Abstract
A 105-kilobase bacterial artificial chromosome (BAC) clone from the ovate-containing region of tomato chromosome 2 was sequenced and annotated. The tomato BAC sequence was then compared, gene by gene, with the sequenced portions of the Arabidopsis thaliana genome. Rather than matching a single portion of the Arabidopsis genome, the tomato clone shows conservation of gene content and order with four different segments of Arabidopsis chromosomes 2–5. The gene order and content of these individual Arabidopsis segments indicate that they derived from a common ancestral segment through two or more rounds of large-scale genome duplication events—possibly polyploidy. One of these duplication events is ancient and may predate the divergence of the Arabidopsis and tomato lineages. The other is more recent and is estimated to have occurred after the divergence of tomato and Arabidopsis ≈112 million years ago. Together, these data suggest that, on the scale of BAC-sized segments of DNA, chromosomal rearrangements (e.g., inversions and translocations) have been only a minor factor in the divergence of genome organization among plants. Rather, the dominating factors have been repeated rounds of large-scale genome duplication followed by selective gene loss. We hypothesize that these processes have led to the network of synteny revealed between tomato and Arabidopsis and predict that such networks of synteny will be common when making comparisons among higher plant taxa (e.g., families).
Keywords: genome evolution, polyploidy
Arabidopsis is a model diploid plant species, ideal for sequencing because of its small genome (1). Currently, more than 80% of the genome has been sequenced (http://www.arabidopsis.org/agi.html), and the entire genome should be completed later this year, revealing the complete gene repertoire of a higher plant and providing insights into plant genome organization (2). However, the full potential of the Arabidopsis sequence will be realized only when its genome structure, gene content, and gene functions can be understood in relationship to its own evolutionary history and to that of other plant species. It is through comparative genomics that researchers will deduce the mechanisms and pathways by which plant genes and genomes have diverged to give the diversity of form, function, and adaptation that now characterize the world's flora. On the practical side, it is expected that the genomic sequence of Arabidopsis can be used to predict gene content and gene function in crop species, most of whose genomes are too large for genomic sequencing any time in the near future.
There are two underlying assumptions required for extrapolating genomic information from Arabidopsis to other plant species. (i) Arabidopsis and all higher plants have inherited gene order and gene content, with modifications, through common ancestry. (ii) The individual genes, now present in modern day plant species, can be used to reconstruct ancestral gene order and content. These assumptions have already been tested and largely verified for species within plant families. For example, in the grass family (Poaceae), which contains such familiar crops as corn, wheat, rice, and millet, gene order has been conserved in large blocks, often comprising entire chromosomes or chromosome arms (3). Comparative sequencing and cross-hybridization of cDNA clones in the grasses have also demonstrated that gene content and often gene function are also conserved (4, 5). Similar results have been obtained with studies in the nightshade family (potato, tomato, and pepper; refs. 6 and 7). However, in the mustard family (Brassicaceae), which includes Arabidopsis, cabbage, and broccoli, genomes seem to have evolved differently from the grasses or nightshades. Although gene content is conserved, the genomes of mustard species are often highly rearranged relative to one another; gene duplications are common, and at least some of them are due to polyploidy (8, 9).
Although comparisons among genomes have been quite common for species within plant families, comparisons between plant families have been rare and fraught with technical difficulties. Specifically, reduced gene similarities between plant families have made comparative mapping, via common probes and Southern hybridization, problematic if not impossible. Nonetheless, research by Paterson et al. (10) suggests that blocks of linked genes are conserved across higher plant families and even between the highly divergent monocots and dicots. Comparative sequencing data of Arabidopsis and rice have been used both to refute (11) and to support (12) this hypothesis.
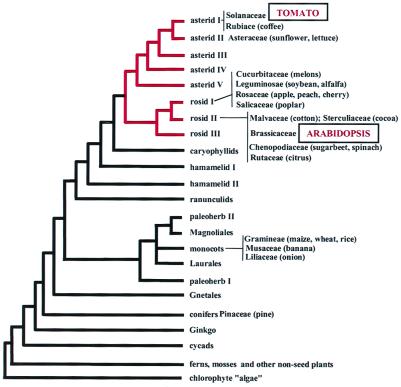
Tomato and Arabidopsis belong to two different families (Solanaceae and Brassicaceae, respectively) that diverged early in the radiation of dicotyledonous plants (Fig. 1). As determined by fossil evidence, the two families separated more than 90 million years ago (MYA) (13). Mitochondrial DNA sequence comparisons place the divergence at 112–156 MYA (14). Because of their early divergence, a comparison of the tomato and Arabidopsis genomes should provide a glimpse of gene and genome evolution since the radiation of dicotyledonous plants and provide information relevant to the large number of species (including many crop plants) that fall within the tomato–Arabidopsis clade (Fig. 1). With these considerations in mind, we have compared, via sequencing and computational analysis, the gene content and gene order of a 105-kilobase (kb) segment of tomato chromosome 2 to its homoeologous counterparts in the Arabidopsis genome.
Figure 1.
Dendrogram depicting phylogenetic relationships of higher plant taxa. Red lines indicate clade containing tomato and Arabidopsis. Contained within this clade are many familiar crop species (in parentheses). Figure based on figure 2 in ref. 43.
