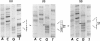Abstract
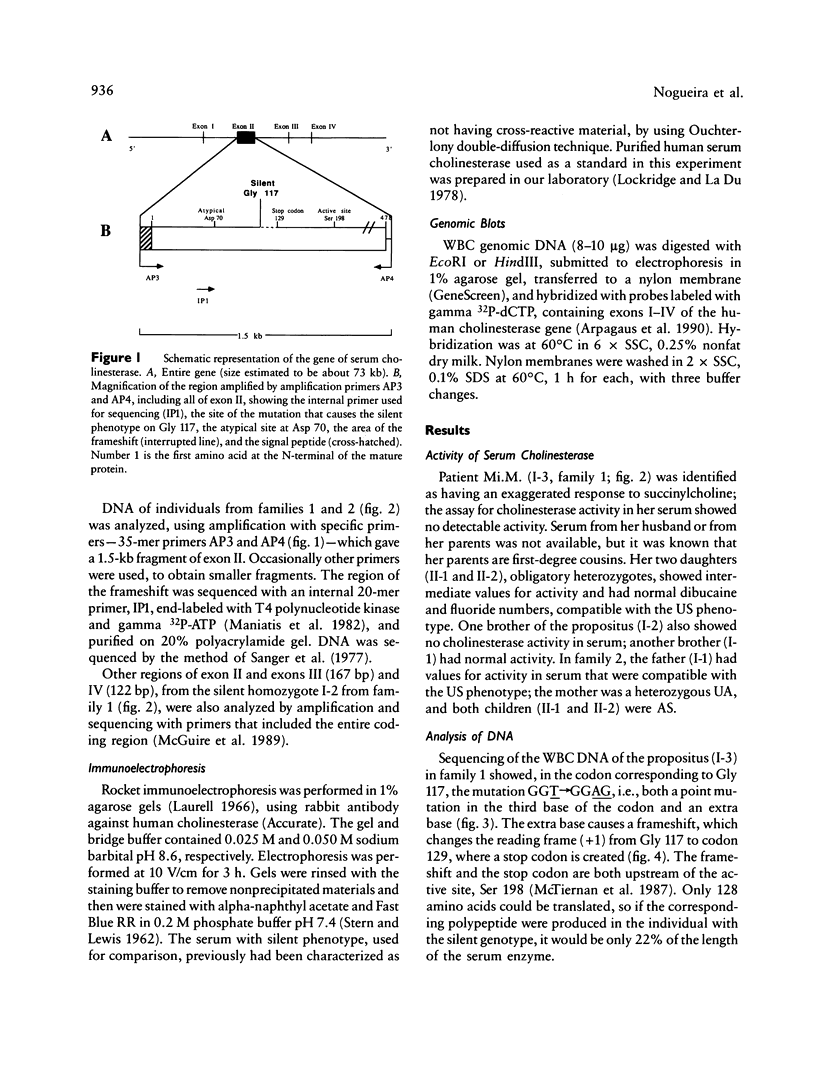
A frameshift mutation that causes a silent phenotype for human serum cholinesterase was identified in the DNA of seven individuals of two unrelated families. The mutation, identified using the polymerase chain reaction, causes a shift in the reading frame from Gly 117, where GGT (Gly)----GGAG (Gly+ 1 base) to a new stop codon created at position 129. This alteration is upstream of the active site (Ser 198), and, if any protein were made, it would represent only 22% of the mature enzyme found in normal serum. Results of analysis of the enzymatic activities in serum agreed with the genotypes inferred from the nucleotide sequence. Rocket immunoelectrophoresis using alpha-naphthyl acetate to detect enzymatic activity showed an absence of cross-reactive material, as expected. One additional individual with a silent phenotype did not show the same frameshift mutation. This was not unexpected, since there must be considerable molecular heterogeneity involved in causes for the silent cholinesterase phenotype. This is the first report of a molecular mechanism underlying the silent phenotype for serum cholinesterase. The analytical approach used was similar to the one we recently employed to identify the mutation that causes the atypical cholinesterase variant.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Agarwal D. P., Srivastava L. M., Goedde H. W. A note on suxamethonium sensitivity and serum cholinesterase variants. Hum Genet. 1976 Apr 15;32(1):85–88. doi: 10.1007/BF00569981. [DOI] [PubMed] [Google Scholar]
- Arnason A., Jensson O., Gudmundsson S. Serum esterases of Icelanders. I. A "silent" pseudocholinesterase gene in an Icelandic family. Clin Genet. 1975 May-Jun;7(5):405–412. doi: 10.1111/j.1399-0004.1975.tb00349.x. [DOI] [PubMed] [Google Scholar]
- Arpagaus M., Kott M., Vatsis K. P., Bartels C. F., La Du B. N., Lockridge O. Structure of the gene for human butyrylcholinesterase. Evidence for a single copy. Biochemistry. 1990 Jan 9;29(1):124–131. doi: 10.1021/bi00453a015. [DOI] [PubMed] [Google Scholar]
- Bateman J. F., Lamande S. R., Dahl H. H., Chan D., Mascara T., Cole W. G. A frameshift mutation results in a truncated nonfunctional carboxyl-terminal pro alpha 1(I) propeptide of type I collagen in osteogenesis imperfecta. J Biol Chem. 1989 Jul 5;264(19):10960–10964. [PubMed] [Google Scholar]
- Chan V., Chan T. K., Kan Y. W., Todd D. A novel beta-thalassemia frameshift mutation (codon 14/15), detectable by direct visualization of abnormal restriction fragment in amplified genomic DNA. Blood. 1988 Oct;72(4):1420–1423. [PubMed] [Google Scholar]
- Das P. K. Further evidence on the heterogeneity of "silent" serum cholinesterase variants. Hum Hered. 1973;23(1):88–96. doi: 10.1159/000152559. [DOI] [PubMed] [Google Scholar]
