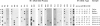Abstract
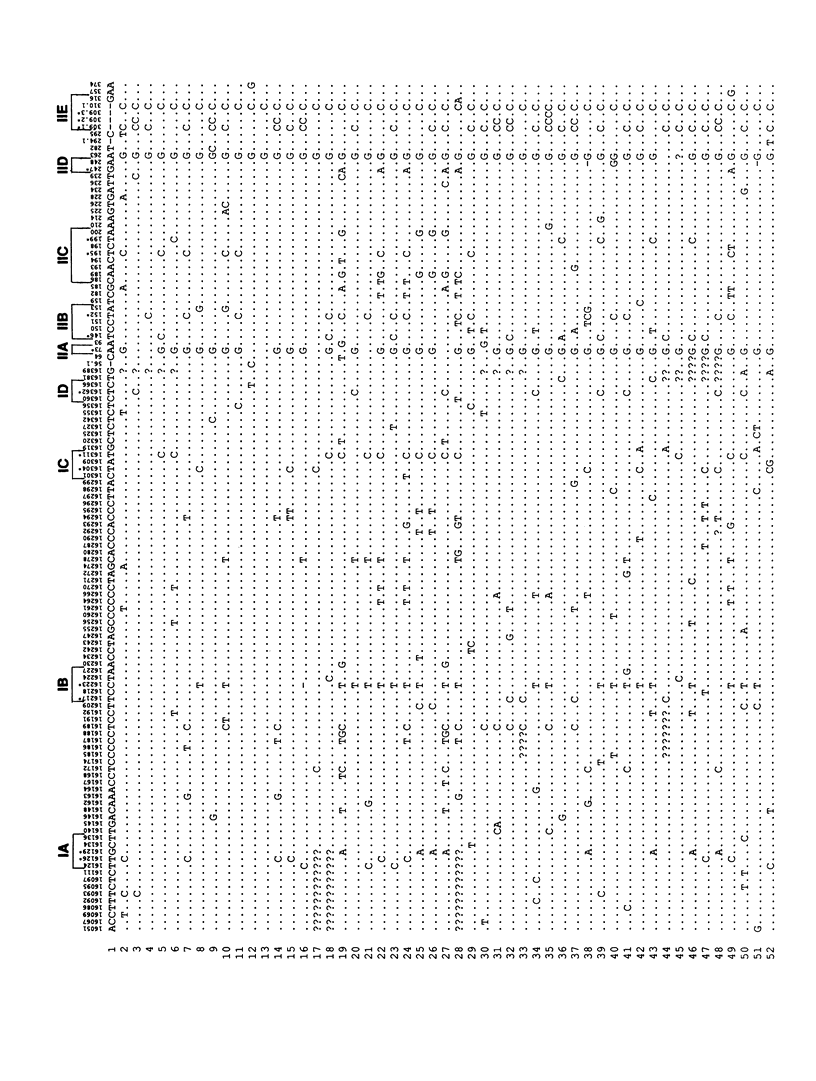
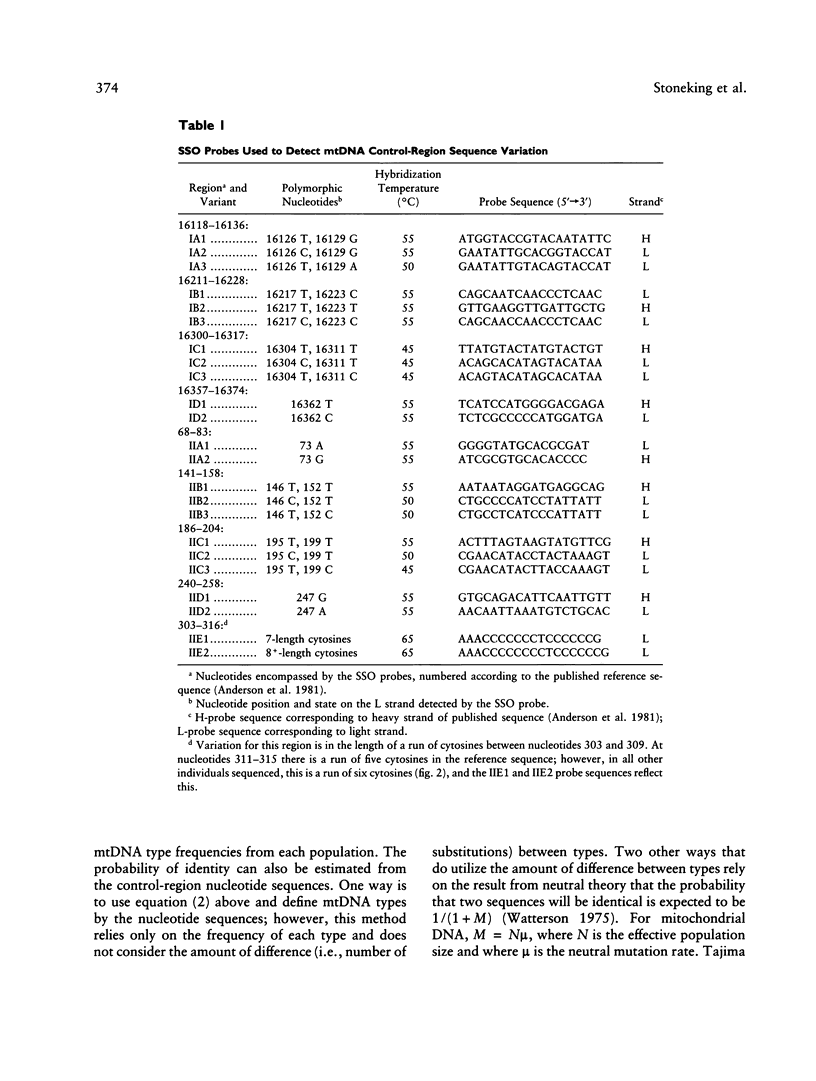
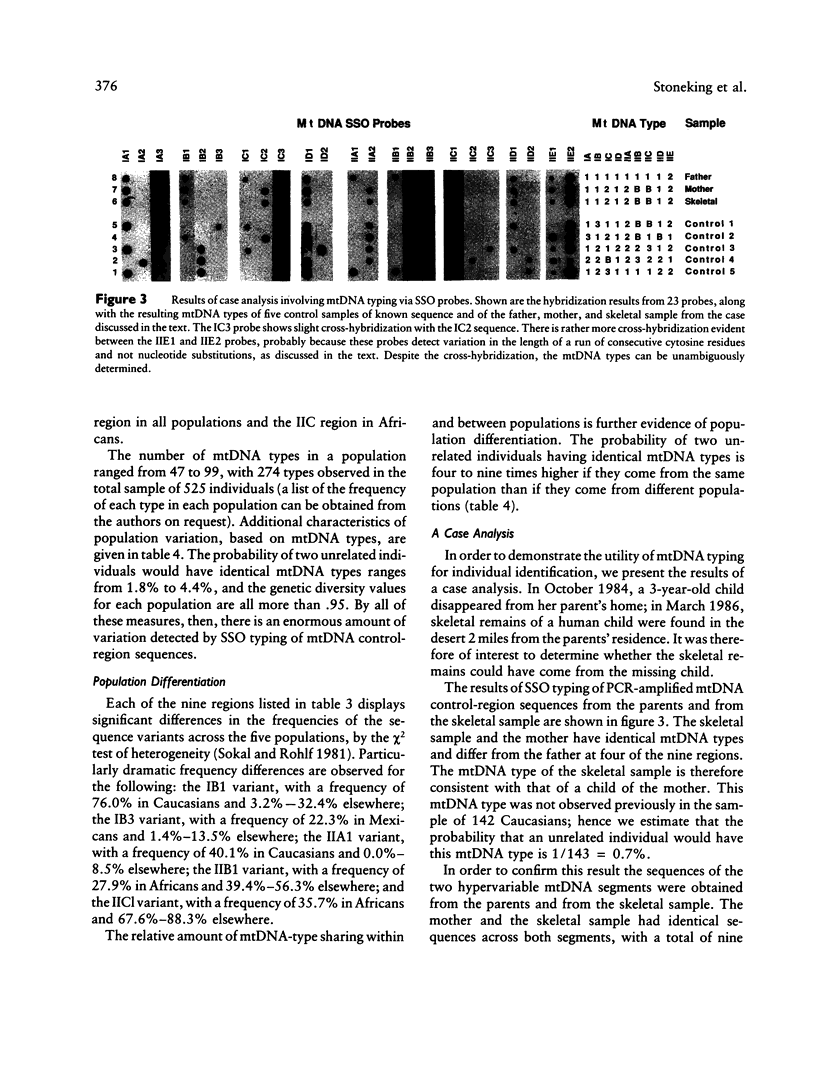
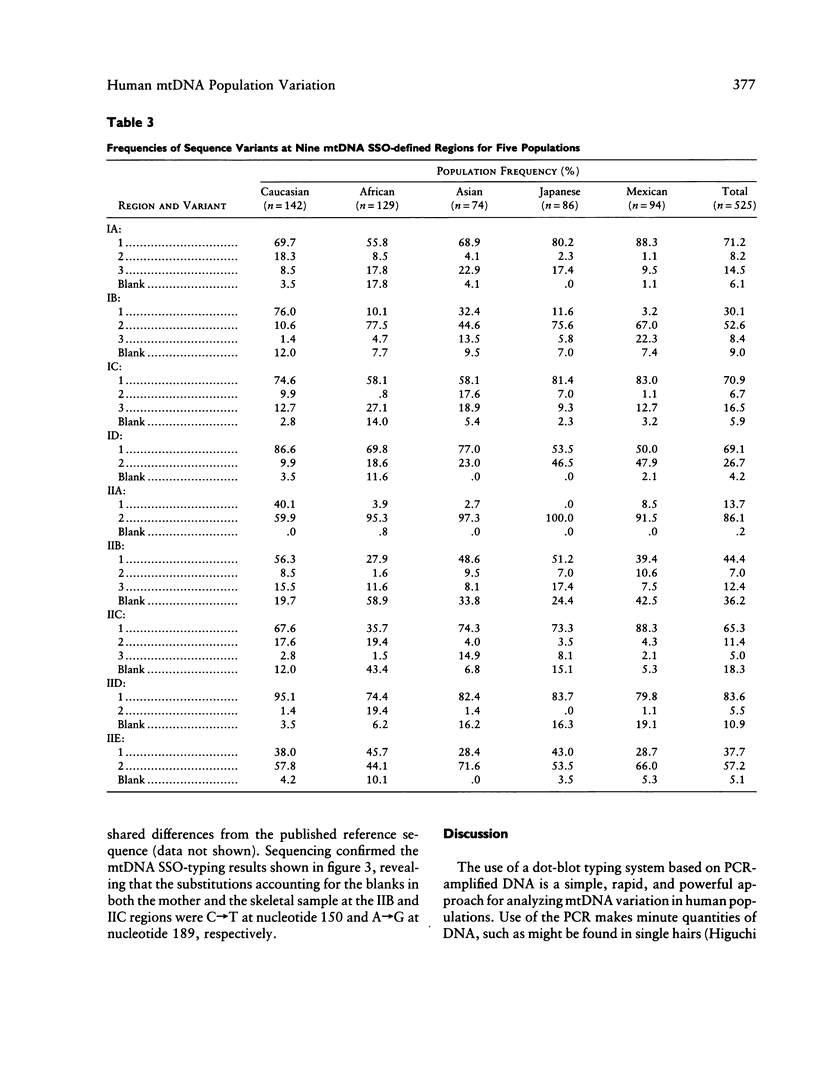
A method for detecting sequence variation of hypervariable segments of the mtDNA control region was developed. The technique uses hybridization of sequence-specific oligonucleotide (SSO) probes to DNA sequences that have been amplified by PCR. The nucleotide sequences of the two hypervariable segments of the mtDNA control region from 52 individuals were determined; these sequences were then used to define nine regions suitable for SSO typing. A total of 23 SSO probes were used to detect sequence variants at these nine regions in 525 individuals from five ethnic groups (African, Asian, Caucasian, Japanese, and Mexican). The SSO typing revealed an enormous amount of variability, with 274 mtDNA types observed among these 525 individuals and with diversity values, for each population, exceeding .95. For each of the nine mtDNA regions significant differences in the frequencies of sequence variants were observed between these five populations. The mtDNA SSO-typing system was successfully applied to a case involving individual identification of skeletal remains; the probability of a random match was approximately 0.7%. The potential useful applications of this mtDNA SSO-typing system thus include the analysis of individual identity as well as population genetic studies.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F. Sequence and organization of the human mitochondrial genome. Nature. 1981 Apr 9;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Aquadro C. F., Greenberg B. D. Human mitochondrial DNA variation and evolution: analysis of nucleotide sequences from seven individuals. Genetics. 1983 Feb;103(2):287–312. doi: 10.1093/genetics/103.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazs I., Baird M., Clyne M., Meade E. Human population genetic studies of five hypervariable DNA loci. Am J Hum Genet. 1989 Feb;44(2):182–190. [PMC free article] [PubMed] [Google Scholar]