Materials and Methods
A bacterial artificial chromosome (BAC19; hereafter referred to as Tomato II) from the ovate-containing region of tomato chromosome 2 was isolated previously (H.-M.K., unpublished work), subjected to shotgun sequencing, and assembled with the phrap software package (15, 16). The resulting Tomato II sequence was 105,308 bp assembled from 3,257 reads with a 7-fold average depth of coverage and a minimum 3-fold depth of coverage. The nonvector ends of the contig were verified by end sequencing the BAC clone. The completed sequence was then analyzed for putative ORFs by using the gene prediction programs genscan (17) and genemark.hmm (18) with Arabidopsis settings. Further verification of the predicted ORFs was provided by blast searches (version 2.0.11; ref. 19) against the expressed sequence tag database (dbEST; refs. 20 and 21). By using tblastx and tblastn with the BLOSUM62 substitution matrix, both the complete BAC sequence and each putative tomato ORF were searched against the tiling path of the Arabidopsis genome sequence (http://www.arabidopsis.org/). The positions of tblastx matches were confined to predicted tomato and Arabidopsis ORFs, except for one match that involved only part of T6. The threshold for reporting a match of a tomato ORF to a specific Arabidopsis BAC was an expect value of <E−20. The translated blastprogram was chosen over other versions of blast, because homologous amino acid sequences were detected more easily over the large evolutionary distance separating Arabidopsis and tomato.
On finding Arabidopsis sequences with high scoring matches to individual tomato ORFs, the predicted coding sequences of the Arabidopsis ORFs were isolated from the GenBank annotation. These included Arabidopsis accession numbers (with clone names in parentheses) AC006135 (F24H14), AL132979 (T3A5), Z99708 (C7A10), and ABO18119 (MSN2). ABO18119 (MSN2) had no annotation in GenBank and was analyzed for predicted ORFs with genscan and genemark.hmm. In addition, the selected Arabidopsis regions were matched (via blastp and tblastx) against each other. The results of these analyses are summarized in Figs. 2 and 3.
Figure 2.
ORF matching between Tomato II (Top) and ORFs from segments of Arabidopsis chromosomes 2–5. AthII, AC006135/AC006439 (F24H14/T30D6); AthIII, AL132979 (T3A5); AthIV, Z99708 (C7A10); AthV, AB018119 (MSN2). Dashed lines indicate significant homology matches between specific ORFs. Arrows show orientation of ORFs. Numbers indicate specific ORFs. Open circles indicate that an ORF has a corresponding EST.
Figure 3.
Graphical representation of predicted gene content/order of the ancestral chromosome segment from which Tomato II, AthII, AthIII, AthIV, and AthV descended. Brackets indicate regions where ancestral gene order could not be deduced. Homoeologous ORF matches between Tomato II and segments of the Arabidopsis genome. See Table 1 and Fig. 2 for detailed information on ORFs.
clustalw (22) was used to construct pairwise global protein alignments between ORFs having a significant blast alignment (expect value <E−10). The number of nonsynonymous nucleotide substitutions per site (dN) was calculated for the regions of the coding sequences corresponding the aligned peptide substrings by using a codon-based substitution model (23) as implemented in the paml software package (14). The substitution matrix followed a proportional model, and the four-state discrete gamma distribution of rate variation among sites, the autocorrelation in rate between sites, and the transition–transversion ratio were all estimated from the data. The observed distribution of pairwise dN values was strongly bimodal, with <5% of the values between 0.5 and 1. Therefore, an upper threshold of dN = 1 was chosen to avoid including what were presumably spurious alignments. The connected components in the graph connecting ORFs with pairwise dN < 1 were extracted and realigned, again by using clustalw. Pairwise divergence values were recalculated, as before, excluding residues with gaps. In application of the molecular clock, divergence times were calculated as t = dN/(2r), where r is the rate of nonsynonymous substitutions per lineage per site per year.
All computational analyses (except BAC assembly) were performed on Velocity, a 256-processor Dell/Intel cluster running Microsoft Windows NT/2000 at the Cornell Theory Center (http://www.tc.cornell.edu).
Results and Discussion
The tomato genome has not been sequenced; therefore, it was important to select, for comparative sequencing, a portion of the genome for which the putative orthologous counterpart of Arabidopsis had already been sequenced. This selection was accomplished through a combination of strategy and fortuity. The ovate gene, controlling fruit shape, resides on chromosome 2 of tomato and has been the focus of developmental/genetic mapping studies, and several BAC clones containing this locus have been isolated (Fig. 2; ref. 24; H.-M.K., J.L., and S.D.T., unpublished work). Moreover, several ORFs near the ovate locus had been shown to have homologous matches to sequences in Arabidopsis accession Z99708 (C7A10) from chromosome 4 (H.-M.K., unpublished work). As a result of this finding, it was decided to sequence the entirety of a single 105-kb BAC (Tomato II) from the ovate region of tomato chromosome 2 (Fig. 2).
Estimates of Gene Density and Total Gene Number in Tomato.
