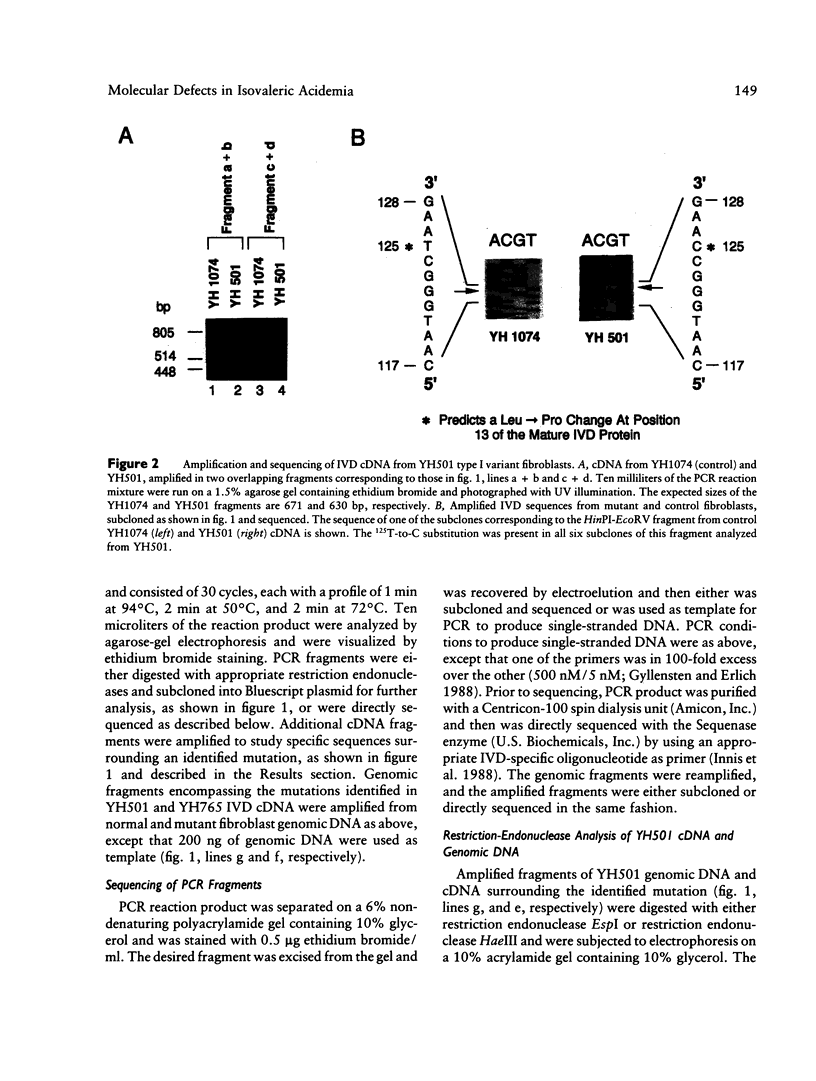
Abstract
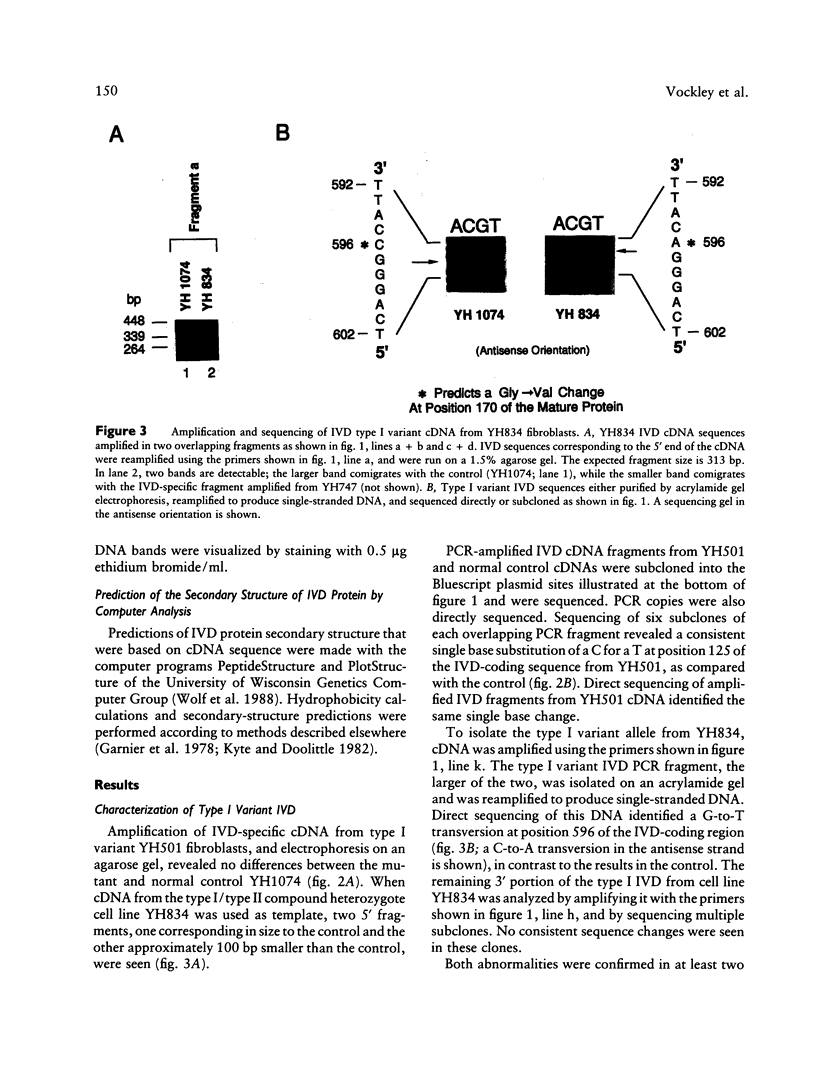
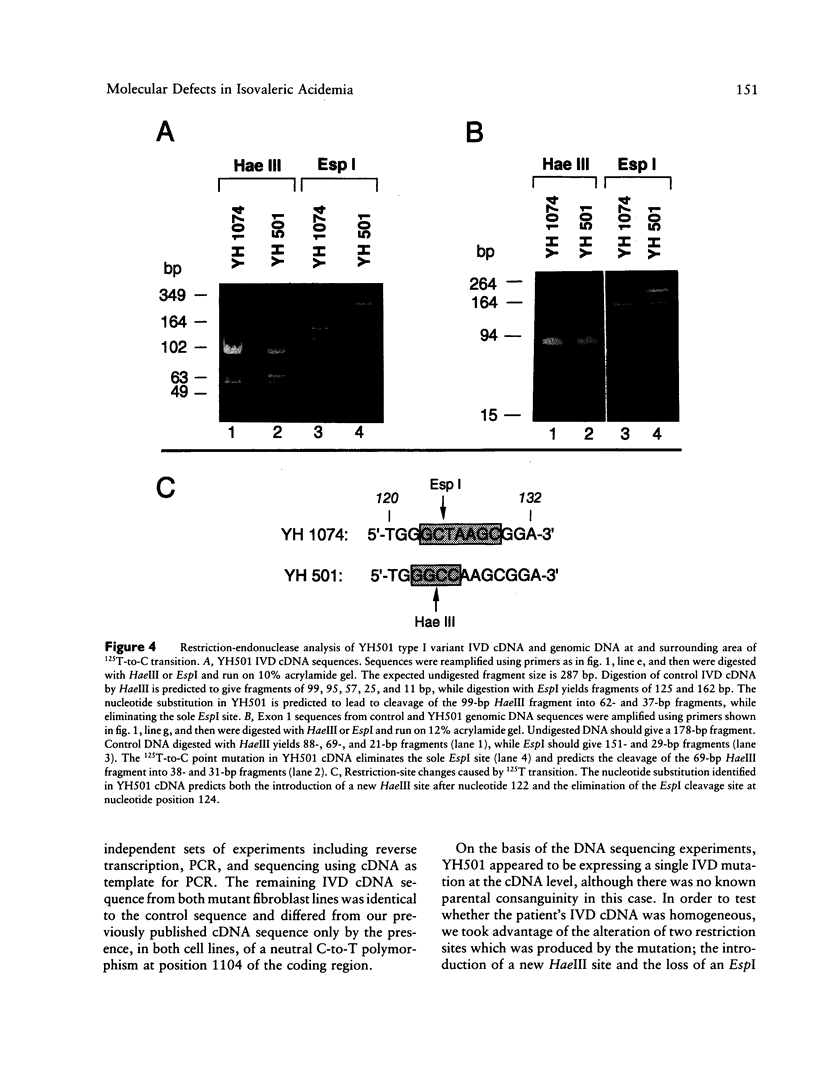
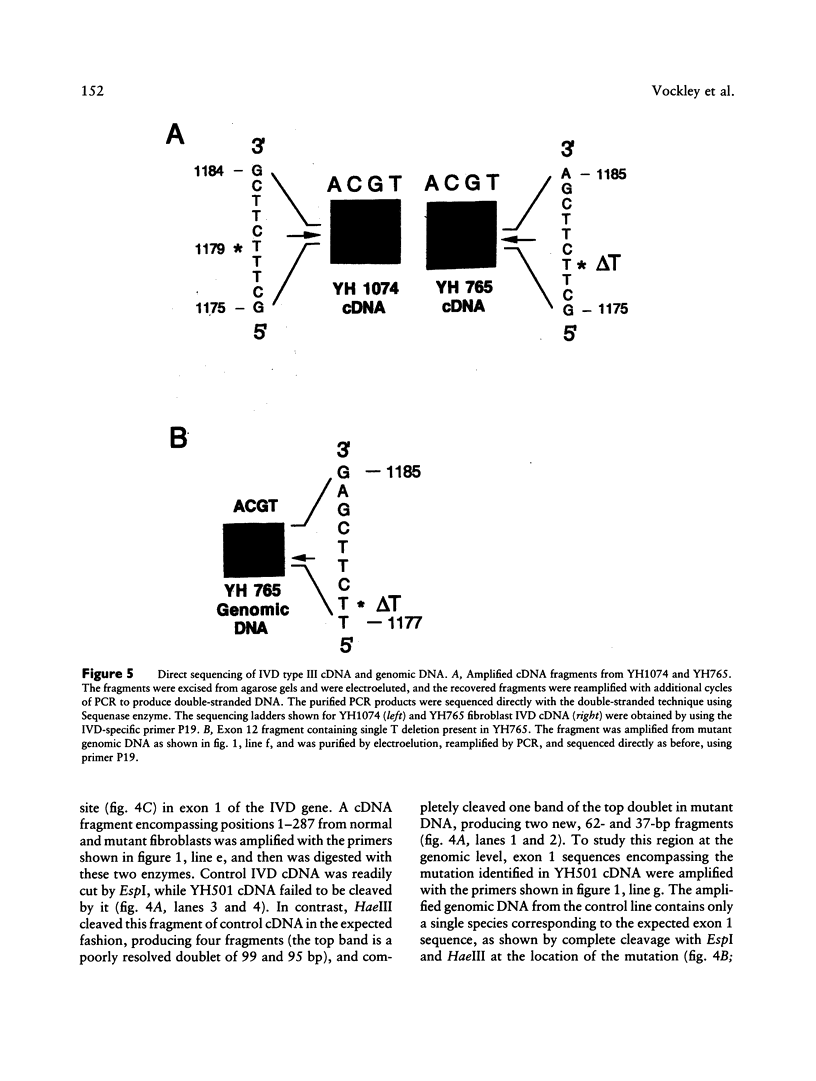
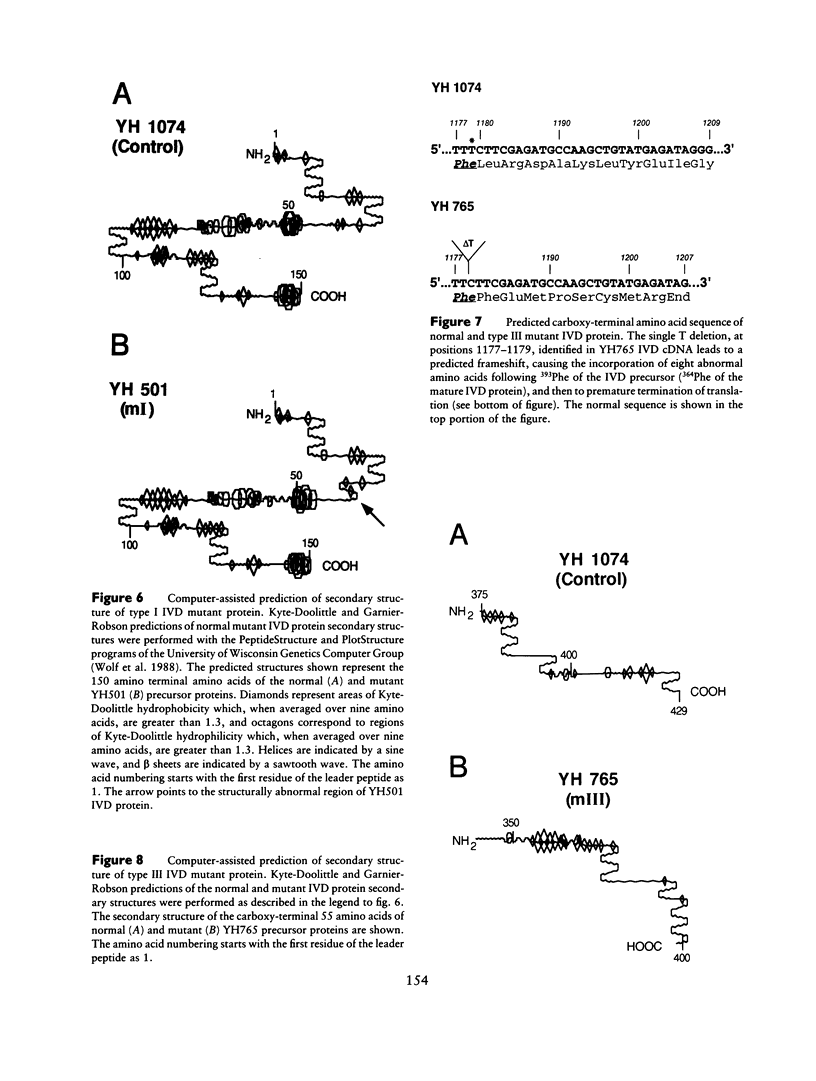
Isovaleric acidemia (IVA) is an inborn error of leucine metabolism and is caused by a genetically determined deficiency of isovaleryl-CoA dehydrogenase (IVD), a mitochondrial matrix enzyme. IVD is produced as a 45-kDa precursor and then is transported into the mitochondria, where it is processed to its mature 43-kDa size. Previous [35S]methionine-labeling studies of fibroblasts from IVA patients have revealed at least five classes of mutations within the IVD gene. In size, IVD precursor and mature proteins produced by class I mutants are indistinguishable from their normal counterparts. Class II, III, and IV mutants make IVD precursor proteins which are 23 kDa smaller than normal. Subsequent processing in class III and IV mutants is normal but proceeds inefficiently in class II mutants. Class V mutants make no detectable IVD protein. In order to further study these mutations at the molecular level, the IVD coding region from mutant fibroblast cDNA was amplified by the PCR and was analyzed by DNA sequencing. cDNA