Abstract
In wild-type cells, the 3′ poly(A) structure is necessary for translation of mRNA and for mRNA stability. The superkiller 2 (ski2), ski3, ski6, ski7, and ski8 mutations enhance the expression of the poly(A)− mRNAs of yeast RNA viruses. Ski2p is a DEVH-box RNA helicase and Slh1p resembles Ski2p. Both repress L-A double-stranded RNA (dsRNA) virus copy number, further suggesting that their functions may overlap. We find that slh1Δ ski2Δ double mutants are healthy (in the absence of viruses) and show normal rates of turnover of several cellular mRNAs. The slh1Δ ski2Δ strains translate electroporated nonpoly(A) mRNA with the same kinetics as polyA+ mRNA. Thus, the translation apparatus is inherently capable of efficiently using nonpoly(A) mRNA even in the presence of normal amounts of competing poly(A)+ mRNA, but is normally prevented from doing so by the combined action of the nonessential proteins Ski2p and Slh1p.
The 5′ cap (7mGpppXp … ) and 3′ poly(A) structures of eukaryotic mRNAs have important roles in promoting mRNA stability in the nucleus, its transport to the cytoplasm, and its translation and stability in the cytoplasm. These structures produce a synergistic effect on translation rates. Electroporation of mRNA into living cells (1, 2) and in vitro translation (3) indicate a 50-fold preference for polyA+ (A+) over poly(A)− (A−) mRNA and a 24-fold preference for cap+ (C+) over cap− (C−) mRNA. Although poly(A) tails are essential for translation in wild-type cells, cells depleted of A+ mRNA can use mRNAs with ≤ 25 residues of poly(A) tail (ref. 4 and Discussion). The 3′ poly(A) structure also protects mRNA from degradation by 5′ decapping and subsequent 5′ → 3′ degradation by Xrn1p/Ski1p (5).
The superkiller 2 (ski2), ski3, ski6, ski7, and ski8 mutants, isolated because they show elevated expression of the A− mRNAs of the cytoplasmic L-A and M1 viruses, also overexpress electroporated A− mRNA. The ski2 mutant also overexpresses cellular A− mRNA made from a RNA polymerase I promoter (6–9). Ski2p is a nonessential 1,286 residue protein with motifs characteristic of RNA helicases and nucleolar proteins (8). ski2 strains also have elevated copy number of another dsRNA virus called L-BC and a single-stranded RNA replicon called 20S RNA (10, 11), both of which lack 3′ poly(A) (12, 13). The ski2Δ (as well as ski3Δ, ski7Δ, and ski8Δ) cells express electroporated C+ A− mRNAs 20-fold better than do wild-type cells, but still use A+ mRNAs threefold more efficiently (9, 14). No effect of ski mutations was seen on C− A+ mRNAs. Blocking the predominant 5′ → 3′ mRNA degradation pathway also enables detection of decreased 3′ → 5′ degradation of mRNA in ski2Δ strains but whether this effect is secondary to a primary effect on translation was not determined (15). Two human homologs of Ski2p have been identified (16–18), one of which is both nucleolar and associated with 40S ribosomal subunits and with polysomes (19).
YGR271w (which we call SLH1 for SKI2-like helicase) encodes a nonessential 1,967-residue protein with two copies of the helicase motifs, and substantial homology to Ski2p outside the helicase domains as well, suggesting that their functions might overlap (20). As in ski2 mutants, the copy number of L-A dsRNA was increased in the slh1Δ strain (20).
We show here that ski2Δ slh1Δ double mutants express mRNAs lacking the 3′ poly(A) structure fully as efficiently and for as long a duration as they express A+ mRNA. These strains are nonetheless healthy in the absence of the L-A and M viruses. This finding shows that, even in the presence of the normal complement of competing cellular A+ mRNAs, the translation apparatus does not inherently require (or even prefer) the 3′ poly(A) structure to translate a mRNA. Rather, this requirement is imposed by the combined action of Ski2p and Slh1p.
Materials and Methods
Sacccharomyces cerevisiae strains 3221 (MATa his3 trp1 ura3 L-A-o M-o Gal+), 3515 (3221 ski2∷HIS3), and 4696–1B (MATα his3 ski6–2 Gal+), were described (8, 21). Strain 4709 (MATα his3 trp1 ura3 ski2∷HIS3 ski6–2 Gal+) was derived from a cross between 3515 and 4696–1B. The slh1∷URA3 disruption was constructed in strains 3221 and 3515 with the disruption plasmid described (20), forming strains 4106 and 4107, respectively. The Ura+ colonies were examined by colony PCR, confirming the absence of the normal gene and presence of the disruption construct. Cytoduction from strain 3166 (MATα kar1–1 leu1 thr1 L-A M1) and 3431(MATα leu1 kar1–1 L-A M-o) was used to introduce either L-A and M1 or L-A alone into ρo derivatives of strains 3221, 3515, 4106, and 4107.