- Endo H., Hasegawa K., Narisawa K., Tada K., Kagawa Y., Ohta S. Defective gene in lactic acidosis: abnormal pyruvate dehydrogenase E1 alpha-subunit caused by a frame shift. Am J Hum Genet. 1989 Mar;44(3):358–364. [PMC free article] [PubMed] [Google Scholar]
- Evans R. T., Magill P. J. Evidence for mutation being the source of the abnormal gene for plasma cholinesterase. J Med Genet. 1974 Jun;11(2):117–120. doi: 10.1136/jmg.11.2.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh P. J., Schmeissner U., Hofer M., Miller J. H. Genetic studies of the lac repressor. VII. On the molecular nature of spontaneous hotspots in the lacI gene of Escherichia coli. J Mol Biol. 1978 Dec 25;126(4):847–857. doi: 10.1016/0022-2836(78)90023-2. [DOI] [PubMed] [Google Scholar]
- Goedde H. W., Altland K. Evidence for different "silent genes" in the human serum pseudocholinesterase polymorphism. Ann N Y Acad Sci. 1968 Jun 14;151(1):540–544. doi: 10.1111/j.1749-6632.1968.tb11913.x. [DOI] [PubMed] [Google Scholar]
- Goedde H. W., Gehring D., Hofmann R. A. On the problem of a "silent gene" in pseudocholinesterase polymorphism. Biochim Biophys Acta. 1965 Sep 13;107(2):391–393. doi: 10.1016/0304-4165(65)90149-2. [DOI] [PubMed] [Google Scholar]
- Gutsche B. B., Scott E. M., Wright R. C. Hereditary deficiency of pseudocholinesterase in Eskimos. Nature. 1967 Jul 15;215(5098):322–323. doi: 10.1038/215322b0. [DOI] [PubMed] [Google Scholar]
- HARRIS H., WHITTAKER M. Differential inhibition of human serum cholinesterase with fluoride: recognition of two new phenotypes. Nature. 1961 Jul 29;191:496–498. doi: 10.1038/191496a0. [DOI] [PubMed] [Google Scholar]
- HARRIS H., WHITTAKER M., LEHMANN H., SILK E. The pseudocholinesterase variants. Esterase levels and dibucaine numbers in families selected through suxamethonium sensitive individuals. Acta Genet Stat Med. 1960;10:1–16. doi: 10.1159/000151112. [DOI] [PubMed] [Google Scholar]
- HART S. M., MITCHELL J. V. Suxamethonium in the absence of pseudocholinesterase. Br J Anaesth. 1962 Mar;34:207–209. doi: 10.1093/bja/34.3.207. [DOI] [PubMed] [Google Scholar]
- HODGKIN W., GIBLETT E. R., LEVINE H., BAUER W., MOTULSKY A. G. COMPLETE PSEUDOCHOLINESTERASE DEFICIENCY: GENETIC AND IMMUNOLOGIC CHARACTERIZATION. J Clin Invest. 1965 Mar;44:486–493. doi: 10.1172/JCI105162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KALOW W., GENEST K. A method for the detection of atypical forms of human serum cholinesterase; determination of dibucaine numbers. Can J Biochem Physiol. 1957 Jun;35(6):339–346. doi: 10.1139/y57-041. [DOI] [PubMed] [Google Scholar]
- KALOW W., LINDSAY H. A. A comparison of optical and manometric methods for the assay of human serum cholinesterase. Can J Biochem Physiol. 1955 Jul;33(4):568–574. [PubMed] [Google Scholar]
