Abstract
Myxococcus xanthus develops species-specific multicellular fruiting bodies. Starting from a uniform mat of cells, some cells enter into nascent fruiting body aggregates, whereas other cells remain outside. The cells within the fruiting body differentiate from rods into spherical, heat-resistant spores, whereas the cells outside the aggregates, called peripheral cells, remain rod-shaped. Early developmentally regulated genes are expressed in peripheral cells as well as by cells in the fruiting bodies. By contrast, late developmental genes are only expressed by cells within the nascent fruiting bodies. The data show that peripheral cells begin to develop, but are unable to express genes that are switched on later than about 6 h after the start of development. All of the genes whose expression is limited to the fruiting body are dependent on C-signaling either directly or indirectly, whereas the genes that are equally expressed in peripheral rods and in fruiting body cells are not. One of the C-signal-dependent and spatially patterned operons is called dev, and the dev operon has been implicated in the process of sporulation. It is proposed that expression of certain genes, including those of the dev operon, is limited to the nascent fruiting body because fruiting body cells engage in a high level of C-signaling. Peripheral cells do less C-signaling than fruiting body cells, because they have a different spatial arrangement and are at lower density. As a consequence, peripheral cells fail to express the late genes necessary for spore differentiation.
Keywords: spatial pattern, positive feedback, Myxobacteria, cell–cell interaction
How spatial patterns of differentiated cells arise is a central issue for animal and plant development. Myxococcus xanthus and other myxobacteria differentiate spores in response to nutrient deprivation. Although most bacteria sporulate individually, myxobacteria build large structured masses of spores, called fruiting bodies. Under nutrient-rich conditions, M. xanthus grows and divides as rod-shaped cells. When its development is induced by starvation, a hundred thousand cells contribute to building a fruiting body, whose shape is species-specific. Cells that have entered into the fruiting body finally differentiate into environmentally resistant myxospores, which can survive years without nutrients. However, not all of the starvation-induced cells become spores. Cells within fruiting bodies become spores, cells outside and between these multicellular structures remain rod-shaped and nonresistant (1). These cells, called peripheral rod cells, never become spores, despite their synthesis of two sporulation proteins, Tps and C (1). Dworkin and Gibson (2) showed that every cell innately has the capacity to become a spore. A difference in the developmental fate of peripheral rods and of fruiting body cells constitutes a spatial pattern that needs to be explained.
Most patterns involve cell-to-cell signaling, and sporulation depends on C-signaling. Ordinarily, each cell is simultaneously a transmitter and a receiver of the C-signal. The CsgA protein, on the surface of one cell, induces three processes in a contiguous cell (3–5). One consequence of signal reception is an enhancement of csgA expression. After initial episodes of C-signaling, an increase in the amount of CsgA protein per responding cell is observed (6, 7). Although the cell's receptor for C-signal has not yet been identified, it has been established that CsgA protein on one cell engenders an activating modification (probably phosphorylation) of the FruA response regulator protein in the other cell (8, 9). Activation of FruA signals the frz phospho-relay (10) to methylate FrzCD (11, 12), and to adjust the movement parameters of responding cells (13). Increased speed, increased movement intervals, and decreased stop frequency are the consequences, and these changes allow the cells to congregate into mound-shaped aggregates. Finally, activated FruA induces sporulation. However, it has been shown that sporulation requires a higher intensity of C-signaling than aggregation (6, 14). C-signal-dependent csgA expression has a lower threshold than aggregation or sporulation. As C-signaling intensity rises with csgA expression, it first reaches the threshold for aggregation, then, enhanced by positive feedback, it finally reaches the threshold for sporulation.
An operon was discovered several years ago that is expressed at high level, but only in a fraction of developing cells. This operon was originally identified in a search for developmentally regulated genes using the transposable element, Tn5lac (15). Tn5lac has a promoterless lacZ preceded by translation stops in all three reading frames (16). Expression in single cells was measured, using fluorescence activated cell sorting (FACS) to reveal the cell-to-cell distribution of β-galactosidase activity (17). Most developmentally regulated Tn5lac strains, like Ω4499 and Ω4506 (15), showed unimodal expression of β-galactosidase. Having been induced to develop for a given time, all their cells were expressing lacZ to similar levels distributed continuously about a single strain-characteristic modal value. But Tn5lac insertion Ω4473 showed bimodal expression. Some individual cells expressed the locus at its maximum level, whereas others showed little or no expression. The bimodal expression indicated that a gene marked by the insertion Ω4473 was differentially expressed in two cell populations—one at high level, the other at low. The “high” regulatory state of individual developing cells was maintained at least 3 h in the sorted cells (17). A second Tn5lac insertion, called Ω4414, was found at a different position within the same operon as Ω4473 (15, 18). The critical role of this operon in development was indicated by the fact that either insertion causes more than a 1,000-fold reduction in the number of viable spores. The operon identified by Ω4473 and Ω4414, which was named dev, is developmentally regulated, and its expression begins around the time of aggregation (18).