The luciferase mRNA expression plasmids T7 LUC (A−)and T7 LUC A50 [poly(A)50] have been described (1). For RNA synthesis, T7 LUC was linearized with SmaI and T7 LUC A50 was linearized with DraI. C− transcripts were synthesized with the MEGAscript transcription kit (Ambion, Austin, TX) whereas C+ transcripts were synthesized with the Ambion mMessage mMachine transcription kit. Electroporation of luciferase RNAs was performed as described (2).
For RNA extraction, yeast strains were grown in 10 ml of liquid culture to an OD600 of 0.6. Cycloheximide was added to 100 μg/ml and the cells were centrifuged and washed in 1 ml of water containing 100 μg/ml cycloheximide. The cell pellets were resuspended in 50 μl of SDS buffer (0.5 M NaCl/50 mM Tris⋅HCl pH 7.4/0.2% SDS/10 mM EDTA/1.5 mM β-mercaptoethanol) and 50 μl of phenol:chloroform:isoamyl alcohol (25:24:1). Glass beads were added and the tubes were vortexed three times for 1 min each (on ice in between). After lysis, 100 μl of SDS buffer was added, and the tubes were vortexed and centrifuged. The supernatant was removed to a new tube, 100 μl of SDS buffer was added to the beads, and the extraction was repeated. The clarified supernatants were combined and extracted with phenol:chloroform:isoamyl alcohol (25:24:1). Total RNAs were precipitated with 2 vol of ethanol. Similar results were obtained by using a similar procedure without cycloheximide addition.
To measure the effect of ski2Δ slh1Δ on mRNA decay, 200 ml of mutant and wild-type cells were grown to OD600 = 0.6. Transcription was inhibited by the addition of thiolutin (a kind gift of Edmund Hafner, Pfizer, Groton, CT) as described (22), and aliquots of cells were removed at intervals. RNA was extracted by using the RNeasy Mini RNA extraction kit (Qiagen, Chatsworth, CA) after breaking cells with glass beads. Extracted RNA was analyzed by the Northern blot method. The membranes were hybridized with either 5′-32P-labeled oligonucleotide probes complementary to regions of the coding sequences of ACT1 and PGK1 (23) or to a RNA probe complementary to 18S rRNA.
Results
Ski2Δ slh1Δ Mutants Are Healthy in the Absence of L-A and M.
To determine the functional relation of SLH1 to SKI2 and their combined effects on poly(A)-dependent expression, we made a series of isogenic single and double mutants. In the absence of L-A and M1 dsRNA viruses, the growth rates of either ski2Δ or slh1Δ or the double mutants were normal on rich media at 15 or 30°C (Fig. 1), but the double mutants were slightly slow growing at 37°C. We found that the known cold- and heat-sensitivity of growth of ski2 mutants in the presence of L-A and M1 (7, 24) were more severe in the ski2Δ slh1Δ double mutant. The near normal growth of the ski2Δ slh1Δ L-A-o M-o double mutant suggests that Slh1p is not a component of the 5′ → 3′ mRNA decay pathway because such mutations are synthetic lethal with ski2Δ (15, 25).
Figure 1.
Normal growth at 30°C of ski2Δ slh1Δ cells. Isogenic wild type, ski2Δ, slh1Δ, and ski2Δ slh1Δ mutants were grown on yeast extract/peptone/dextrose plates at 30°C for 3 days.
Polysome gradients of extracts of the isogenic wild type, ski2Δ, slh1Δ, and ski2Δ slh1Δ strains were all normal, with comparable amounts and distributions of polysomes, no imbalance of free 60S and 40S subunits, and no halfmers (data not shown). This result, and the normal growth rates at 30°C indicate that these strains had normal amounts of endogenous mRNAs.
Ski2Δ slh1Δ Mutants Have Derepressed Copy Number of Two dsRNA Viruses That Make Nonpoly(A) mRNA.