- Cann R. L., Brown W. M., Wilson A. C. Polymorphic sites and the mechanism of evolution in human mitochondrial DNA. Genetics. 1984 Mar;106(3):479–499. doi: 10.1093/genetics/106.3.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann R. L., Stoneking M., Wilson A. C. Mitochondrial DNA and human evolution. Nature. 1987 Jan 1;325(6099):31–36. doi: 10.1038/325031a0. [DOI] [PubMed] [Google Scholar]
- Cohen J. E. DNA fingerprinting for forensic identification: potential effects on data interpretation of subpopulation heterogeneity and band number variability. Am J Hum Genet. 1990 Feb;46(2):358–368. [PMC free article] [PubMed] [Google Scholar]
- Giles R. E., Blanc H., Cann H. M., Wallace D. C. Maternal inheritance of human mitochondrial DNA. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6715–6719. doi: 10.1073/pnas.77.11.6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedde H. W., Singh S., Agarwal D. P., Fritze G., Stapel K., Paik Y. K. Genotyping of mitochondrial aldehyde dehydrogenase in blood samples using allele-specific oligonucleotides: comparison with phenotyping in hair roots. Hum Genet. 1989 Mar;81(4):305–307. doi: 10.1007/BF00283679. [DOI] [PubMed] [Google Scholar]
- Gyllensten U. B., Erlich H. A. Generation of single-stranded DNA by the polymerase chain reaction and its application to direct sequencing of the HLA-DQA locus. Proc Natl Acad Sci U S A. 1988 Oct;85(20):7652–7656. doi: 10.1073/pnas.85.20.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht N. B., Liem H., Kleene K. C., Distel R. J., Ho S. M. Maternal inheritance of the mouse mitochondrial genome is not mediated by a loss or gross alteration of the paternal mitochondrial DNA or by methylation of the oocyte mitochondrial DNA. Dev Biol. 1984 Apr;102(2):452–461. doi: 10.1016/0012-1606(84)90210-0. [DOI] [PubMed] [Google Scholar]