Because dev mutants are defective in sporulation, and because expression of the dev operon is either high or low in individual cells later in development, we have tested the possibility that dev expression is related to spore localization within the fruiting body. To find which cells express dev during development, we created a transcriptional fusion between the dev operon and the gfp gene, which encodes the green fluorescence protein. The observed spatial distribution of fluorescence of this fusion strain, indeed, shows localization to the fruiting body, and no expression in the peripheral rods. Examination of many developmentally regulated genes revealed a clear pattern: early genes were equally expressed in peripheral rods and fruiting bodies, whereas late genes were preferentially expressed in the fruiting body. Moreover, all of the differentially expressed genes depend on C-signaling.
Methods
Bacteria and Plasmids.
Strains and plasmids used are listed in Table 1. The Escherichia coli C strain C-2421 (19), used for cloning, was grown in LB medium containing 0.5% NaCl. Ampicillin was used at a concentration of 50 μg/ml or kanamycin at a concentration of 50 μg/ml to select for maintenance of plasmids. M. xanthus strains were grown in CYE (casitone yeast extract) liquid medium (20) or on CYE agar plates supplemented with 40 μg/ml kanamycin when necessary. Galactose resistance was selected by plating dilutions of mid exponential phase cells on CYE plates containing 1% galactose.
Table 1.
Myxobacterial strains and plasmids
| Bacteria or plasmid | Characteristics | Ref. |
|---|---|---|
| DK1622 | M. xanthus standard strain | 51 |
| DK5517 | DK1622 Ω4414 tetr dev∷pLT19 | 18 |
| DK7536 | DK1622 Ω7536 | 40 |
| DK10469 | DK1622 pilA∷lacZ, pilA+ | |
| pLT4 | 7.8 kb SalI fragment containing part of the dev operon | 18 |
| pBJ113 | pUC118 containing kanr and galK; Plasmid used for constructing gene replacements. Derived from pKG2 | 26 |
| pBJ113 ckgfp | pBJ113 containing a transcriptional fusion of the dev operon and gfp | This work |
| pBJ130 | pPLT4 with the BamHI to EcoRI sites missing from the polylinker region | This work |
| pBJgfp1-2 | An Xbal amber linker ligated into pSPgfp-1 | This work |
| pSPgfp-1 | gfp ligated into pSP72 | This work |
Protocols for minipreparations of DNA, restriction digests, ligations, and transformations are those standardly used (21). Chromosomal DNA was isolated as previously described (22). Plasmids were introduced into M. xanthus by electroporation, as described (23).
Fruiting body development was carried out on TPM (Tris phosphate magnesium) agar plates (15). Cells were grown to 100 Klett density units (KU), sedimented, and washed with an equal volume of TPM. The cells were pelleted again and resuspended in TPM to a density of 1,000 KU. Aliquots of 20 μl of this suspension were spotted on TPM agar plates and were incubated at 32°C. β-galactosidase activity was measured as described (15). Development was also carried out in submerged culture (24). Sporulation efficiencies (mutant relative to wild type) were measured after 5 days of development in submerged culture. Cultures were harvested, suspended in 1 ml of TPM buffer, sonicated, and heated to kill cells that had not become spores. Survivors were then plated onto CYE plates for germination and growth at 32°C for 4 or 5 days.
Separation of Fruiting Bodies and Peripheral Rods.
The procedure described by O'Connor and Zusman (1) was modified by inducing development on TPM agar instead of CF agar, and the harvested cells were centrifuged at 50 × g for 5 min twice to help limit the number of fruiting body cells remaining in suspension. Finally, 400-μl aliquots of fruiting body cells and peripheral rods were quick-frozen in liquid nitrogen and stored at −80oC until they were extracted for β-galactosidase assays.
For Western blotting, cells were harvested after 18 h of development on TPM agar plates. Separated peripheral rods and fruiting bodies were suspended in SDS lysis buffer. Samples were fractionated on a 12% polyacrylamide gel, transferred to Immobilon P (Millipore) by using a semidry blotting apparatus. The blot was probed with anti-CsgA antibody, followed by peroxidase-conjugated goat anti-rabbit IgG (Roche Molecular Biochemicals). The antiserum to CsgA was the gift of T. Kruse, S. Lobendanz, N. Berthelsen, and L. Sogaard-Andersen (unpublished observations) of Odense University (Denmark). The blots were developed with a chemiluminescence reagent (NEN Life Science Products) and exposed to Amersham autoradiography Hyperfilm-MP. Each sample was tested four times.
Construction of a dev-gfp Transcriptional Fusion.