By using the gene finding programs genscan and genemark.hmm, 17 putative ORFs were identified in Tomato II (Table 1; Fig. 2). The average density for this segment of the tomato genome was thus calculated to be 1 gene per 6.2 kb. This gene density is not much less than the densities calculated for the Arabidopsis genome: 1 gene per 4.4 kb (chromosome 2; ref. 25) and 1 gene per 4.6 kb (chromosome 4; ref. 26). This finding is surprising, considering that the tomato genome contains more than 900 megabases of DNA compared with ≈120 megabases for Arabidopsis—more than a 7-fold difference (27). If we extrapolate the gene density of Tomato II to the entire tomato genome, we estimate the total gene content of tomato to be 145,000 genes, considerably greater than the 20,000–25,000 estimated for Arabidopsis (1, 2).
Table 1.
Tomato II BAC with position of ORFs
| ORF | Identification of closest match | Position, bp | No. of predicted introns | Coding length, bp | EST |
|---|---|---|---|---|---|
| T1 | No significant matches | 1,539–5,644 | 2 | 135 | None |
| T2 | No significant matches | 5,681–8,880 | 0 | 1,077 | None |
| T3 | Transcription factor TFIIB | 9,251–13,712 | 2 | 1,278 | None |
| T4 | No significant matches | 14,367–18,474 | 1 | 489 | cLED17H21 |
| T5 | No significant matches | 20,068–21,037 | 1 | 777 | None |
| T6 | No significant matches | 23,259–30,134 | 1 | 636 | None |
| T7 | Arabidopsis adenylosuccinate synthetase | 38,746–41,450 | 3 | 1,503 | cLER16J17 |
| T8/T9 | Arabidopsis thaliana membrane-associated salt-inducible protein-like | 42,122–43,424/ | 1/0 | 504/337 | cLES15K9 |
| 44,667–46,811 | |||||
| T10 | Solanum tuberosum UDP-glucose pyrophosphorylase | 47,682–59,038 | 12 | 2,052 | cLED4L20 |
| T11 | Nicotiana plumbaginifolia mRNA for U2 snRNP auxiliary factor, large | 60,651–68,325 | 10 | 1,599 | cLEC9M14 |
| T12 | Zea mays zinc finger protein | 71,937–76,330 | 3 | 1,560 | cLES11M19 |
| T13 | Pumpkin mRNA for MP27 and MP32 | 76,385–83,857 | 0 | 1,500 | None |
| T14 | Oryza sativa Scarecrow-like protein (Scl1) | 90,655–92,603 | 0 | 1,611 | None |
| T15 | Lycopersicon esculentum GBF4 mRNA for G box binding protein | 92,627–101,097 | 10 | 1,311 | None |
| T16 | No significant matches | 101,358–103,458 | 1 | 174 | None |
| T17 | Arabidopsis thaliana mRNA for ATN1 protein kinase | 103,555–105,106 | 2 | 587 | None |
Tomato (and all species in the genus Lycopersicon) is considered to be a diploid (2n = 24) with normal bivalent pairing and normal Mendelian segregation ratios. Moreover, the haploid chromosome number (n = 12) is common for species throughout the family Solanaceae. Hence, the higher gene number in tomato cannot be explained by recent polyploidy. However, the possibility that tomato is an ancient polyploid (and therefore has significant gene duplication) cannot be excluded. Another likely explanation for the predicted high gene number is that the gene density of Tomato II may not be representative of the entire tomato genome. Considerable heterogeneity in gene density may exist among different portions of the tomato genome. It is already known that tomato has substantial segments of repetitive DNA, both at telomeres and in the centromeric heterochromatic DNA (28), both of which are likely to have a much lower gene density than the euchromatic region from which Tomato II was isolated. However, we cannot exclude the possibility that tomato and other solanaceous species do have an overall gene content significantly greater than Arabidopsis.
ORFs on Tomato II Match Multiple Sites in the Arabidopsis Genome.
Of the 17 predicted ORFs on Tomato II, 4 (24%) had no matches with any Arabidopsis BAC at the established statistical threshold, suggesting one of several possibilities. (i) The counterparts to these genes were deleted in the Arabidopsis lineage after tomato and Arabidopsis diverged. (ii) These ORFs represent fast-evolving genes; hence, the Arabidopsis homologs are no longer recognizable. (iii) These genes have matches in the regions of the Arabidopsis genome that have not yet been sequenced. (iv) Some of the putative tomato ORFs do not constitute functional genes but are artifacts of the gene-finding algorithm. This latter explanation cannot hold true in all cases, because one of the tomato ORFs (T10), with no Arabidopsis match, does have a corresponding tomato EST, indicating that it is an expressed gene (Table 1; Fig. 2). The remaining three ORFs have no corresponding EST, nor do they have a significant match to any other sequences in GenBank, a result consistent with them being rapidly evolving genes, spurious ORFs, or ORFs corresponding to an unsequenced portion of the Arabidopsis genome. The remaining 13 ORFs on Tomato II have significant matches (at the amino acid level) with ORFs from the Arabidopsis genome (Figs. 2 and 3). Of these Tomato II ORFs, 12 had cross-matches to one or more of four different Arabidopsis BAC/P1 accessions corresponding to four different chromosomal regions (chromosomes 2–5) forming a network of microsynteny (Figs. 2 and 3).
