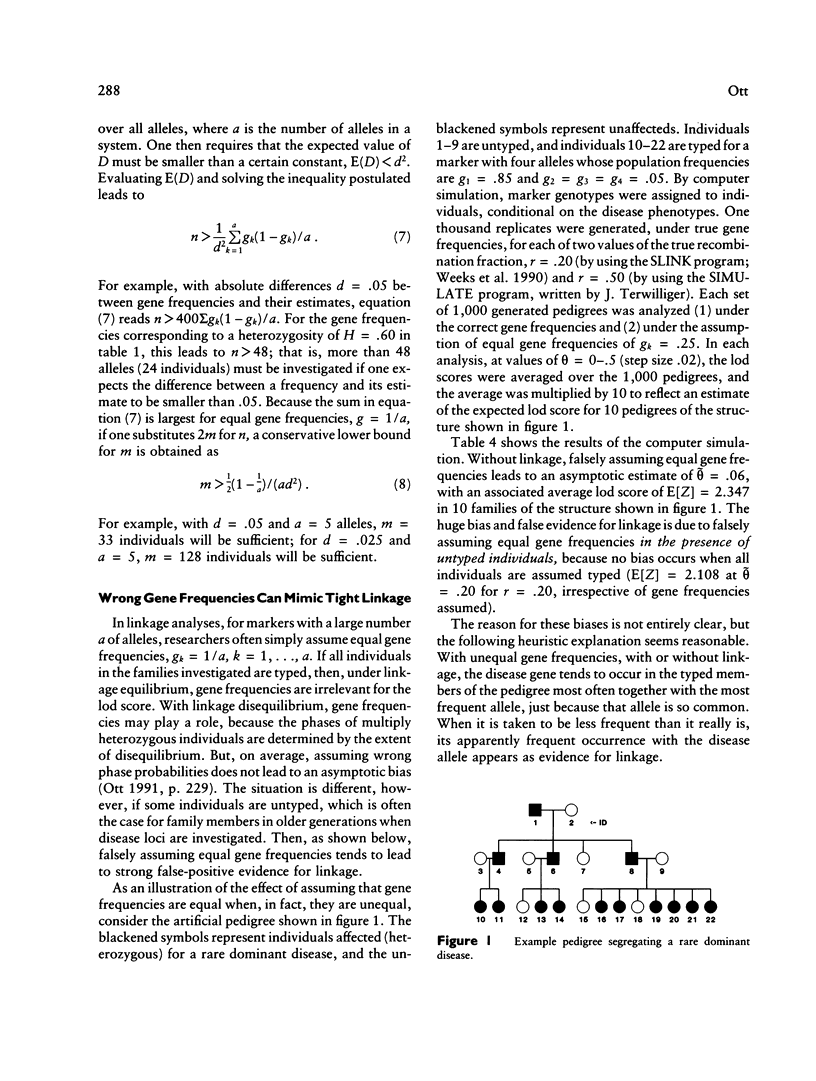
Before new markers are thoroughly characterized, they are usually screened for high polymorphism on the basis of a small panel of individuals. Four commonly used screening strategies are compared in terms of their power to correctly classify a marker as having heterozygosity of 70% or higher. A small number of typed individuals (10, say) are shown to provide good discrimination power between low- and high-heterozygosity markers when the markers have a small number of alleles. Characterizing markers in more detail requires larger sample sizes (e.g., at least 80-100 individuals) if there is to be a high probability of detecting most or all alleles. For linkage analyses involving highly polymorphic markers, the practice of arbitrarily assuming equal gene frequencies can cause serious trouble. In the presence of untyped individuals, when gene frequencies are unequal but are assumed to be equal in the analysis, recombination-fraction estimates tend to be badly biased, leading to strong false-positive evidence for linkage.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boehnke M. Allele frequency estimation from data on relatives. Am J Hum Genet. 1991 Jan;48(1):22–25. [PMC free article] [PubMed] [Google Scholar]
- Devlin B., Risch N., Roeder K. No excess of homozygosity at loci used for DNA fingerprinting. Science. 1990 Sep 21;249(4975):1416–1420. doi: 10.1126/science.2205919. [DOI] [PubMed] [Google Scholar]
- Ott J. A simple scheme for the analysis of HLA linkages in pedigrees. Ann Hum Genet. 1978 Oct;42(2):255–257. doi: 10.1111/j.1469-1809.1978.tb00657.x. [DOI] [PubMed] [Google Scholar]