The pgfpmut3 plasmid (25) was cleaved with XbaI and HindIII, the DNA ends treated with the Klenow polymerase, and the 800-bp gfp-containing fragment was isolated. This fragment was ligated into a plasmid, pSP72, which had been cleaved with SacI and the DNA made blunt with Klenow polymerase. The ligated plasmid is designated pSPgfp-1 (Table 1). Next, stop codons were introduced in all three frames upstream of the gfp gene by ligating a XbaI amber linker into the EcoRI site of pSP72gfp-1, which had been treated with the Klenow polymerase, to create pBJgfp1–2. To fuse gfp to the dev operon, pBJgfp1–2 was cleaved with PstI, the DNA ends made blunt with Klenow polymerase, and then cleaved with ClaI. The resulting gfp-containing fragment was ligated into pBJ130 (Table 1) that had been cleaved with KpnI, the DNA ends treated with Klenow polymerase, and then cleaved with ClaI. From the resulting plasmid (pBJ130ckgfp), a dev-gfp fusion fragment was removed with ClaI and KpnI, and ligated into pBJ113. The resulting plasmid, pBJ113ckgfp, was electroporated into DK1622, and kanamycin-resistant colonies were selected. By using the galK gene in pBJ113 as a counter selectable marker, galactose resistant colonies were obtained, which were screened for the loss of the wild-type dev locus and the presence of the fusion dev∷gfp allele (26).
Microscopic Examination of Cells Within Fruiting Bodies.
Cells labeled with the green fluorescent protein (GFP) were allowed to develop as spots on TPM agar as described for development. After 12 and 24 h of development, cells were photographed through a Nikon Eclipse E800 microscope equipped with a Chroma FITC-HYQ filter to visualize GFP-labeled cells. Images of developing cells and fruiting bodies were captured with an Optronics Engineering (Goleta, CA) DE1–750T RGB video camera and recorded on an optical disk cartridge. To examine the fluorescence of individual GFP-labeled cells within developing fruiting bodies, a glass coverslip was placed on top of cells after they developed on TPM agar for 12 h, crushing the fruiting bodies and laterally spreading individual cells out from the mass.
Spatial Distribution of Peripheral Rod Cells.
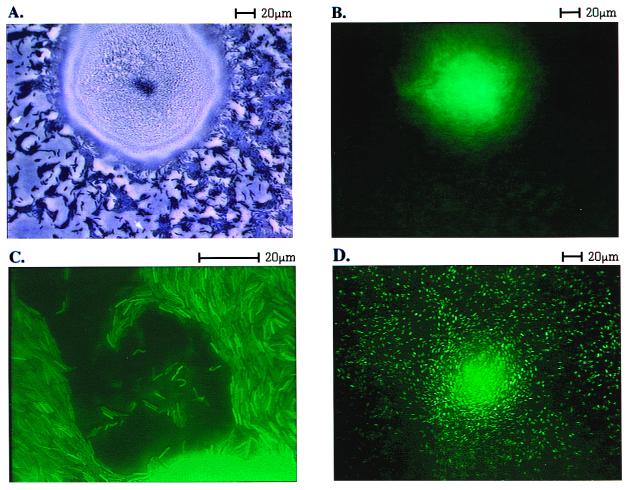
Photomicrographs such as that shown in Fig. 1A were analyzed to determine the way peripheral rod cells are arranged. A number of single cells are evident in the field, and one is indicated by an arrow pointing down. All 14 well-isolated single cells in this field were traced, and the average area of a single cell was measured. On average, their lengths were 10 times their widths, in agreement with other phase-contrast images of M. xanthus cells (27). The cells in direct contact with the edge of the fruiting body and cells in contact with those cells were set aside. The remaining cells are clustered in multicellular islands, a representative island is indicated by an arrow pointing up in Fig. 1A. Phase contrast optics show no gaps within the islands. It is therefore assumed that the cells are close packed side-by-side and in end-to-end contact. To estimate the arrangement of such cells, fields like that of Fig. 1A were projected onto paper, and the outline of each island was traced. It is evident by comparing the opacity of single cells and islands in Fig. 1A that the islands are only one cell layer thick. Each island was approximated as a rectangle, which was then tiled with average single rods, oriented with their long axes parallel to the long axis of the rectangle. For each tiling, the number of free cell ends (not in contact with another cell end) was counted. The ratio of free ends to total ends was calculated for each island. The ratios were weighted by the estimated number of cells in the island, to estimate the average ratio, free ends/total ends for the entire field of cells.
Figure 1.
Spatial localization of dev expression, measured in a dev∷gfp transcriptional fusion strain. (A) Bright field microscopy with phase contrast after 24 h of development of the dev∷gfp strain showing a fruiting body surrounded by peripheral rod cells. An arrow points up to indicate an island of cells; an arrow points down to indicate a single cell. (B) Same microscopic field as in A now using fluorescence. (C) A different dev∷gfp fruiting body that has been flattened under a coverslip to spread out the cells from within the mass, and only fruiting body cells are present in this field. It was photographed at 3× higher magnification than A or B. (D) Fruiting body and adjacent peripheral rods formed by a pilA∷gfp strain photographed in fluorescence mode.
Results
The dev∷gfp Fusion Is Expressed in Fruiting Bodies, Not in Peripheral Rods.