Evidence for Multiple Rounds of Large-Scale Duplication in the Arabidopsis Lineage.
The matches between Tomato II and four different segments of the Arabidopsis genome must be due to multiple rounds of duplication in the Arabidopsis lineage and cannot be explained by simple chromosome rearrangements (e.g., inversions, translocations, and transpositions). The reason for this assertion is that a number of the ORFs anchoring the Arabidopsis segments to Tomato II are in duplicate or triplicate in Arabidopsis (Figs. 2 and 3). Moreover, each of these Arabidopsis segments are anchored to each other through a network of matching homoeologous ORFs (Figs. 2 and 3). The duplication of the Arabidopsis chromosome 2 and 4 segments has been noted already and extends over more than 4.6 megabases (26, 29). Based on the results reported herein, this chromosome 2 and 4 duplication can now be extended to segments of chromosomes 3 and 5 (Figs. 2 and 3). We propose that these four matching segments of Arabidopsis represent the vestiges of at least two ancient, large-scale duplication events in the Arabidopsis genome and that Tomato II is a homoeologous counterpart to these in the tomato genome lineage (Figs. 2 and 3).
Conservation of Gene Order in Tomato and Duplicate Segments of the Arabidopsis Genome.
The homoeologous ORFs that anchor the Arabidopsis segments to one another and to Tomato II appear in the same order in all segments (Figs. 2 and 3), suggesting (i) that all segments descended from a common template, (ii) that this ancestral template predates the divergence of tomato and Arabidopsis, and (iii) that the order of genes in this ancestral template has been largely conserved in both the Arabidopsis and tomato lineages. However, each segment has its own subset of ordered, conserved ORFs interspersed with deleted ORFs or ORFs that have no recognizable counterparts in the other segments.
Evidence for Accelerated Gene Loss in the Duplicated, Syntenic Regions of Arabidopsis.
Although Tomato II can be anchored (via conserved ORFs) with all four Arabidopsis regions, individually, none of the Arabidopsis segments contains the full set of matching ORFs (Figs. 2 and 3). The number of ORF matches between Tomato II and individual Arabidopsis regions was seven matches for AthIV, five matches for AthII, three matches for AthV, and two matches for AthIII (Figs. 1 and 2). However, together, these four BACs account for matches to 12 of the 17 ORFs in Tomato II (Fig. 2). These results are consistent with all segments having diverged from a common template as described above. Moreover, we interpret the results to reflect two additional properties of the evolutionary history of Arabidopsis and tomato. (i) The different gene content of the Arabidopsis and Tomato II homoeologous segments reflects progressive gene loss after duplication as seen in yeast (30). (ii) Deletion of individual genes must have occurred more frequently than rearrangements (e.g., inversions and translocations), because the latter would have resulted in changes in gene order that were not observed in this study.
Comparison of Length and Gene Content of Conserved Syntenic Intervals Between Tomato II Homoeologous Segments in Arabidopsis.
It was possible to compare the overall length (in base pairs) of intervals flanked by syntenic ORFs in Tomato II and the corresponding Arabidopsis segments. For example, ORFs T3 and T15 on Tomato II have corresponding matches with AthIV.3 and AthIV.11 (Figs. 2 and 3). The AthIV.3 to AthIV.11 interval is 28 kb and contains nine predicted ORFs. The corresponding interval in tomato (T3 to T15), is 92 kb and contains 13 predicted ORFs. Similar comparisons were made between syntenic intervals on Tomato II and each of the other three corresponding Arabidopsis regions (AthII, AthIII, and AthV; Fig. 2). On average, the tomato intervals were approximately five times longer than those of Arabidopsis (tomato average = 65 kb; Arabidopsis = 15 kb). The tomato intervals also contained more predicted coding regions than their Arabidopsis counterparts (9.8 for tomato versus 4.8 for Arabidopsis).
Although each of the syntenic intervals in Arabidopsis contains fewer predicted ORFs than the corresponding tomato interval, the majority of the ORFs on Tomato II do have counterparts in one of the four matching Arabidopsis segments (Figs. 2 and 3). Hence, each of the four Arabidopsis segments is deficient in more than one of the ORFs found on Tomato II, but together, the four segments have retained conserved matches to most of the tomato ORFs (Figs. 2 and 3). Therefore, Arabidopsis does not seem to have an overall diminished gene repertoire (compared with tomato and for the regions examined), but rather, the matching counterparts to Tomato II ORFs are scattered throughout the four different syntenic segments of Arabidopsis. A possible explanation for this phenomenon is that, subsequent to large-scale duplication events (leading to the four matching segments AthII, AthIII, AthIV, and AthV), selected members of the resulting duplicated gene sets were eliminated progressively in the Arabidopsis lineage. If this hypothesis proves correct, then the gene content/organization of Tomato II more closely matches that of the ancestral dicot genome than does any one of the matching Arabidopsis segments.
Evidence for a Transposition Event into Tomato II?
