Abstract
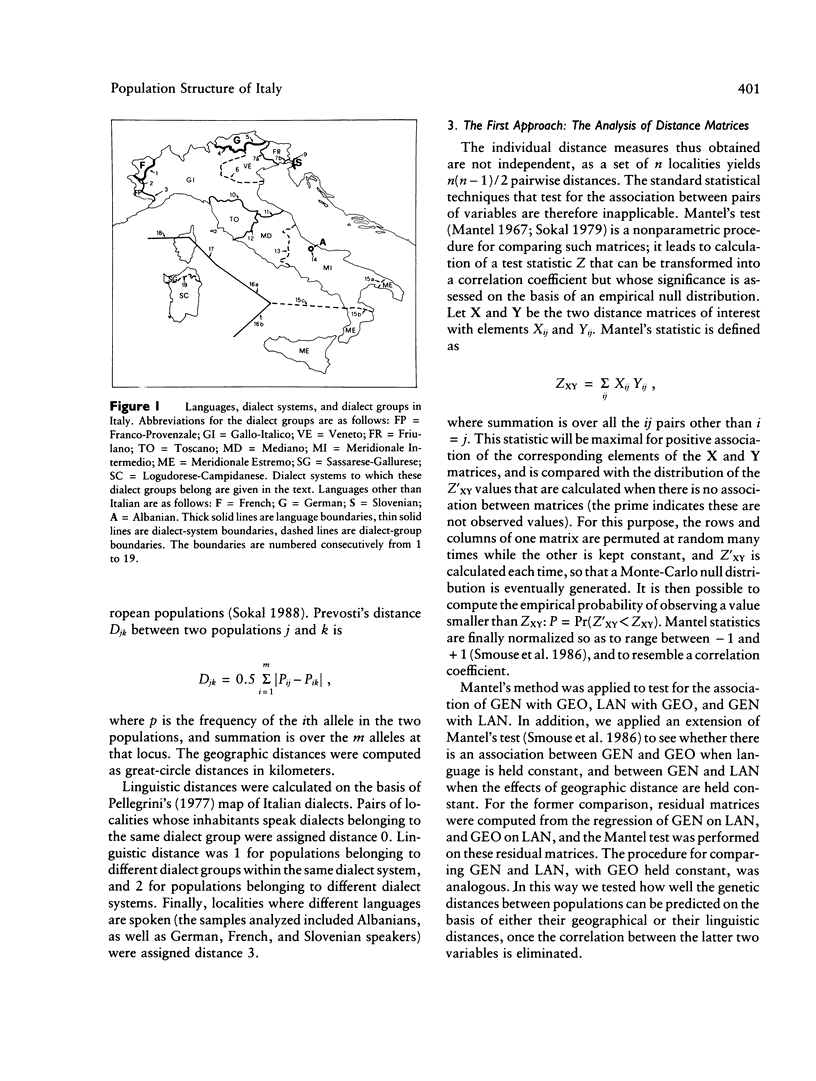
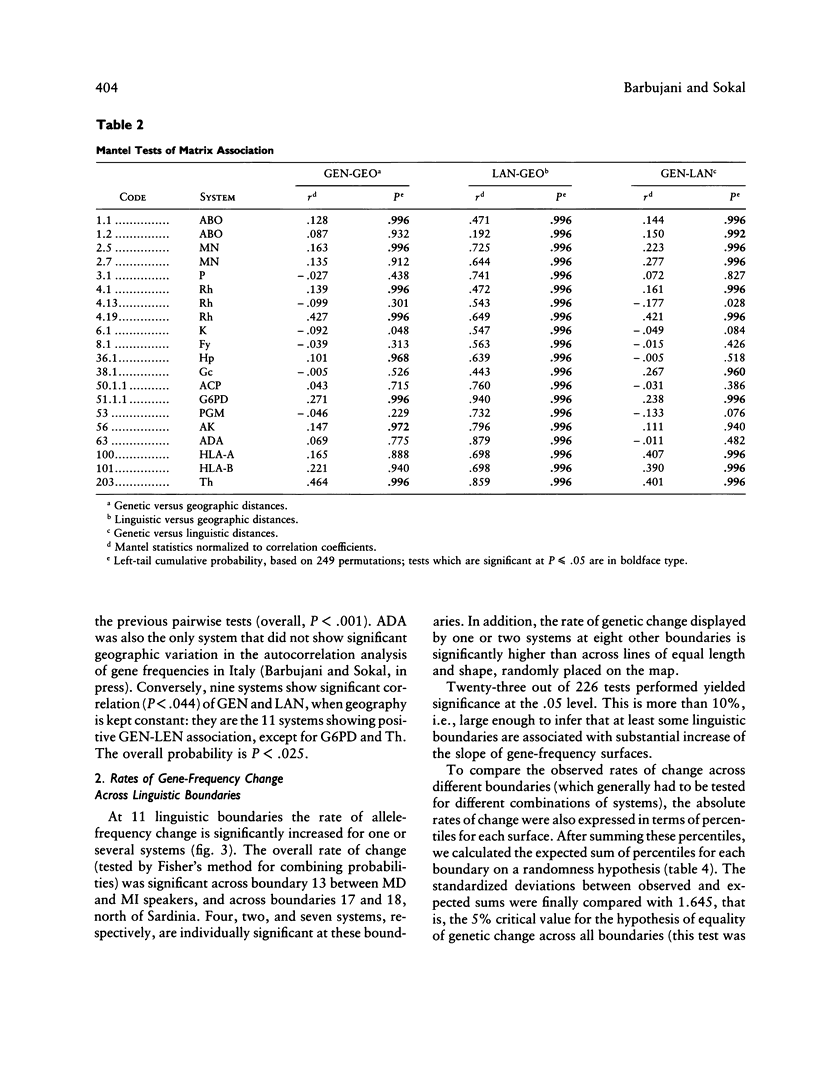
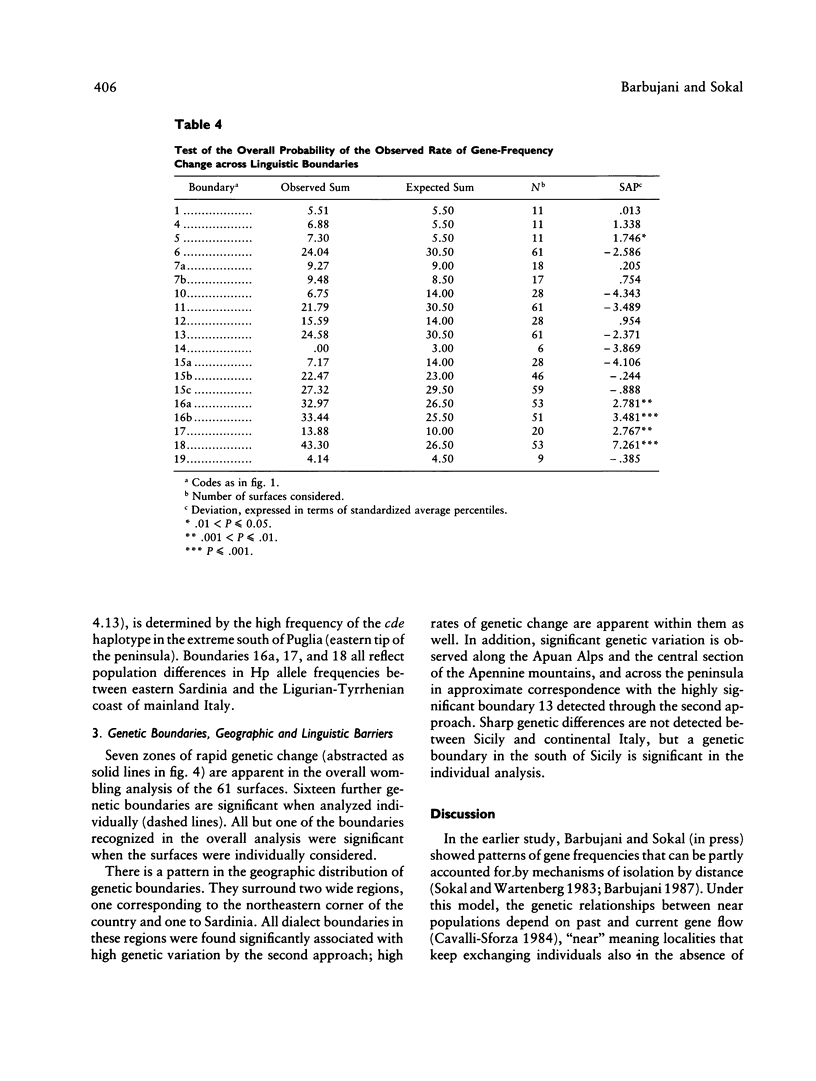
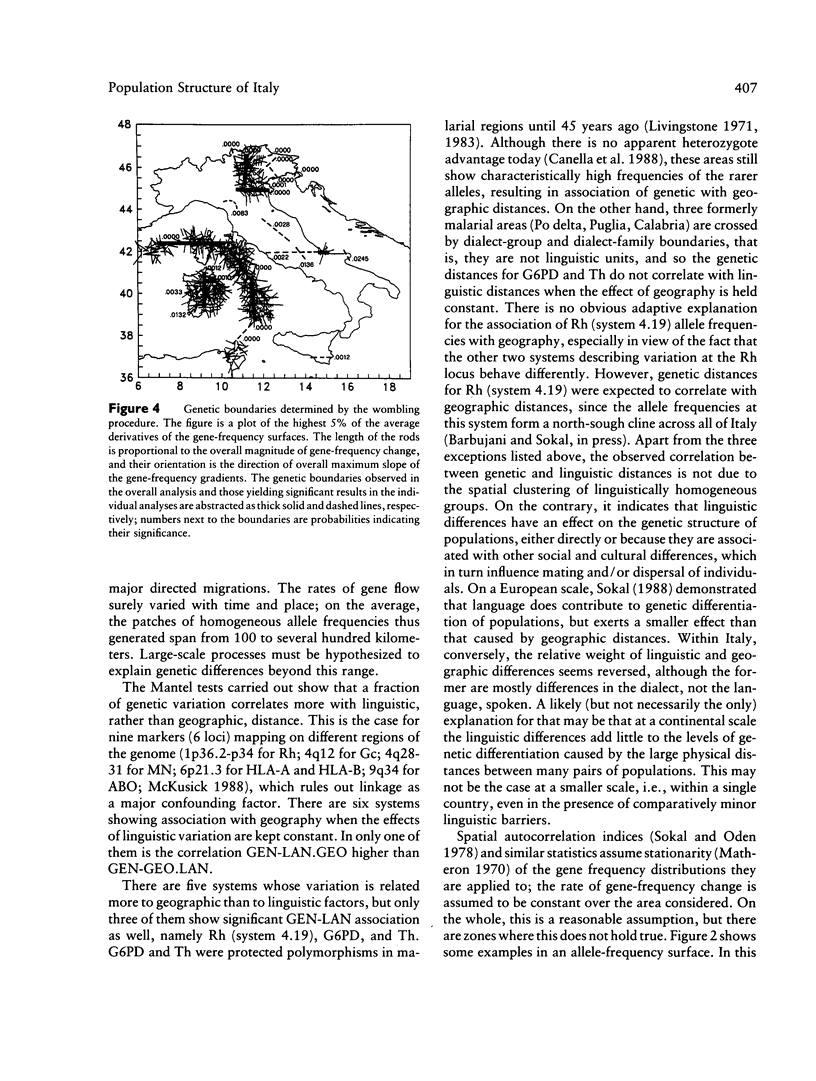
Three approaches were employed to evaluate the relative importance of geographic and linguistic factors in maintaining genetic differentiation of Italian populations as shown by blood groups and erythrocyte and serum markers. Genetic distances are closer to linguistic than to geographic distances. Gene-frequency change across 12 linguistic boundaries is significantly more rapid than at random locations. The zones of sharp genetic variation correspond to physical barriers to gene flow and to boundaries between dialect families, which overlap widely. However, two linguistically differentiated populations appear genetically differentiated despite the absence of physical obstacles to gene flow around them. The Po River is associated with abrupt genetic change only in the area where it corresponds to a dialect boundary. At most loci the genetic population structure seems affected by linguistic rather than geographic factors; exceptions are the systems that were subject to malarial selection in geographically close but linguistically heterogeneous localities. Gene flow appears to homogenize gene frequencies within regions corresponding to dialect families but not between them, leading to the patchy distributions of allele frequencies that were detected in an earlier study.