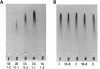
- KALOW W., STARON N. On distribution and inheritance of atypical forms of human serum cholinesterase, as indicated by dibucaine numbers. Can J Biochem Physiol. 1957 Dec;35(12):1305–1320. [PubMed] [Google Scholar]
- Kazazian H. H., Jr, Boehm C. D. Molecular basis and prenatal diagnosis of beta-thalassemia. Blood. 1988 Oct;72(4):1107–1116. [PubMed] [Google Scholar]
- Kunkel T. A. Frameshift mutagenesis by eucaryotic DNA polymerases in vitro. J Biol Chem. 1986 Oct 15;261(29):13581–13587. [PubMed] [Google Scholar]
- LEHMANN H., RYAN E. The familial incidence of low pseudocholinesterase level. Lancet. 1956 Jul 21;271(6934):124–124. doi: 10.1016/s0140-6736(56)90869-8. [DOI] [PubMed] [Google Scholar]
- LIDDELL J., LEHMANN H., SILK E. A 'silent' pseudo-cholinesterase gene. Nature. 1962 Feb 10;193:561–562. doi: 10.1038/193561a0. [DOI] [PubMed] [Google Scholar]
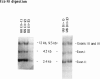
- Laurell C. B. Quantitative estimation of proteins by electrophoresis in agarose gel containing antibodies. Anal Biochem. 1966 Apr;15(1):45–52. doi: 10.1016/0003-2697(66)90246-6. [DOI] [PubMed] [Google Scholar]
- Lockridge O., La Du B. N. Comparison of atypical and usual human serum cholinesterase. Purification, number of active sites, substrate affinity, and turnover number. J Biol Chem. 1978 Jan 25;253(2):361–366. [PubMed] [Google Scholar]
- Lubin A. H., Garry P. J., Owen G. M., Prince L. C., Dietz A. A. Further variation of the "silent" cholinesterase gene. Biochem Med. 1973 Aug;8(1):160–165. doi: 10.1016/0006-2944(73)90019-7. [DOI] [PubMed] [Google Scholar]
- McGuire M. C., Nogueira C. P., Bartels C. F., Lightstone H., Hajra A., Van der Spek A. F., Lockridge O., La Du B. N. Identification of the structural mutation responsible for the dibucaine-resistant (atypical) variant form of human serum cholinesterase. Proc Natl Acad Sci U S A. 1989 Feb;86(3):953–957. doi: 10.1073/pnas.86.3.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McTiernan C., Adkins S., Chatonnet A., Vaughan T. A., Bartels C. F., Kott M., Rosenberry T. L., La Du B. N., Lockridge O. Brain cDNA clone for human cholinesterase. Proc Natl Acad Sci U S A. 1987 Oct;84(19):6682–6686. doi: 10.1073/pnas.84.19.6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura O., Hirosawa S., Kato A., Aoki N. Molecular basis for congenital deficiency of alpha 2-plasmin inhibitor. A frameshift mutation leading to elongation of the deduced amino acid sequence. J Clin Invest. 1989 May;83(5):1598–1604. doi: 10.1172/JCI114057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nukiwa T., Takahashi H., Brantly M., Courtney M., Crystal R. G. alpha 1-Antitrypsin nullGranite Falls, a nonexpressing alpha 1-antitrypsin gene associated with a frameshift to stop mutation in a coding exon. J Biol Chem. 1987 Sep 5;262(25):11999–12004. [PubMed] [Google Scholar]