from class I mutant alleles from two of seven class I mutant cell lines each contained a different missense mutation. In cDNA from a class III mutant, a single base deletion at position 1179 of the coding region was identified which leads to a shift in reading frame, predicting the incorporation of eight abnormal amino acids followed by a premature termination codon. Sequencing of amplified IVD cDNA from a type V mutant has failed to identify any abnormalities. It most probably is deficient in translation of the IVD mRNA. A new class of IVD mutant allele which appears to be transcriptionally defective (type VI) was also identified. Additional study of this set of IVD mutations should add both to our knowledge of the biosynthetic pathway of mitochondrial proteins and to our understanding of the clinical heterogeneity seen in IVA.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barker D., Schafer M., White R. Restriction sites containing CpG show a higher frequency of polymorphism in human DNA. Cell. 1984 Jan;36(1):131–138. doi: 10.1016/0092-8674(84)90081-3. [DOI] [PubMed] [Google Scholar]
- Finocchiaro G., Ito M., Ikeda Y., Tanaka K. Molecular cloning and nucleotide sequence of cDNAs encoding the alpha-subunit of human electron transfer flavoprotein. J Biol Chem. 1988 Oct 25;263(30):15773–15780. [PubMed] [Google Scholar]
- Garnier J., Osguthorpe D. J., Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978 Mar 25;120(1):97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- Gyllensten U. B., Erlich H. A. Generation of single-stranded DNA by the polymerase chain reaction and its application to direct sequencing of the HLA-DQA locus. Proc Natl Acad Sci U S A. 1988 Oct;85(20):7652–7656. doi: 10.1073/pnas.85.20.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y., Dabrowski C., Tanaka K. Separation and properties of five distinct acyl-CoA dehydrogenases from rat liver mitochondria. Identification of a new 2-methyl branched chain acyl-CoA dehydrogenase. J Biol Chem. 1983 Jan 25;258(2):1066–1076. [PubMed] [Google Scholar]
- Ikeda Y., Keese S. M., Fenton W. A., Tanaka K. Biosynthesis of four rat liver mitochondrial acyl-CoA dehydrogenases: in vitro synthesis, import into mitochondria, and processing of their precursors in a cell-free system and in cultured cells. Arch Biochem Biophys. 1987 Feb 1;252(2):662–674. doi: 10.1016/0003-9861(87)90072-5. [DOI] [PubMed] [Google Scholar]