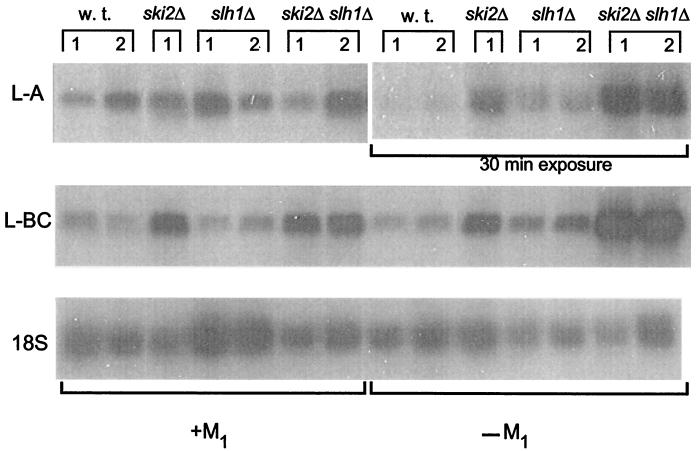
The copy number of the dsRNA viruses reflects the efficiency of expression of their C− A− mRNAs (8). The slh1Δ mutation increased the L-A copy number in the absence of M1 (20) or in its presence but had little effect on L-BC (Fig. 2). The ski2Δ mutation increased L-A and L-BC copy number (Fig. 2; ref. 10). In the absence of M1, the ski2Δ slh1Δ double mutant has higher levels of L-A and L-BC dsRNA than either single mutant (Fig. 2).
Figure 2.
Elevated L-A and L-BC viral dsRNA copy number in slh1 ski2 strains. Two colonies of each strain were grown. Total RNA (5 μg) was loaded onto a 1.2% formaldehyde-agarose gel. The size-fractionated RNAs were transferred to Hybond-N+ membranes (Amersham International) and hybridized under high-stringency conditions to radiolabeled RNA probes corresponding to 18S ribosomal RNA and the ds L-A or L-BC RNAs. After hybridization, the membranes were exposed to film for 16 h unless otherwise noted.
Nonpoly(A) mRNA Is Translated As Well As Poly(A)+ mRNA in ski2Δ slh1Δ Cells.
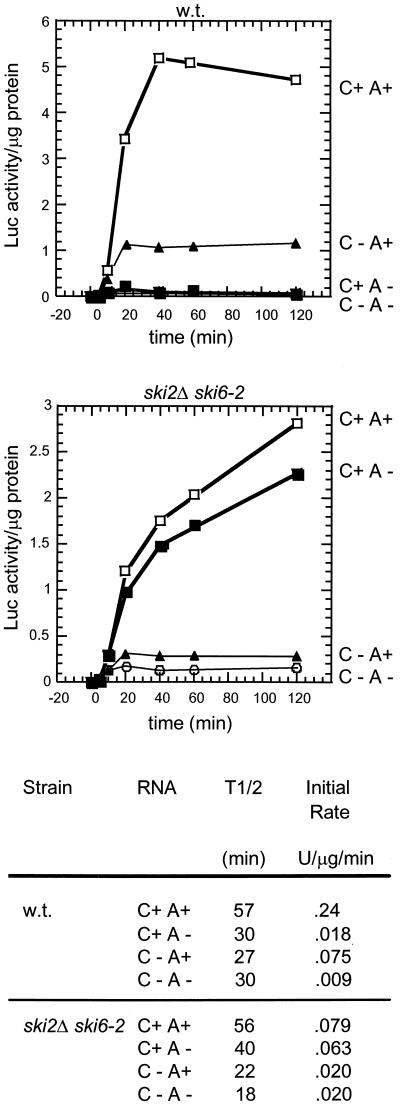
The mRNA electroporation method is uniquely suitable for examining the effects of 5′ cap and 3′ poly(A) on mRNA expression (1, 2, 9, 26). We examined the effect of slh1Δ either alone or in combination with ski2Δ on the expression of electroporated mRNAs with or without the cap or poly(A) (Fig. 3); slh1Δ alone showed only a small effect (< twofold) on expression of electroporated mRNAs with little selectivity. In contrast, ski2Δ specifically derepressed the expression of A− mRNAs, affecting mainly the initial rate of expression, but also modestly increasing the duration of expression (Fig. 3, ref. 9). Nonetheless, the ski2Δ strain expressed A+ mRNA significantly better than nonpoly(A) mRNA. In contrast, the ski2Δ slh1Δ double mutant expressed nonpoly(A) mRNA fully as well as the otherwise identical A+ mRNA (Fig. 3). Both the initial rate, indicative of translation rate, and the duration of expression, a measure of mRNA stability, were indistinguishable.