Is there a difference in the regulatory state of peripheral rods and fruiting body cells? A clue was offered by previous work on the dev operon (18), which was found to be expressed only in a fraction of developing cells (17). Whether dev might be expressed in the peripheral rods, in the cells within the nascent fruiting body, or in a fraction of each type was not answered by those experiments. To distinguish among the three possibilities, we constructed a transcriptional fusion of the dev operon to gfp, and tracked its expression with fluorescence microscopy at 24 h of development when spores are beginning to form. Fig. 1A shows a single nascent fruiting body aggregate densely packed with cells, as shown by its opacity. The fruiting body is surrounded by peripheral rod cells, most of which are clustered in islands of 5–100 cells (one indicated by an arrow pointing up in Fig. 1A). Fig. 1B is a photomicrograph showing the fluorescent image of the same field. Evidently, fluorescence is localized to the fruiting body; there is none in the many thousands of peripheral rods observed in this (or other such) experiments. Despite their origin in the same population of developing cells and close spatial proximity, cells of a nascent fruiting body are able to express the dev operon, whereas their neighboring peripheral rods are not.
Two control experiments were carried out to be certain that individual peripheral rods would have been visibly fluorescent had they been expressing dev. In one, a nascent fruiting body built of dev∷gfp cells was gently flattened by pressing it beneath a coverslip. Flattening tends to spread cells laterally from their initial position inside an aggregate, leaving cells that were neighbors within the fruiting body near to each other, and allowing individual cells to be resolved by bringing them into the plane of focus of a high magnification (×60) lens with a shallow depth of field. Fig. 1C shows cells spread outward from a fruiting body. Several plumes of cells have spread away from the nascent fruiting body, which is still evident as a bright glow along the lower right edge of Fig. 1C. After spreading, individual cells from the fruiting body are seen to be fluorescent over their entire length and width. Individual dev∷gfp cells and cells in side-by-side apposition can each be seen distinctly by their fluorescence.
As a second positive control, a pilA∷gfp fusion strain was starved to develop fruiting bodies. The pilA gene encodes the structural subunit (pilin) of the pili of M. xanthus, which is expressed in all cells during growth and throughout development (28, 29). Fluorescence microscopy of pilA∷gfp cells mixed 1:50 with wild-type (DK1622) cells showed high level expression of individual peripheral rods as well as of nascent fruiting body aggregates (Fig. 1D). The pilA∷gfp culture had been diluted 1:50 with wild-type (DK1622) cells lacking gfp to decrease the total fluorescence and its resulting glare. The fruiting body is evident as a densely speckled mass in the center, whereas the periphery shows many individual cells within the islands of mostly nonfluorescent cells. The pilA∷gfp rods are, like the dev∷gfp cells, uniformly fluorescent. The ability to see individual fluorescing dev∷gfp cells when they are released from within the fruiting body, and the fluorescence of individual pilA∷gfp peripheral rods argues that, had peripheral rods been expressing the dev-gfp fusion to an extent comparable to fruiting body cells, their fluorescence would have been evident in Fig. 1B.
Differentially Expressed Genes Depend on the Same Signal.
Is localized expression unique to dev? The answer evidently is no, because four other reporters that show localized expression are revealed by the data of Table 2. Expression of the gene reporters Ω7536, Ω4403, Ω4435, Ω4459, and Ω4414 (dev) are localized to the fruiting bodies, with activity ratios (fruiting bodies: peripheral rods) in the range 3.5–14.4. By contrast, four others, pilA, Ω4455, Ω4469, and Ω4521, are equally expressed in fruiting bodies and in peripheral rods. Within measurement error their activity ratios are 1, which is to say they are expressed equally throughout the developing population. No reporter was expressed only in peripheral rods. The pilA∷lacZ β-galactosidase assay data in Table 2 confirm the experiment with pilA∷gfp (Fig. 1D), and the Ω4414 data independently confirm the localization seen in Fig. 1B, noting that Ω4414 is fused at a different site in dev from that of gfp. It is striking that all of the localized genes are C-signal dependent, whereas no members of the nonlocalized group are C-signal dependent (Table 2). O'Connor and Zusman (1) had measured expression of several developmentally regulated proteins by western blotting and found them to be expressed in both peripheral rods and fruiting bodies. Among these proteins was protein S, which is A-signal dependent (30). A second A-signal-dependent gene, Ω4521 (31), which is nonlocalized is also shown in Table 2. The expression and failure to localize of these two genes argues that A-signaling precedes localization.
Table 2.