In Arabidopsis, the homoeologous matches to the ORFs on Tomato II are nearly all contained in the AthII, AthIII, AthIV, and AthV network described above. The only exception was Tomato II ORF T7 (Fig. 2). For T7, no significant match was found within the AthII, AthIII, AthIV, and AthV network. However, a very strong match (tblastx E value = E−144) was found with an Arabidopsis ORF on a segment of chromosome 3 (BAC F15B8) that is 2.5 megabases from AthIII. The simplest interpretation of this finding is that T7 was transposed into the Tomato II segment after tomato and Arabidopsis diverged. Such a transposition may have been transposon-mediated—a mechanism well documented in plants (31, 32).
Alignments of Multiple Ortholog Sets—Evidence That Most Introns Predate the Divergence of Arabidopsis and Tomato.
By using the sets of syntenic ORFs, it is possible to determine how well intron positions have been conserved since the divergence of the tomato and Arabidopsis genomes. Comparison of each tomato ORF and its best corresponding match in Arabidopsis revealed that of 56 introns (analysis restricted to regions with clear amino acid alignments), 42 (21 pairs) or 75% are in corresponding positions. The 25% of introns not in common were as likely to occur in Arabidopsis as in tomato. These results indicate that tomato genes, on average, do not have more introns than their Arabidopsis counterparts; therefore, intron number cannot account for the difference in DNA content between the two species. Moreover, the position of most introns probably was established before the divergence of tomato and Arabidopsis, which is estimated to be more than 100 MYA (13, 14).
Consistent Bias Toward Longer Introns and Intergenic Spaces—Evidence for Less Efficient Monitoring/Removal of Noncoding DNA in the Tomato Lineage?
Although intron number and position are largely conserved between tomato–Arabidopsis homologs, individual introns were, on average, twice as long in tomato as in their Arabidopsis counterparts: tomato average = 387 bp; Arabidopsis average = 143 bp; (P < 0.001, based on paired t test). To compare intergenic spacer lengths, it was necessary to find consecutive ORFs in Tomato II that had clear, conserved counterparts in individual Arabidopsis segments. Only two such intervals were identified (Tomato II.3–Tomato II.4 versus AthIV.3–AthIV.4; and Tomato II.13–Tomato II.14 versus AthIV.8–AthIV.9; Fig. 2). In both instances, the intergenic spacers were longer (23% and 75%, respectively) in tomato than in their Arabidopsis counterparts. Further, when all inter-ORF spacers in Tomato II are compared with all inter-ORF spacers in AthII, AthIII, AthIV, and AthV, the average spacer length of tomato spaces was 37% greater than that of Arabidopsis: (tomato average = 3,085 bp, n = 16, SD = 2,583; Arabidopsis average = 2244 bp, n = 120, SD = 2,088). Thus, both types of comparisons indicate that inter-ORF (or intergenic) spacers are longer in tomato than in Arabidopsis.
That both introns and intergenic spacers are longer in tomato than in Arabidopsis suggests that there is an overall difference between the two lineages in the rates of accumulation or deletion of noncoding DNA. The greatly increased fraction of nongenic DNA in maize relative to rice seems to be due to the explosion of transposable elements in the maize lineage (33). However, there is no apparent excess of transposable elements in the tomato noncoding DNA. Differences in deletion rate seem to explain some of the variation in genome size in insects (34) and is more consistent with our observations in this study. Differences in the rate of accumulation or deletion of noncoding DNA may reflect positive selection for optimal cell size, as advocated in ref. 35, or may be due to other biochemical or life history pressures. In that regard, it is worth noting that tomato is a perennial in its native habitat, whereas Arabidopsis is a weedy annual.
Duplicated ORFs Within Segments.
Within the region examined on Tomato II, a single tandem duplication was observed (ORFs T8 and T9; Figs. 2 and 3). The divergence estimate for T8–T9 is saturated for dN, suggesting that this tandem duplication preceded the divergence of any of the five chromosomal segments seen in this study. Other ancient tandem duplications were found in AthII (ORFs 14 and 15), AthIII (ORFs 10 and 11), and AthV (ORFs 5–7; Figs. 2 and 3).
Matches Between ORFs in Inverted Orientation.
As described earlier, gene order is largely conserved in homoeologous regions of the tomato and Arabidopsis genomes (Figs. 2 and 3). Interestingly, in several instances, the corresponding homoeologous ORFs (between Tomato II and the matching Arabidopsis segments) were in reversed orientation (opposite strands; Fig. 2). Of the 12 ORF:ORF matches that link Tomato II to Arabidopsis, three (TomatoII.4:AthIV.4; TomatoII.11:AthIV.7; and TomatoII.12:AthV.11) have reverse orientations despite the fact that they reside in otherwise conserved syntenous regions (Figs. 2 and 3). It has been shown previously in Caenorhabditis elegans and Drosophila that a significant portion of adjacent gene duplications are in opposite orientation (36, 37). It is therefore plausible that the inverted ORF matches found in the current study resulted from inverted gene duplication followed by loss of the copy in the original orientation.
Use of the Molecular Clock to Date the Large-Scale Duplication Events.