- Helmuth R., Fildes N., Blake E., Luce M. C., Chimera J., Madej R., Gorodezky C., Stoneking M., Schmill N., Klitz W. HLA-DQ alpha allele and genotype frequencies in various human populations, determined by using enzymatic amplification and oligonucleotide probes. Am J Hum Genet. 1990 Sep;47(3):515–523. [PMC free article] [PubMed] [Google Scholar]
- Higuchi R., von Beroldingen C. H., Sensabaugh G. F., Erlich H. A. DNA typing from single hairs. Nature. 1988 Apr 7;332(6164):543–546. doi: 10.1038/332543a0. [DOI] [PubMed] [Google Scholar]
- Horai S., Hayasaka K. Intraspecific nucleotide sequence differences in the major noncoding region of human mitochondrial DNA. Am J Hum Genet. 1990 Apr;46(4):828–842. [PMC free article] [PubMed] [Google Scholar]
- Ikuta S., Takagi K., Wallace R. B., Itakura K. Dissociation kinetics of 19 base paired oligonucleotide-DNA duplexes containing different single mismatched base pairs. Nucleic Acids Res. 1987 Jan 26;15(2):797–811. doi: 10.1093/nar/15.2.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys A. J., Wilson V., Thein S. L. Individual-specific 'fingerprints' of human DNA. Nature. 1985 Jul 4;316(6023):76–79. doi: 10.1038/316076a0. [DOI] [PubMed] [Google Scholar]
- Lander E. S. DNA fingerprinting on trial. Nature. 1989 Jun 15;339(6225):501–505. doi: 10.1038/339501a0. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Leppert M., O'Connell P., Wolff R., Holm T., Culver M., Martin C., Fujimoto E., Hoff M., Kumlin E. Variable number of tandem repeat (VNTR) markers for human gene mapping. Science. 1987 Mar 27;235(4796):1616–1622. doi: 10.1126/science.3029872. [DOI] [PubMed] [Google Scholar]