Full text
PDF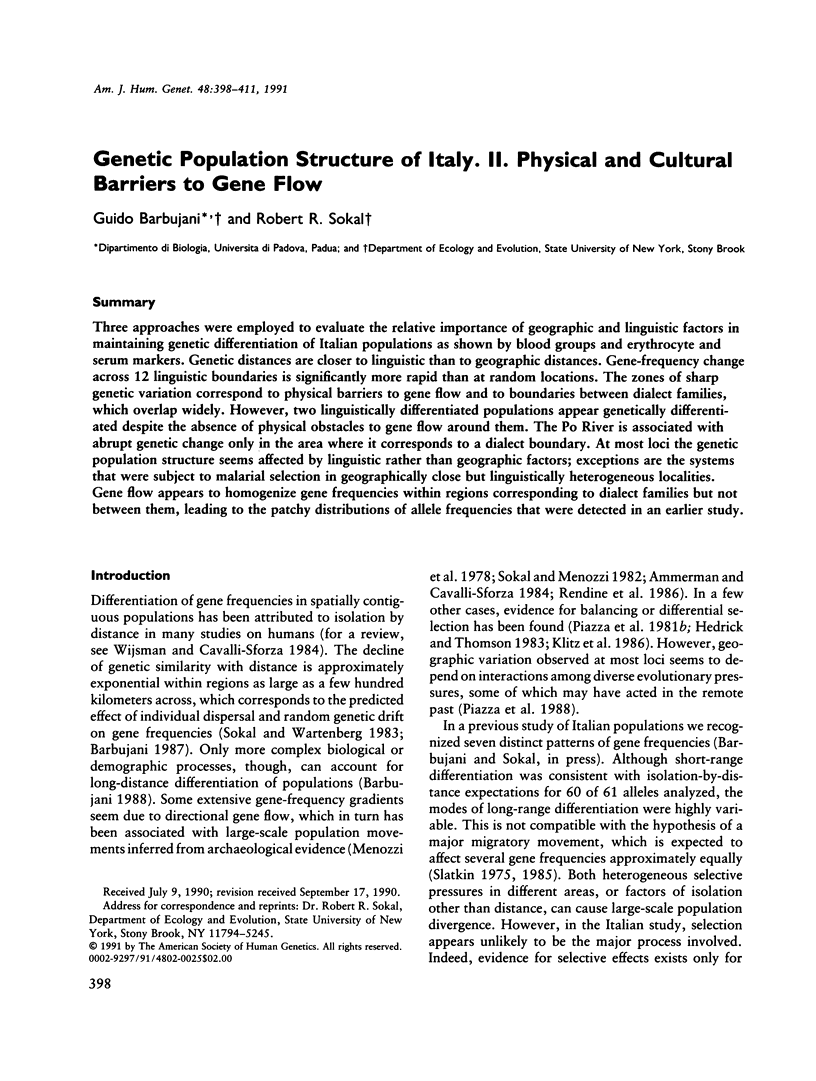









Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barbujani G., Sokal R. R. Zones of sharp genetic change in Europe are also linguistic boundaries. Proc Natl Acad Sci U S A. 1990 Mar;87(5):1816–1819. doi: 10.1073/pnas.87.5.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrantes R., Smouse P. E., Mohrenweiser H. W., Gershowitz H., Azofeifa J., Arias T. D., Neel J. V. Microevolution in lower Central America: genetic characterization of the Chibcha-speaking groups of Costa Rica and Panama, and a consensus taxonomy based on genetic and linguistic affinity. Am J Hum Genet. 1990 Jan;46(1):63–84. [PMC free article] [PubMed] [Google Scholar]
- Barton N. H., Jones J. S. Evolution. The language of the genes. Nature. 1990 Aug 2;346(6283):415–416. doi: 10.1038/346415a0. [DOI] [PubMed] [Google Scholar]
- Beretta M., Mazzetti P., Frosina G., Schilirò G., Russo A., Russo G., Barrai I. Population structure of eastern Sicily. Hum Hered. 1986;36(6):379–387. doi: 10.1159/000153662. [DOI] [PubMed] [Google Scholar]
- Canella R., Barbujani G., Cucchi P., Siniscalco M., Vullo C., Barrai I. Biological performance in beta-thal heterozygotes and normals: results of a longitudinal comparison in a former malarial environment. Ann Hum Genet. 1987 Oct;51(Pt 4):337–343. doi: 10.1111/j.1469-1809.1987.tb01068.x. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza L. L., Edwards A. W. Phylogenetic analysis. Models and estimation procedures. Am J Hum Genet. 1967 May;19(3 Pt 1):233–257. [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza L. L., Piazza A., Menozzi P., Mountain J. Reconstruction of human evolution: bringing together genetic, archaeological, and linguistic data. Proc Natl Acad Sci U S A. 1988 Aug;85(16):6002–6006. doi: 10.1073/pnas.85.16.6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza L. L. Population structure and human evolution. Proc R Soc Lond B Biol Sci. 1966 Mar 22;164(995):362–379. doi: 10.1098/rspb.1966.0038. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. How can we infer geography and history from gene frequencies? J Theor Biol. 1982 May 7;96(1):9–20. doi: 10.1016/0022-5193(82)90152-7. [DOI] [PubMed] [Google Scholar]
- Friedlaender J. S., Sgaramella-Zonta L. A., Kidd K. K., Lai L. Y., Clark P., Walsh R. J. Biological divergences in south-central Bougainville: an analysis of blood polymorphism gene frequencies and anthropometric measurements utilizing tree models, and a comparison of these variables with linguistic, geographic, and migrational "distances". Am J Hum Genet. 1971 May;23(3):253–270. [PMC free article] [PubMed] [Google Scholar]