Because no ortholog is shared among all segments (Figs. 2 and 3), the topology and branch lengths of the phylogenetic tree connecting the five homoeologous regions must necessarily be inferred by combining evidence from different sets of orthologs. The median distance matrix (Table 2) suggests that AthII and AthIV as well as AthIII and AthV are two monophyletic clades that diverged at roughly the same point in time (Fig. 3). It seems plausible that these two duplications resulted from a single whole-genome duplication event (e.g., polyploidization). The grouping of these two clades in the phylogeny is also supported by the large number of shared ancestral genes within each putative clade (Fig. 3).
Table 2.
Median nonsynonymous divergence among corresponding ORFS on Tomato II BAC and selected Arabidopsis segments
| Segment | AthII | AthIII | AthIV | AthV |
|---|---|---|---|---|
| Tomato II | 0.43/2 | 0.42/2 | 0.26/7 | 0.27/3 |
| AthII | na | 0.21/4 | na | |
| AthIII | na | 0.21/3 | ||
| AthIV | na |
Numerator, median nonsynonymous divergence; denominator, number of pairwise ORF comparisons (see Table 1). na, not applicable.
We assume a clock-like rate of nucleotide substitution to date the duplication and divergence events, but calculated dates must be treated with caution. We have used dN, even though nonsynonymous substitutions are typically more erratic than synonymous substitutions, because dS is saturated for nearly all comparisons. Calibration of the clock for plant nuclear genes is based on only a handful of pairwise comparisons—six in ref. 38 and nine in ref. 39—both based on a single fossil event, the divergence of the pooid and oryzoid grasses 50–70 MYA. Extrapolation to more divergent comparisons and the use of alternative methods of calculation introduce additional uncertainties. Nonetheless, taking 9.4E − 10 year−1 as the typical rate of nonsynonymous substitution for plant nuclear genes (39), the median dN value of 0.21 yields an estimated divergence time of approximately 112 MYA for both pairs of sister segments.
The branching order among tomato and the two putative Arabidopsis clades is not clear. However, because there are two sets of orthologous genes that link Tomato II with AthII and AthIV and another two that link Tomato II with AthIII and AthV, we can calculate estimates of the branch length between the more recent Arabidopsis duplication and the common ancestor of Arabidopsis and tomato. This length, assuming an ultrametric tree, is L = (dT,A1 + dT,A2 − 2dA1,A2)/4, where T is the ortholog on the tomato segment, A1 and A2 are the corresponding orthologs on the two Arabidopsis segments, and dX,Y denotes the dN estimate between orthologs X and Y. We can then estimate the ratio of the older to the more recent branch length by using the equation R = 2L/dA1,A2. Using these formulae, we obtain estimates of L = 0.06–0.12, and R = 0.3–0.8, with overlapping ranges for estimates from the two different clades. This overlap implies that the first duplication in Arabidopsis occurred either shortly before or sometime after the divergence of the two species; however, the exact branching order is unknown. The median and mean value of R are both ≈0.6, which suggests that the speciation event occurred roughly 70 MY before the most recent duplication within Arabidopsis or 180 MYA. Taking the median of all divergence values between all orthologous matches between tomato and the four Arabidopsis segments yields an alternative tomato–Arabidopsis divergence estimate of about 150 MYA. These numbers are comparable to the estimate of 112–156 MYA obtained in ref. 14 with mitochondrial sequence data. The inferred phylogenetic relationships and divergence estimates are summarized in Fig. 3.
In summary, we are led to hypothesize two large-scale duplication events in the antecedents of the Arabidopsis genome, the results of which are the segments of chromosome 2–5 under study (Figs. 2 and 3). The best documented mechanism capable of generating such large-scale duplications in plants is polyploidy. It is estimated that up to 70% of all living plant species are of polyploid origin (40, 41). Given the large phylogenetic distance between tomato and Arabidopsis, it seems plausible that two polyploidization events may have occurred in the lineage of Arabidopsis resulting in the four large-scale duplications (i.e., AthII, AthIII, AthIV, and AthV) described in this article. It is important to note the possibility that Tomato II also has duplicate, matching segments within its genome (also as a result of polyploid events); however, there is no evidence bearing on this issue at the current time.
Predictions of a Polyploidy Model for the Origin of the Duplications.
The model presented above and in Fig. 3 makes some very clear testable predictions. First, if two rounds of polyploidy in the Arabidopsis lineage are the cause of these reported duplications, then further analyses should reveal similar networks of homoeologous sets of segments elsewhere in Arabidopsis. In this regard, a recent comparative mapping study between soybean and Arabidopsis presents evidence for patterns of duplications in Arabidopsis compatible with a polyploidization model (42). Second, if at least one of these proposed polyploid events occurred before the divergence of the tomato and Arabidopsis lineages, then tomato (and many other dicotyledonous plants) should show vestiges of the duplication event(s) in their genomes. Third, the model predicts that Arabidopsis and most flowering plants are likely ancient polyploids, and as comparisons are made across greater and greater phylogenetic distances, the likelihood increases that polyploidy has occurred (subsequent to speciation) in the lineage of one or both species being compared. If this prediction is correct, comparisons across families of plants will not result in matches between single homoeologous segments but rather in matches among sets of homologous genes and duplicated gene segments.
Estimating the Gene Number of the Ancestral Dicot Genome.