- Ikeda Y., Keese S. M., Tanaka K. Molecular heterogeneity of variant isovaleryl-CoA dehydrogenase from cultured isovaleric acidemia fibroblasts. Proc Natl Acad Sci U S A. 1985 Oct;82(20):7081–7085. doi: 10.1073/pnas.82.20.7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y., Tanaka K. Purification and characterization of isovaleryl coenzyme A dehydrogenase from rat liver mitochondria. J Biol Chem. 1983 Jan 25;258(2):1077–1085. [PubMed] [Google Scholar]
- Innis M. A., Myambo K. B., Gelfand D. H., Brow M. A. DNA sequencing with Thermus aquaticus DNA polymerase and direct sequencing of polymerase chain reaction-amplified DNA. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9436–9440. doi: 10.1073/pnas.85.24.9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. J., Wu J. Structure of the medium-chain acyl-CoA dehydrogenase from pig liver mitochondria at 3-A resolution. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6677–6681. doi: 10.1073/pnas.85.18.6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Matsubara Y., Indo Y., Naito E., Ozasa H., Glassberg R., Vockley J., Ikeda Y., Kraus J., Tanaka K. Molecular cloning and nucleotide sequence of cDNAs encoding the precursors of rat long chain acyl-coenzyme A, short chain acyl-coenzyme A, and isovaleryl-coenzyme A dehydrogenases. Sequence homology of four enzymes of the acyl-CoA dehydrogenase family. J Biol Chem. 1989 Sep 25;264(27):16321–16331. [PubMed] [Google Scholar]
- Matsubara Y., Ito M., Glassberg R., Satyabhama S., Ikeda Y., Tanaka K. Nucleotide sequence of messenger RNA encoding human isovaleryl-coenzyme A dehydrogenase and its expression in isovaleric acidemia fibroblasts. J Clin Invest. 1990 Apr;85(4):1058–1064. doi: 10.1172/JCI114536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhead W. J., Tanaka K. Demonstration of a specific mitochondrial isovaleryl-CoA dehydrogenase deficiency in fibroblasts from patients with isovaleric acidemia. Proc Natl Acad Sci U S A. 1980 Jan;77(1):580–583. doi: 10.1073/pnas.77.1.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Budd M. A., Efron M. L., Isselbacher K. J. Isovaleric acidemia: a new genetic defect of leucine metabolism. Proc Natl Acad Sci U S A. 1966 Jul;56(1):236–242. doi: 10.1073/pnas.56.1.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf H., Modrow S., Motz M., Jameson B. A., Hermann G., Förtsch B. An integrated family of amino acid sequence analysis programs. Comput Appl Biosci. 1988 Mar;4(1):187–191. doi: 10.1093/bioinformatics/4.1.187. [DOI] [PubMed] [Google Scholar]