- Okada Y., Streisinger G., Owen J. E., Newton J., Tsugita A., Inouye M. Molecular basis of a mutational hot spot in the lysozyme gene of bacteriophage T4. Nature. 1972 Apr 14;236(5346):338–341. doi: 10.1038/236338a0. [DOI] [PubMed] [Google Scholar]
- Pihlajaniemi T., Dickson L. A., Pope F. M., Korhonen V. R., Nicholls A., Prockop D. J., Myers J. C. Osteogenesis imperfecta: cloning of a pro-alpha 2(I) collagen gene with a frameshift mutation. J Biol Chem. 1984 Nov 10;259(21):12941–12944. [PubMed] [Google Scholar]
- Prody C. A., Dreyfus P., Zamir R., Zakut H., Soreq H. De novo amplification within a "silent" human cholinesterase gene in a family subjected to prolonged exposure to organophosphorous insecticides. Proc Natl Acad Sci U S A. 1989 Jan;86(2):690–694. doi: 10.1073/pnas.86.2.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao P. R., Gopalam K. B. High incidence of the silent allele at cholinesterase locus I in Vysyas of Andhra Pradesh (S. India). Hum Genet. 1979 Nov 1;52(1):139–141. doi: 10.1007/BF00284608. [DOI] [PubMed] [Google Scholar]
- Rubinstein H. M., Dietz A. A., Hodges L. K., Lubrano T., Czebotar V. Silent cholinesterase gene: variations in the properties of serum enzyme in apparent homozygotes. J Clin Invest. 1970 Mar;49(3):479–486. doi: 10.1172/JCI106257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SIMPSON N. E., KALOW W. THE "SILENT" GENE FOR SERUM CHOLINESTERASE. Am J Hum Genet. 1964 Jun;16:180–188. [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott E. M. Properties of the trace enzyme in human serum cholinesterase deficiency. Biochem Biophys Res Commun. 1970 Mar 12;38(5):902–906. doi: 10.1016/0006-291x(70)90806-5. [DOI] [PubMed] [Google Scholar]
- Scott E. M., Wright R. C. A third type of serum cholinesterase deficiency in Eskimos. Am J Hum Genet. 1976 May;28(3):253–256. [PMC free article] [PubMed] [Google Scholar]
- Sifers R. N., Brashears-Macatee S., Kidd V. J., Muensch H., Woo S. L. A frameshift mutation results in a truncated alpha 1-antitrypsin that is retained within the rough endoplasmic reticulum. J Biol Chem. 1988 May 25;263(15):7330–7335. [PubMed] [Google Scholar]
- Simpson N. E. Genetics of esterases in man. Ann N Y Acad Sci. 1968 Jul 31;151(2):699–709. doi: 10.1111/j.1749-6632.1968.tb48251.x. [DOI] [PubMed] [Google Scholar]
- Streisinger G., Okada Y., Emrich J., Newton J., Tsugita A., Terzaghi E., Inouye M. Frameshift mutations and the genetic code. This paper is dedicated to Professor Theodosius Dobzhansky on the occasion of his 66th birthday. Cold Spring Harb Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]