If polyploidy was a factor in the evolution of the Arabidopsis genome, the gene number for the progenitor of Arabidopsis and tomato (and hence many other higher plant families) could have been considerably less than the 20,000–25,000 genes estimated for Arabidopsis (1, 2). Assuming that the pattern of postduplication gene deletion observed in this study is typical of the Arabidopsis genome as a whole and that transposition of genes into and out of the four homoeologous blocks is negligible, we can estimate the number of genes present in the preduplication ancestor. There are 14 present-day genes shared among all four duplicated segments in the region bracketed by ORF.H and ORF.N in Fig. 3 (counting AthIV.8 and AthII.11 as 0.5 because of their ambiguous positions). The estimated number of ancestral genes is the number of paralogous components plus the number of present-day genes with no matches or 7.5. Thus, we estimate that the ancestral genome, before the duplications (and before the divergence of Arabidopsis and tomato) to have been approximately one-half the number of genes seen in the present-day Arabidopsis genome.
Conclusions
The cumulative results from the above analyses of the Tomato II BAC and its corresponding counterparts in Arabidopsis, suggest the following modes of genome evolution. At least two rounds of large-scale duplication (possibly polyploidy) occurred in the lineage leading to Arabidopsis. One of those duplication events is ancient and possibly predates the radiation of dicotyledonous plants; the other likely occurred after tomato and Arabidopsis diverged (≈150 MYA). Moreover, on the scale of a BAC sized clone, gene order has been well conserved between Arabidopsis and tomato. Hence, on this scale, chromosomal rearrangements (e.g., inversions and translocations) have likely played a minor role in the divergence of genome organization among plants. Rather, the dominating factors have been repeated rounds of large-scale genome duplication followed by selective gene loss. Gene loss rates (per segment) seem to have been greater in the Arabidopsis lineage than in the tomato lineage.
Finally, results from this study indicate that syntenic relationships can be detected between the Arabidopsis genome and the genomes of more divergent families of plants based on gene homologies but that matches tying Arabidopsis to other plant genomes will not likely be based on single ortholog pairs but rather networks of homologous genes created by multiple rounds of genome duplication followed by gene divergence and gene loss. Establishing genome relationships among divergent plant families on a gene-for-gene basis may therefore be more complicated than originally expected. However, such analyses will eventually allow for an understanding of the events and mechanisms that have molded higher plant genome evolution and the exchange of sequence and functional information among species. Also, if Arabidopsis is indeed an ancient polyploid, then the Arabidopsis genome project will provide the first in-depth look at the structural and functional consequences of polyploidization in plants over very long periods of evolutionary time.
Acknowledgments
We acknowledge Oleg Iartchouk and Craig Deloughery for the sequencing of the Tomato II BAC and Andreas Matern for help in use of assembly and annotation programs. Thanks to Charles Aquadro, Jeff Doyle, and Anne Frary for helpful discussions and comments. This research was funded by a grant from the Cereon Corporation and by National Science Foundation Grant DBI-9872617.
Abbreviations
- kb
kilobase
- MYA
million years ago
- BAC
bacterial artificial chromosome
- EST
expressed sequence tag
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF273333).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.160271297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.160271297
References
- 1.Meinke D W, Cherry J M, Dean C D, Rounsley S, Koornneef M. Science. 1998;282:662–682. doi: 10.1126/science.282.5389.662. [DOI] [PubMed] [Google Scholar]
- 2.Somerville C, Somerville S. Science. 1999;285:380–383. doi: 10.1126/science.285.5426.380. [DOI] [PubMed] [Google Scholar]
- 3.Gale M D, Devos K M. Science. 1998;282:656–659. doi: 10.1126/science.282.5389.656. [DOI] [PubMed] [Google Scholar]
- 4.Van Deynze A E, Sorrells M E, Park W D, Ayres N M, Fu H, Cartinhour S W, Paul E, McCouch S R. Theor Appl Genet. 1998;97:356–369. [Google Scholar]