Expression of Tn5lacZ fusions in developing cells
| Strain | Expression
|
β-galactosidase specific activity*
|
Activity ratio§ | ||
|---|---|---|---|---|---|
| Start† | C-depend‡ | Fruiting bodies | Peripheral rods | ||
| pilA | 0 | − | 600 ± 52 | 512 ± 43 | 1.2 |
| Ω4521 | 2 | − | 632 ± 27 | 525 ± 79 | 1.2 |
| Ω4455 | 3 | − | 160 ± 16 | 152 ± 18 | 1.0 |
| Ω4469 | 4 | − | 647 ± 58 | 800 ± 85 | 0.8 |
| Ω4414 (dev) | 6 | + | 470 ± 48 | 107 ± 36 | 4.4 |
| Ω7536 | 8 | + | 347 ± 55 | 24 ± 2 | 14.4 |
| Ω4403 | 11 | + | 323 ± 14 | 66 ± 23 | 4.9 |
| Ω4459 | 15 | + | 295 ± 14 | 34 ± 11 | 8.7 |
| Ω4435 | 20 | + | 170 ± 40 | 48 ± 5 | 3.5 |
β-galactosidase specific activity for separated fruiting body cells and peripheral rods carrying the indicated Tn5lacZ fusions (15) was determined after 24 h of development on TPM starvation agar. Mean specific activity ± SD were determined from three independent experiments.
Hours after the initiation of development by starvation when each reporter starts its expression.
+ indicates that expression depends on C-signaling, and − indicates that it does not (33).
β-galactosidase specific activity in fruiting bodies divided by that in peripheral rods.
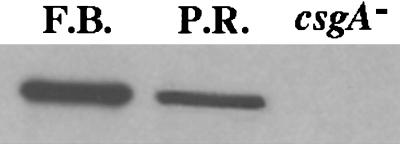
Because all of the genes and reporters that are localized to the fruiting body are C-signal-dependent, the levels of C-factor were measured in extracts of separated peripheral rods and fruiting body cells. SDS gels of the cell extracts were probed with CsgA-specific antibody, csgA being the gene that encodes C-factor (32). The third lane in Fig. 2 is an extract of a csgA mutant culture, and shows the specificity of the antibody. Photometric scanning of the blots shown in Fig. 2 reveals that fruiting body cells are expressing at least two to three times more C-factor than peripheral rods, equal amounts of total cell protein having been added to each lane. Because csgA expression in the whole culture rises 2- to 3-fold as a result of a C-signal-dependent positive feedback (6, 7), the possibility was considered that conditions necessary for C-signaling are less favorable in the peripheral rods than in fruiting body cells.
Figure 2.
CsgA-protein levels in nascent fruiting bodies and peripheral rods. Fruiting bodies (F.B.) and peripheral rods (P.R.) were isolated from TPM agar plates at 18 h of development as described in Materials and Methods. Protein extracts of each were separated by gel electrophoresis, and the proteins were exposed to CsgA-specific antibody, as described (7). An extract of a csgA mutant strain was similarly treated to test the specificity of the anti-CsgA antibody. The same total amount of protein was loaded into each lane.
Opportunities for C-Signaling Among the Peripheral Rods.
After induction by starvation, fruiting body development unfolds progressively from A-signaling to C-signaling (33). Because A-signal-dependent genes tps and Ω4521 are expressed normally in the peripheral rod cells (ref. 1, and Table 2), the peripheral rods have progressed normally through the early stages of development up to C-signaling. Previous work has shown that cell-to-cell transfer of C-signal is critically dependent on end-to-end contacts between signaling cells (34, 35). Individual M. xanthus cells, like those in Fig. 1A, have an average length to width ratio of 10:1. When elongated cells pack into islands, as do the peripheral rods in Fig. 1A, it is evident that the cells are frequently in side-by-side contact with each other, and this is supported by many microscopic studies in which the movement of cells within islands has been observed (27, 36). However, opportunities for end-to-end contact among peripheral rods have never been evaluated. Accordingly, the number of potential end-to-end contacts were counted in photomicrographs such as that shown in Fig. 1A. The cell arrangement and number were estimated from each island's size and shape, assuming that the island area is filled with a single layer of close packed rods having the size of Myxococcus cells. The maximum possible number of end-to-end contacts was obtained by assuming that all of the cells are aligned with their long axes parallel to the long axis of an island. (Alignment perpendicular to the long axis would give fewer such contacts. Mixed alignments would disrupt a regular lattice, and there would be fewer end-to-end contacts in such cases, as well.) The results of these measurements and calculations were compiled for the field shown in Fig. 1A, which has a total of 2,211 peripheral rods in 55 distinct islands, ranging in size from 6 to 198 cells. Other fields of peripheral rods, not quantified, were observed to have a qualitatively similar distribution of islands as the one in Fig. 1A. The overall average fraction of free cell-ends (ends that are not in contact with other ends) for the 2,211 cells was estimated as 0.35 ± 0.19, or about 1 end in 3. By contrast, the fraction of free cell ends in the outer domain of a nascent fruiting body appears to be much closer to zero (37, 38), a point to be taken up in Discussion.
Sporulation.
We argue that localized expression exists to ensure that spores form within the fruiting body. The dev locus is required for normal sporulation. Viable spore levels as low as 0.2% of wild type are observed in a dev insertion strain (Table 3), and even lower levels have been reported in other dev mutant strains (18). There is at least a 3- to 4-fold reduction in the number of spheroidal spore-like bodies visible in the light microscope, clearly indicating that the mutant has a defect in spore formation. The severe germination defect also evident in the data of Table 3 is understood as a consequence of the failure to form a proper spore.