Figure 3.
ski2Δ slh1Δ strains translate A+ and A− mRNAs alike. Isogenic strains 3221 (wild type), 3515 (ski2Δ), 4106 (slh1Δ), and 4107 (ski2Δslh1Δ) were electroporated (1.3 × 108 cells) with 2 μg of RNA. Cells were maintained at 25°C and assayed for luciferase activity at the indicated timepoints as described (9). Protein concentrations of all lysates were measured by the Bio-Rad protein assay kit. The functional half-lives of each mRNA were calculated by determining the time required for each to produce 50% maximal luciferase activity (1). The maximum rate of luciferase synthesis was the maximum rate change of the activity curves (generally during the first 20–40 min of the assay). The results shown are an average of three experiments. The maximum luciferase translation rate (<0.4 units/μg cell protein/min) is about 10−6 part of total protein synthesis in these cells. (□) C+ A+; (■) C+ A−; (●) C− A+; and (○) C− A−.
Nonpoly(A) mRNA Is Translated As Well As Poly(A)+ mRNA in ski2Δ ski6–2 Cells.
Unlike SKI2, SKI3, SKI7, and SKI8, the SKI6 gene is essential, and temperature-sensitive ski6–2 mutants show defective 60S ribosomal subunits and slow growth at 30°C. In ski6–2 mutants, elevated viral dsRNA copy number and enhanced expression of A− mRNAs are observed even at the permissive temperature (7, 21). Ski6p/Rrp41p is a component of the “exosome” complex of 3′ → 5′ exoribonucleases (27) involved in rRNA processing (21, 27). The ski6–2 effect on expression of nonpoly(A) mRNAs may be mediated by effects on translation (21) or mRNA stability (15) or both.
Combining ski2Δ and ski6–2 mutations had an effect similar to that observed in the ski2Δ slh1Δ strain. Although the slow growth of ski6–2 strains produces reduced translation rates with all mRNAs, including C+ A+ mRNA, the kinetics of expression of C+ A− mRNA was essentially the same as that of C+ A+ mRNA in the double mutant (Fig. 4). Compared with the wild-type strain, the double mutant showed a small increase in the duration of expression of C+ A− mRNA and a dramatic increase in the initial rate of expression.
Figure 4.
ski2Δ ski6–2 strains translate A+ and A− mRNAs alike. Strains 3221 (wild type) and 4709 (ski2Δ ski6–2) were electroporated with 2 μg of luciferase mRNA and assayed for luciferase production as in Fig. 2.
Decay of ACT1 and PGK1 mRNA Is Normal in ski2Δ slh1Δ Cells.
The decay of ACT1 and PGK1 mRNA was examined in isogenic wild-type and ski2Δ slh1Δ cells by arresting new mRNA synthesis with thiolutin (ref. 22; Materials and Methods). Northern blot analysis was used to measure the amounts of ACT1 and PGK1 mRNA at various times, normalizing the blots by probing as well for 18S rRNA. We found no difference between the wild type and the double mutant in the decay rates of either mRNA (data not shown), suggesting that Ski2p and Slh1p do not function in the degradation pathway for these mRNAs.
Discussion
Our data indicate that the translation apparatus is inherently capable of efficiently using mRNAs lacking the 3′ poly(A) structure, even in the presence of the competing A+ cellular mRNA. The nonessential systems involving Ski2p and Slh1p normally prevent the utilization of such mRNAs. These systems work on the naturally A− mRNAs produced by the L-A and L-BC viruses, as indicated by elevated viral copy number in the single mutants and particularly in the double mutant.
The roles of Ski2p and Ski6p in mRNA expression are controversial, with one group proposing translation as the primary effect and turnover effects as secondary (9, 21) whereas another group suggests 3′ → 5′ mRNA degradation is the primary effect (15). The main mRNA turnover system is the 5′ → 3′ system involving Xrn1p/Ski1p (28). Only by blocking this pathway in cis or in trans can an effect of Ski2p or Ski6p on mRNA stability be detected (15). In our experiments, the 5′ → 3′ degradation pathway is not blocked, but we nonetheless see a dramatic effect of the ski2Δ and ski6–2 single mutations, and the double ski2Δ slh1Δ or ski2Δ ski6–2 mutations on the ability to translate nonpoly(A) mRNAs. A+ and A− mRNAs are translated at the same rate for the same duration in the ski2Δ slh1Δ strain.