- 5.Chen M, SanMiguel P, de Oliveira A C, Woo S-S, Zhang H, Wing R A, Bennetzen J L. Proc Natl Acad Sci USA. 1997;94:3431–3435. doi: 10.1073/pnas.94.7.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanksley S D, Ganal M W, Prince J P, deVicente M C, Bonierbale M W, Broun P, Fulton T M, Giovanonni J J, Grandillo S, Martin G B, et al. Genetics. 1992;132:1141–1160. doi: 10.1093/genetics/132.4.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Livingstone K D, Lackney V K, Blauth J R, van Wijk R, Jahn M K. Genetics. 1999;152:1183–1202. doi: 10.1093/genetics/152.3.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lagercrantz U, Lydiate D J. Genetics. 1996;144:1903–1910. doi: 10.1093/genetics/144.4.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lagercrantz U. Genetics. 1998;150:1217–1228. doi: 10.1093/genetics/150.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paterson A H, Lan T H, Reischmann K P, Chang C, Lin Y R, Liu S C, Burow M D, Kowalski S P, Katsar C S, DelMonte T A, et al. Nat Genet. 1996;14:380–382. doi: 10.1038/ng1296-380. [DOI] [PubMed] [Google Scholar]
- 11.Devos K M, Beales J, Nagamura Y, Sasaki T. Genome Res. 1999;9:825–829. doi: 10.1101/gr.9.9.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Dodeweerd A-M, Hall C R, Bent E G, Johnson S J, Bevan M W, Bancroft I. Genome. 1999;42:887–892. [PubMed] [Google Scholar]
- 13.Gandolfo M A, Nixon K C, Crepet W L. Am J Bot. 1998;85:964–974. [PubMed] [Google Scholar]
- 14.Yang Y W, Lai K N, Tai P Y, Li W H. J Mol Evol. 1999;48:597–604. doi: 10.1007/pl00006502. [DOI] [PubMed] [Google Scholar]
- 15.Gordon D, Abajian C, Green P. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 16.Ewing B, Hillier L, Wendl M, Green P. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 17.Burge C, Karlin S. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 18.Lukashin A, Borodovsky M. Nucleic Acids Res. 1998;26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altschul S, Madden T, Schaffer A, Zhang J H, Zhang Z, Miller W, Lipman D. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benson D A, Karsch-Mizrachi I, Lipman D J, Ostell J, Rapp B A, Wheeler D L. Nucleic Acids Res. 2000;28:15–18. doi: 10.1093/nar/28.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boguski M S, Lowe T M, Tolstoshev C M. Nat Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- 22.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldman N, Yang Z. Mol Biol Evol. 1994;11:725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- 24.Ku H K, Tanksley S D. Theor Appl Genet. 1999;9:844–850. [Google Scholar]
- 25.Lin X, Kaul S, Rounsley S, Shea T P, Benito M I, Town C D, Fujii C Y, Mason T, Bowman C L, Barnstead M, et al. Nature (London) 1999;402:761–768. doi: 10.1038/45471. [DOI] [PubMed] [Google Scholar]
- 26.Mayer K, Schuller C, Wambutt R, Murphy G, Volckaert G, Pohl T, Dusterhoft A, Stiekema W, Entian K D, Terryn N, et al. Nature (London) 1999;402:769–777. [Google Scholar]
- 27.Arumuganathan K, Earle E. Plant Mol Biol Rep. 1991;9:208–218. [Google Scholar]
- 28.Ganal M W, Lapitan N L V, Tanksley D. Mol Gen Genet. 1988;213:262–268. [Google Scholar]
- 29.Terryn N, Heijnen L, De Keyser A, Van Asseldonck M, De Clercq R, Verbakel H, Gielen J, Zabeau M, Villarroel R, Jesse T, et al. FEBS Lett. 1999;445:237–245. doi: 10.1016/s0014-5793(99)00097-6. [DOI] [PubMed] [Google Scholar]
- 30.Keogh T S, Seioghe C, Wolfe K H. Yeast. 1998;14:443–457. doi: 10.1002/(SICI)1097-0061(19980330)14:5<443::AID-YEA243>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 31.Kunze R, Saedler H, Lonnig W E. In: Advances in Botanical Research. Callow J A, editor. Vol. 27. San Diego: Academic; 1997. pp. 332–470. [Google Scholar]
- 32.Lonnig W E, Saedler H. Gene. 1997;205:245–253. doi: 10.1016/s0378-1119(97)00397-1. [DOI] [PubMed] [Google Scholar]
- 33.SanMiguel P, Gaut B S, Tikhonov A, Nakajima Y, Bennetzen J L. Genetics. 1998;20:43–45. doi: 10.1038/1695. [DOI] [PubMed] [Google Scholar]
- 34.Petrov D A, Sangster T A, Johnston J S, Hartl D L, Shaw K L. Science. 2000;287:1060–1062. doi: 10.1126/science.287.5455.1060. [DOI] [PubMed] [Google Scholar]
- 35.Beaton M J, Cavalier-Smith T. J Mol Evol. 1999;48:555–564. [Google Scholar]
- 36.Semple C, Wolfe K H. J Mol Evol. 1999;48:555–564. doi: 10.1007/pl00006498. [DOI] [PubMed] [Google Scholar]
- 37.Rubin G M, Yandell M D, Wortman J R, Gabor Miklos G L, Nelson C R, Hariharan I K, Fortini M E, Li P W, Apweiler R, Fleischmann W, et al. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolfe K H, Sharp P M, Li W H. J Mol Evol. 1989;29:208–211. [Google Scholar]
- 39.Gaut B S. Evol Biol. 1998;30:93–120. [Google Scholar]
- 40.Masterson J. Science. 1994;264:421–424. doi: 10.1126/science.264.5157.421. [DOI] [PubMed] [Google Scholar]
- 41.Wendel J F. Plant Mol Biol. 2000;42:225–249. [PubMed] [Google Scholar]
- 42.Grant D, Cregan P, Shoemaker R. Proc Natl Acad Sci USA. 2000;97:4168–4173. doi: 10.1073/pnas.070430597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chase M W, Soltis D E, Olmstead R G, Morgan D, Les D H, Mishler B D, Duvall M R, Price R A, Hills H G, Qiu Y-L, et al. Ann Mo Bot Gard. 1993;80:528–580. [Google Scholar]