Table 3.
Sporulation of dev mutants in submerged culture
| Strain | Percent wild-type sporulation
|
|
|---|---|---|
| Morphological spores* | Viable spores† | |
| DK1622 | 100.0 ± (2.9) | 100.0 ± (3.4) |
| DK4473 | 12.6 ± (3.0) | 5.4 ± (2.1) |
| DK5508 | 28.0 ± (1.3) | 0.2 ± (0.2) |
| DK11239 | 20.7 ± (1.1) | 1.3 ± (0.4) |
Development in submerged culture for 5 days. Each assay was made three times. The mean values for three independent assays with the standard deviations in parentheses are shown.
Values were determined by counting the number of sonication-resistant, spherical-shaped cells in a Petroff-Hausser chamber observed by phase-control microscopy.
Viable spore assay is described in Materials and Methods.
Discussion
Cellular Responses to C-Signaling.
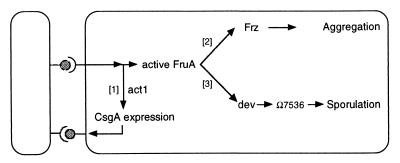
We suggest that the spatially localized expression pattern of sporulation genes is the product of differences between peripheral rod cells and nascent fruiting body cells in their dynamics of C-signaling. The pattern can be understood in terms of the C-signaling response pathway, diagramed in Fig. 3. Evidence supporting the circuit of Fig. 3 was described in the introduction, as well as the evidence for different response thresholds for its branches [1], [2], and [3].
Figure 3.
The C-signal transduction pathway. Both cells have the same transduction circuit, but for clarity the circuit is shown only in the cell on the right. Evidence for the pathway is described in the text.
CsgA protein, found on the cell surface (represented by lollipops in Fig. 3), transmits the C-signal (3, 5). C-signaling enhances csgA expression as a consequence of branch [1] Fig. 3. Following initial episodes of C-signaling, an increase in the amount of CsgA protein per cell is observed (6, 7). When such a signal-intensified cell transmits to another cell, the amount of activated FruA rises in the latter until it activates the frz phospho-relay, on branch [2] Fig. 3 (9, 39). The frz phospho-relay modulates cell movement parameters (13), leading cells to associate into mounds. Cells from the region surrounding a mound stream directly into that mound, a phenomenon clearly recorded in the time lapse films of H. Reichenbach (36). Inside the mound, the cells continue their circulation, but now in closed orbits, giving the mounds their spherical symmetry. Circulation is clear from the pulsations of the mounds (36), and from the tracking of cells (38).
As more and more cells congregate within a mound, the cell density rises, and the number of collisions between cells per unit time increases, further reinforcing the increases in the CsgA levels because of signaling. Once the increase in cell density and the positive feedback have pushed the level of C-signaling to high values, expression of dev is induced and subsequently sporulation, branch [3]. Recently, the Ω7536 operon has been shown to be directly involved in the cell shape change from a rod to a sphere (40). Expression of Ω7536 depends on dev (40). Moreover, dev is also required for expression of two other presumed spore proteins, Ω4401 and Ω4435. Release of β-galactosidase from these fusion strains requires that the spores be opened (ref. 15, and A. Garza, unpublished observations). Based on these findings, the FACS experiments described in the introduction, and the requirement of dev for sporulation, we propose that dev acts as a sporulation switch, activating a family of genes, which includes Ω7536, Ω4401, and Ω4435, functions involved in the transformation of a rod cell into a spherical spore.
Gene Expression Localized to the Nascent Fruiting Body.
The cells in the outer domain of a nascent fruiting body are at high density, regularly close-packed, and aligned (37, 38). Cells in the outer domain are moving in generally circular orbits (36, 37). Together, these observations suggest that the rod-shaped cells are flowing like logs at high density in a rapidly moving stream, and consequently would be nudging each other end-to-end. C-signaling depends on end-to-end contacts; it fails when cells are unable to make them even though they are able to make side-by-side contacts (34, 35, 41). As a consequence, C-signaling will only rise to a high level in the cells of the outer domain of a nascent fruiting body and induce sporulation after the fruiting body cell density rises to some threshold level.
A different state is found among the peripheral rod cells. They are grouped as islands of cells that are separated from each other, and they are only occasionally moving. Island cells are in side-by-side contact with each other (Fig. 1A), and because of the shape of the islands and the gaps between them, an estimated one-third of the ends of island cells are not in contact with other ends. As a consequence, C-signaling occurs at a lower level than in the outer domain of the fruiting body. The measurements of CsgA protein in the peripheral rod cells by Western blot (Fig. 2) show that they do have the signaling protein. However, cells within nascent fruiting bodies are C-signaling at higher intensity than peripheral rods because the former have accumulated higher levels of CsgA protein by positive feedback than the latter. A higher intensity of C-signaling within nascent fruiting bodies and the consequent localized gene expression are the major new experimental and conceptual results of this work.