If Ski2p and Slh1p were solely involved in mRNA turnover, our results would imply that translation in vivo in the wild type only requires the 3′ poly(A) to slow mRNA degradation. If Ski2p and Slhp are involved in the translation process itself, our results imply that these proteins modify the translation apparatus to impose the 3′ poly(A) requirement. In either case, these proteins, and their associated components, play key roles in the treatment by the cell of mRNAs, preventing the utilization of mRNAs lacking the 3′ poly(A), produced as a result of partial degradation or errors in synthesis or splicing or because they are viral mRNAs synthesized without poly(A).
Ski2p Acts Primarily on Translation.
Unless the prevailing 5′ → 3′ mRNA degradation pathway is blocked [in cis by a poly(G) tract or in trans by mutating a gene encoding an element of the pathway], there is no effect of a ski2Δ mutation on mRNA stability (15). Nevertheless, without blocking the 5′ → 3′ mRNA degradation pathway, a ski2Δ mutation dramatically increases the expression of (i) the natural viral nonpoly(A) mRNAs; (ii) nonpoly(A) mRNA made from a RNA polymerase I promoter; and (iii) nonpoly(A) mRNA introduced by electroporation. This indicates that mRNA stability is not the basis of action of Ski2p in these cases. In this study, we have also examined the stability of ACT1 and PGK1 mRNA, and find no difference between the isogenic wild-type and ski2Δ slh1Δ strains.
Comparisons of the kinetics of luciferase synthesis from C+ A− mRNAs in wild-type and ski2Δ (9) or ski2Δ slh1Δ mutant strains (this work) indicate that the functional half-lives are only modestly changed in the mutants, but the initial rates are dramatically increased. This pattern again indicates a primary effect on translation, with an effect on stability that may be secondary. Because inhibition of translation initiation generally leads to a shortened mRNA half-life (29), it is likely that improved translation of a mRNA leads to a lengthened half-life.
The effects of a ski2 mutation on the L-A and M1 viral system also support an action on translation rather than mRNA stability. The M1 dsRNA viral segment encodes the secreted killer toxin. The M1 viral (+) single-stranded RNA lacks a 3′ poly(A) tail and is both the toxin mRNA and an intermediate in replication. Under conditions of excess coat protein availability, a ski2 mutation increases expression from M1 (+) single-stranded RNA, but decreases the copy number of the dsRNA. If the ski2 mutation inactivated an RNA-degrading system, the effect on expression and dsRNA copy number should be in the same direction (8). We suggest that the ski2 mutation diverts viral (+) strands toward translation and therefore away from packaging.
3′ Poly(A) Tails Are Dispensable for Translation.
Other data, in addition to that shown here, support the ability of cells to translate nonpoly(A) mRNA. Proweller and Butler showed that temperature-sensitive mutants in the essential poly(A) polymerase gene (pap1) at the nonpermissive temperature are depleted of total mRNA, but accumulate poly(A)-deficient mRNA that is found on polysomes as large as the corresponding A+ mRNA (4). This indicates that under conditions of mRNA depletion, ribosomes can use mRNA that has at most a short (<25 residues) 3′ poly(A). Mutants in the main 5′ → 3′ exoribonuclease responsible for degrading mRNAs also have some polysome-bound poly(A)-deficient mRNAs, but their relative translation efficiency was not measured (23). Finally, the ability of cells to propagate the L-A and L-BC dsRNA viruses and the 20S and 23S RNA replicons, all of which lack a 3′ poly(A) tail, implies their mRNAs are translated (albeit poorly) without this structure to produce the encoded RNA polymerases and coat proteins (30).
The data presented here shows that a healthy strain, with the normal amount of competing cellular A+ mRNA, can translate a mRNA lacking the 3′poly(A) tail fully as well as the same mRNA having the poly(A) tail, but only if Ski2p and Slh1p are absent.
Acknowledgments
We thank Enzo Martegani for providing the slh1::URA3 plasmid and Ed Hafner and Pfizer for the gift of thiolutin.