- Harding R. M., Sokal R. R. Classification of the European language families by genetic distance. Proc Natl Acad Sci U S A. 1988 Dec;85(23):9370–9372. doi: 10.1073/pnas.85.23.9370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick P. W., Thomson G. Evidence for balancing selection at HLA. Genetics. 1983 Jul;104(3):449–456. doi: 10.1093/genetics/104.3.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klitz W., Thomson G., Baur M. P. Contrasting evolutionary histories among tightly linked HLA loci. Am J Hum Genet. 1986 Sep;39(3):340–349. [PMC free article] [PubMed] [Google Scholar]
- Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967 Feb;27(2):209–220. [PubMed] [Google Scholar]
- Piazza A., Olivetti E., Barbanti M., Reali G., Domenici R., Giari A., Benciolini P., Caenazzo L., Cortivo P., Bestetti A. The distribution of some polymorphisms in Italy. Gene Geogr. 1989 Aug-Dec;3(2-3):69–139. [PubMed] [Google Scholar]
- Piazza A., Rendine S., Zei G., Moroni A., Cavalli-Sforza L. L. Migration rates of human populations from surname distributions. Nature. 1987 Oct 22;329(6141):714–716. doi: 10.1038/329714a0. [DOI] [PubMed] [Google Scholar]
- Salzano F. M., Neel J. V., Gershowitz H., Migliazza E. C. Intra and intertribal genetic variation within a linguistic group: the Ge-speaking indians of Brazil. Am J Phys Anthropol. 1977 Sep;47(2):337–347. doi: 10.1002/ajpa.1330470214. [DOI] [PubMed] [Google Scholar]
- Slatkin M. Gene flow and selection in a two-locus system. Genetics. 1975 Dec;81(4):787–802. doi: 10.1093/genetics/81.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal R. R., Harding R. M., Oden N. L. Spatial patterns of human gene frequencies in Europe. Am J Phys Anthropol. 1989 Nov;80(3):267–294. doi: 10.1002/ajpa.1330800302. [DOI] [PubMed] [Google Scholar]
- Sokal R. R., Oden N. L., Thomson B. A. Genetic changes across language boundaries in Europe. Am J Phys Anthropol. 1988 Jul;76(3):337–361. doi: 10.1002/ajpa.1330760308. [DOI] [PubMed] [Google Scholar]
- Sokal R. R., Wartenberg D. E. A Test of Spatial Autocorrelation Analysis Using an Isolation-by-Distance Model. Genetics. 1983 Sep;105(1):219–237. doi: 10.1093/genetics/105.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WOMBLE W. H. Differential systematics. Science. 1951 Sep 28;114(2961):315–322. doi: 10.1126/science.114.2961.315. [DOI] [PubMed] [Google Scholar]
- White N. G., Parsons P. A. Genetic and socio-cultural differentiation in the aborigines of Arnhem Land, Australia. Am J Phys Anthropol. 1973 Jan;38(1):5–14. doi: 10.1002/ajpa.1330380106. [DOI] [PubMed] [Google Scholar]