Expression of dev and sporulation genes like Ω7536 depends on the highest level of C-signaling in M. xanthus. There are many different species of myxobacteria building widely different fruiting body structures, including some with stalks and multiple branches. No matter what the morphology, spore differentiation appears to be delayed until the cell movements that construct the appropriate multicellular structure have been completed (42–45). Chondromyces apiculatus, which has a stalk and branches, sporulates only after the tips of the branches have achieved proper form (46). Stigmatella aurantiaca, which also has a stalk and branches, has a csgA gene that has also been shown to be important for its sporulation and is related to that of M. xanthus (47). The myxobacterial fruiting body is an organ constructed to disperse its cells (48, 49). A coupling of spore differentiation to fruiting body morphogenesis ensures that spores form only within the fruiting body. This macroscopic spore-filled mass can then be carried to a new nutrient-rich location, by a small animal hunting food in the soil where the fruiting bodies are formed. Indeed, mites have been observed to carry fruiting bodies (50). Elevating and packaging the spores in a fruiting body is thus important for the survival of myxobacteria in nature.
Acknowledgments
We thank T. Kruse, S. Lobendanz, N. Berthelsen, and L. Sogaard-Andersen (Odense University) for the antiserum to CsgA. This investigation was supported by U.S. Public Health Service Grant GM 23441 (to D.K.) from the National Institute of General Medical Sciences, and a postdoctoral fellowship GM 18530 from the National Institute of General Medical Sciences, National Institutes of Health (to B.J.).
Abbreviations
- FACS
fluorescence-activated cell sorter
- GFP
green fluorescent protein
References
- 1.O'Connor K A, Zusman D R. J Bacteriol. 1991;173:3318–3333. doi: 10.1128/jb.173.11.3318-3333.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dworkin M, Gibson S. Science. 1964;146:243–244. doi: 10.1126/science.146.3641.243. [DOI] [PubMed] [Google Scholar]
- 3.Shimkets L J, Rafiee H. J Bacteriol. 1990;172:5299–5306. doi: 10.1128/jb.172.9.5299-5306.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shimkets L, Kaiser D. J Bacteriol. 1982;152:462–470. doi: 10.1128/jb.152.1.462-470.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim S K, Kaiser D. Proc Natl Acad Sci USA. 1990;87:3635–3639. doi: 10.1073/pnas.87.10.3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim S K, Kaiser D. J Bacteriol. 1991;173:1722–1728. doi: 10.1128/jb.173.5.1722-1728.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gorski L, Gronewold T, Kaiser D. J Bacteriol. 2000;182:2438–2444. doi: 10.1128/jb.182.9.2438-2444.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogawa M, Fujitani S, Mao X, Inouye S, Komano T. Mol Microbiol. 1996;22:757–767. doi: 10.1046/j.1365-2958.1996.d01-1725.x. [DOI] [PubMed] [Google Scholar]
- 9.Ellehauge E, Norregaard-Madsen M, Søgaard-Andersen L. Mol Microbiol. 1998;30:807–813. doi: 10.1046/j.1365-2958.1998.01113.x. [DOI] [PubMed] [Google Scholar]
- 10.McCleary W R, Zusman D R. J Bacteriol. 1990;172:6661–6668. doi: 10.1128/jb.172.12.6661-6668.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McBride M, Kohler T, Zusman D. J Bacteriol. 1992;174:4246–4257. doi: 10.1128/jb.174.13.4246-4257.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Søgaard-Andersen L, Slack F, Kimsey H, Kaiser D. Genes Dev. 1996;10:740–754. doi: 10.1101/gad.10.6.740. [DOI] [PubMed] [Google Scholar]
- 13.Jelsbak L, Søgaard-Andersen L. Proc Natl Acad Sci USA. 1999;96:5031–5036. doi: 10.1073/pnas.96.9.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li S, Lee B U, Shimkets L. Genes Dev. 1992;6:401–410. doi: 10.1101/gad.6.3.401. [DOI] [PubMed] [Google Scholar]
- 15.Kroos L, Kuspa A, Kaiser D. Dev Biol. 1986;117:252–266. doi: 10.1016/0012-1606(86)90368-4. [DOI] [PubMed] [Google Scholar]