Abbreviations
- A+
poly(A)+
- A−
poly(A)−
- C+
cap+
- C−
cap−
- ski
superkiller
- ds RNA
double-stranded RNA
References
- 1.Gallie D R. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 2.Everett J G, Gallie D R. Yeast. 1992;8:1007–1014. doi: 10.1002/yea.320081203. [DOI] [PubMed] [Google Scholar]
- 3.Iizuka N, Najita L, Franzusoff A, Sarnow P. Mol Cell Biol. 1994;14:7322–7330. doi: 10.1128/mcb.14.11.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Proweller A, Butler S. Genes Dev. 1994;8:2629–2640. doi: 10.1101/gad.8.21.2629. [DOI] [PubMed] [Google Scholar]
- 5.Decker C J, Parker R. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 6.Toh-e A, Guerry P, Wickner R B. J Bacteriol. 1978;136:1002–1007. doi: 10.1128/jb.136.3.1002-1007.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ridley S P, Sommer S S, Wickner R B. Mol Cell Biol. 1984;4:761–770. doi: 10.1128/mcb.4.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Widner W R, Wickner R B. Mol Cell Biol. 1993;13:4331–4341. doi: 10.1128/mcb.13.7.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masison D C, Blanc A, Ribas J C, Carroll K, Sonenberg N, Wickner R B. Mol Cell Biol. 1995;15:2763–2771. doi: 10.1128/mcb.15.5.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ball S G, Tirtiaux C, Wickner R B. Genetics. 1984;107:199–217. doi: 10.1093/genetics/107.2.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsumoto Y, Fishel R, Wickner R B. Proc Natl Acad Sci USA. 1990;87:7628–7632. doi: 10.1073/pnas.87.19.7628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esteban R, Rodriguez-Cousino N, Esteban L M. Prog Nucleic Acid Res Mol Biol. 1993;46:155–182. doi: 10.1016/s0079-6603(08)61021-1. [DOI] [PubMed] [Google Scholar]
- 13.Park C M, Lopinski J D, Masuda J, Tzeng T H, Bruenn J A. Virology. 1996;216:451–454. doi: 10.1006/viro.1996.0083. [DOI] [PubMed] [Google Scholar]
- 14.Benard L, Carroll K, Valle R C P, Wickner R B. J Virol. 1999;73:2893–2900. doi: 10.1128/jvi.73.4.2893-2900.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacobs-Anderson J S, Parker R. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee S-G, Lee I, Kang C, Song K. Genomics. 1995;25:660–666. doi: 10.1016/0888-7543(95)80008-a. [DOI] [PubMed] [Google Scholar]
- 17.Dangel A W, Shen L, Mendoza A R, Wu L-C, Yu C Y. Nucleic Acids Res. 1995;23:2120–2126. doi: 10.1093/nar/23.12.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nomura N, Miyajima N, Sazuka T, Tanaka A, Kawarabayashi Y, Sato S, Nagase T, Seki N, Ishikawa K, Tabata S. DNA Res. 1994;1:27–35. doi: 10.1093/dnares/1.1.27. [DOI] [PubMed] [Google Scholar]
- 19.Qu X, Yang Z, Zhang S, Shen L, Dangel A W, Hughes J H, Redman K L, Wu L-C, Yu C Y. Nucleic Acids Res. 1998;26:4068–4077. doi: 10.1093/nar/26.17.4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martegani E, Vanoni M, Mauri I, Rudoni S, Saliola M, Alberghina L. Yeast. 1997;13:391–397. doi: 10.1002/(SICI)1097-0061(19970330)13:4<391::AID-YEA92>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 21.Benard L, Carroll K, Valle R C P, Wickner R B. Mol Cell Biol. 1998;18:2688–2696. doi: 10.1128/mcb.18.5.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parker R, Herrick D, Peltz S W, Jacobson A. In: Guide to Yeast Genetics and Molecular Biology. Guthrie C, Fink G R, editors. Vol. 194. San Diego: Academic; 1991. pp. 415–423. [Google Scholar]
- 23.Hsu C L, Stevens A. Mol Cell Biol. 1993;13:4826–4835. doi: 10.1128/mcb.13.8.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Esteban R, Wickner R B. Genetics. 1987;117:399–408. doi: 10.1093/genetics/117.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson A W, Kolodner R D. Mol Cell Biol. 1995;15:2719–2727. doi: 10.1128/mcb.15.5.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Preiss T, Hentze M W. Nature (London) 1998;392:516–520. doi: 10.1038/33192. [DOI] [PubMed] [Google Scholar]
- 27.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 28.Muhlrad D, Decker C J, Parker R. Mol Cell Biol. 1995;15:2145–2156. doi: 10.1128/mcb.15.4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwartz D C, Parker R. Mol Cell Biol. 1999;19:5247–5256. doi: 10.1128/mcb.19.8.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wickner R B. In: Fields Virology. Fields B N, Knipe D M, Howley P M, editors. Vol. 1. New York: Raven; 1996. pp. 557–585. [Google Scholar]