- Robin E. D., Wong R. Mitochondrial DNA molecules and virtual number of mitochondria per cell in mammalian cells. J Cell Physiol. 1988 Sep;136(3):507–513. doi: 10.1002/jcp.1041360316. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Bugawan T. L., Horn G. T., Mullis K. B., Erlich H. A. Analysis of enzymatically amplified beta-globin and HLA-DQ alpha DNA with allele-specific oligonucleotide probes. Nature. 1986 Nov 13;324(6093):163–166. doi: 10.1038/324163a0. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Walsh P. S., Levenson C. H., Erlich H. A. Genetic analysis of amplified DNA with immobilized sequence-specific oligonucleotide probes. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6230–6234. doi: 10.1073/pnas.86.16.6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigilant L., Pennington R., Harpending H., Kocher T. D., Wilson A. C. Mitochondrial DNA sequences in single hairs from a southern African population. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9350–9354. doi: 10.1073/pnas.86.23.9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walberg M. W., Clayton D. A. Sequence and properties of the human KB cell and mouse L cell D-loop regions of mitochondrial DNA. Nucleic Acids Res. 1981 Oct 24;9(20):5411–5421. doi: 10.1093/nar/9.20.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson G. A. On the number of segregating sites in genetical models without recombination. Theor Popul Biol. 1975 Apr;7(2):256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- Whittam T. S., Clark A. G., Stoneking M., Cann R. L., Wilson A. C. Allelic variation in human mitochondrial genes based on patterns of restriction site polymorphism. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9611–9615. doi: 10.1073/pnas.83.24.9611. [DOI] [PMC free article] [PubMed] [Google Scholar]