- 16.Kroos L, Kaiser D. Proc Natl Acad Sci USA. 1984;81:5816–5820. doi: 10.1073/pnas.81.18.5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Russo-Marie F, Roederer M, Sager B, Herzenberg L A, Kaiser D. Proc Natl Acad Sci USA. 1993;90:8194–8198. doi: 10.1073/pnas.90.17.8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thony-Meyer L, Kaiser D. J Bacteriol. 1993;175:7450–7462. doi: 10.1128/jb.175.22.7450-7462.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Julien B, Calendar R. J Bacteriol. 1995;177:3743–3751. doi: 10.1128/jb.177.13.3743-3751.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Campos J, Zusman D. Proc Natl Acad Sci USA. 1975;72:518–522. doi: 10.1073/pnas.72.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 22.Avery L, Kaiser D. Mol Gen Genet. 1983;191:99–109. doi: 10.1007/BF00330896. [DOI] [PubMed] [Google Scholar]
- 23.Kashefi K, Hartzell P L. Mol Microbiol. 1995;15:483–494. doi: 10.1111/j.1365-2958.1995.tb02262.x. [DOI] [PubMed] [Google Scholar]
- 24.Kuner J, Kaiser D. J Bacteriol. 1982;151:458–461. doi: 10.1128/jb.151.1.458-461.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cormack B P, Valdivia R H, Falkow S. Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 26.Ueki T, Inouye S, Inouye M. Gene. 1996;183:153–157. doi: 10.1016/s0378-1119(96)00546-x. [DOI] [PubMed] [Google Scholar]
- 27.Kaiser D, Crosby C. Cell Motil. 1983;3:227–245. [Google Scholar]
- 28.Wu S S, Kaiser D. Mol Microbiol. 1995;18:547–558. doi: 10.1111/j.1365-2958.1995.mmi_18030547.x. [DOI] [PubMed] [Google Scholar]
- 29.Wu S S, Kaiser D. J Bacteriol. 1997;179:7748–7758. doi: 10.1128/jb.179.24.7748-7758.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaiser D, Kroos L. In: Myxobacteria II. Dworkin M, Kaiser D, editors. Washington, DC: Am. Soc. Microbiol.; 1993. pp. 257–283. [Google Scholar]
- 31.Kaplan H B, Kuspa A, Kaiser D. J Bacteriol. 1991;173:1460–1470. doi: 10.1128/jb.173.4.1460-1470.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimkets L J, Gill R E, Kaiser D. Proc Natl Acad Sci USA. 1983;80:1406–1410. doi: 10.1073/pnas.80.5.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kroos L, Kaiser D. Genes Dev. 1987;1:840–854. doi: 10.1101/gad.1.8.840. [DOI] [PubMed] [Google Scholar]
- 34.Kim S K, Kaiser D. Science. 1990;249:926–928. doi: 10.1126/science.2118274. [DOI] [PubMed] [Google Scholar]
- 35.Sager B, Kaiser D. Genes Dev. 1994;8:2793–2804. doi: 10.1101/gad.8.23.2793. [DOI] [PubMed] [Google Scholar]
- 36.Reichenbach H, Heunert H H, Kuczka H. Film C 893. Gottingen, Germany: Inst. Wissensch. Film; 1965. [Google Scholar]
- 37.Sager B, Kaiser D. Genes Dev. 1993;7:1645–1653. doi: 10.1101/gad.7.9.1645. [DOI] [PubMed] [Google Scholar]
- 38.Sager B, Kaiser D. Proc Natl Acad Sci USA. 1993;90:3690–3694. doi: 10.1073/pnas.90.8.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Søgaard-Andersen L, Kaiser D. Proc Natl Acad Sci USA. 1996;93:2675–2679. doi: 10.1073/pnas.93.7.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Licking E, Gorski L, Kaiser D. J Bacteriol. 2000;182:3553–3558. doi: 10.1128/jb.182.12.3553-3558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kroos L, Hartzell P, Stephens K, Kaiser D. Genes Dev. 1988;2:1677–1685. doi: 10.1101/gad.2.12a.1677. [DOI] [PubMed] [Google Scholar]
- 42.Reichenbach H. In: Myxobacteria. Rosenberg E, editor. New York: Springer; 1984. pp. 1–50. [Google Scholar]
- 43.Wireman J, Dworkin M. J Bacteriol. 1977;129:796–802. doi: 10.1128/jb.129.2.798-802.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qualls G T, Stephens K, White D. Dev Biol. 1978;66:270–274. doi: 10.1016/0012-1606(78)90291-9. [DOI] [PubMed] [Google Scholar]
- 45.Kaiser D. Annu Rev Genet. 1986;20:539–566. doi: 10.1146/annurev.ge.20.120186.002543. [DOI] [PubMed] [Google Scholar]
- 46.Reichenbach H. Film E 779/1965. Gottingen, Germany: Inst. Wissensch. Film; 1974. [Google Scholar]
- 47.Neumann B, Pospiech A, Schairer H U. Mol Microbiol. 1993;10:1087–1099. doi: 10.1111/j.1365-2958.1993.tb00979.x. [DOI] [PubMed] [Google Scholar]
- 48.Bonner J T. Am Nat. 1982;119:530–552. [Google Scholar]
- 49.Kaiser D. Trends Genet. 1999;15:273–277. doi: 10.1016/s0168-9525(99)01740-0. [DOI] [PubMed] [Google Scholar]
- 50.Reichenbach H. In: Myxobacteria II. Dworkin M, Kaiser D, editors. Washington, DC: Am. Soc. Microbiol.; 1993. pp. pp.13–62. [Google Scholar]
- 51.Kaiser D. Proc Natl Acad Sci USA. 1979;76:5952–5956. doi: 10.1073/pnas.76.11.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]