Abstract
Progress in molecular genetics makes possible the development of alternative disease control strategies that target the competence of mosquitoes to transmit pathogens. We tested the regulatory region of the vitellogenin (Vg) gene of Aedes aegypti for its ability to express potential antipathogen factors in transgenic mosquitoes. Hermes-mediated transformation was used to integrate a 2.1-kb Vg-promoter fragment driving the expression of the Defensin A (DefA) coding region, one of the major insect immune factors. PCR amplification of genomic DNA and Southern blot analyses, carried out through the ninth generation, showed that the Vg-DefA transgene insertion was stable. The Vg-DefA transgene was strongly activated in the fat body by a blood meal. The mRNA levels reached a maximum at 24-h postblood meal, corresponding to the peak expression time of the endogenous Vg gene. High levels of transgenic defensin were accumulated in the hemolymph of bloodfed female mosquitoes, persisting for 20–22 days after a single blood feeding. Purified transgenic defensin showed antibacterial activity comparable to that of defensin isolated from bacterially challenged control mosquitoes. Thus, we have been able to engineer the genetically stable transgenic mosquito with an element of systemic immunity, which is activated through the blood meal-triggered cascade rather than by infection. This work represents a significant step toward the development of molecular genetic approaches to the control of vector competence in pathogen transmission.
Mosquito-borne diseases are among the most devastating in modern times. Malaria is a particularly threatening disease that already affects millions of people, and the numbers are increasing at an alarming rate. Diseases caused by mosquito-borne viruses, most importantly dengue fever, also are on the rise. The major reasons for this catastrophic situation are the unavailability of effective vaccines for malaria and other mosquito-borne diseases and the development of insecticide and drug resistance by vectors and pathogens, respectively (1–3). Therefore, there is an urgent need to explore every possible option for developing unique control strategies against malaria and other vector-borne diseases.
One approach is to block the transmission of a pathogen by using the transgenic manipulation of its mosquito vector (4). In such an approach, a chimeric gene consisting of a coding region for a factor with an antipathogen activity, driven by a tissue- and stage-specific promoter, would be transformed into the vector. Activation of the transgene would result in the production of the antipathogen factor that could adversely affect mosquito stages of parasite development and consequently block its transmission to the vertebrate host.
Recent progress in insect transgenesis, and in particular, the accomplishment of transformation in the yellow fever mosquito, Aedes aegypti (5–7), has made it possible to test this approach experimentally. However, the availability of vector genes with well-characterized properties with respect to their tissue- and stage-specific expression is limited. Likewise, our knowledge about factors, which could effect development of various pathogens of mosquito-borne diseases, is restricted as well. In this work, we have approached modeling the element of systemic immunity in the mosquito by using transgenic technology via use of a well-characterized, fat body-specific gene and a natural mosquito immune factor. The vitellogenin (Vg) gene encodes the major yolk protein precursor, and its promoter could be ideal for driving the expression of an immune antipathogen factor via a blood meal-triggered regulatory cascade. After a blood meal, Vg gene expression is activated to a high level in the female fat body, a center of systemic innate immunity in insects (8–10). Recently, we have shown that a 2.1-kb 5′-upstream promoter region from the Ae. aegypti Vg gene is sufficient for correct tissue- and stage-specific expression in the mosquito Ae. aegypti and Drosophila (V.K., D. Martin, A.A., M. Mienaltowski, and A.R., unpublished data).
Defensin has been chosen for this work because these immune factors are the predominant group of innate antibacterial immune factors in mosquitoes (11–15). Furthermore, studies using exogenous defensins demonstrated that these antibacterial peptides possess potent anti-Plasmodium activity (16, 17). Moreover, defensin has been implicated in the local innate immune response of the Anopheles gambiae midgut to a Plasmodium infection (18). Thus, in this work, we have used the Ae. aegypti Vg promoter to drive the expression of the Ae. aegypti Defensin A (DefA) gene that encodes the prepro-defensin (12).
The major goal of our present work is to engineer a transgenic mosquito in which turning on the gene(s) encoding antipathogen factors and production of biologically active factor(s) are linked to the blood meal-triggered regulatory cascade. Here, we report the successful germ-line transformation of the mosquito vector Ae. aegypti with the Vg-DefA transgene. We show that the Vg-DefA transgene, integrated into mosquito genome, is strongly activated in the fat body after a blood meal, and a biologically active defensin is accumulated in the hemolymph of the transgenic mosquitoes. This work represents a significant step toward the development of molecular genetic approaches to the control of vector competence in pathogen transmission.
Materials and Methods
Plasmid Construction.
The transformation plasmid vector pH[cn], which includes Hermes transposon inverted repeats from Musca domestica and a marker cinnabar (cn) gene from D. melanogaster, described by Jasinskiene et al. (6), was used for construction of the Vg-DefA vector. A 2.1-kb 5′-upstream region of the Vg gene (9) was first subcloned in the pBlueScript (Stratagene) plasmid. The 450-bp coding region of the DefA gene (nucleotides from +50 to +498) (15) was ligated into a BamHI site at the 3′-end of a 2.1- kb Vg fragment (nucleotides from −2,015 to + 70) (9). The resulting 2.5-kb DNA fragment, designated Vg-DefA, was inserted into the double-digested XbaI–XhoI sites of pH[cn]. The resulting plasmid, pH[cn][Vg-DefA], and the plasmid encoding the Hermes transposase, pHSHH1.9 (19), were purified by a Qiagen kit.
Generation of Transgenic Mosquitoes.
The Ae. aegypti kynurenine hydroxylase-white (khw) mutant strain has a white-eye phenotype (20), which is complemented by the D. melanogaster cn gene, which changes the eye color to red (21). The strain was obtained from F. Collins (University of Notre Dame, Notre Dame, IN). Mosquitoes were reared as previously described (22). Newly laid eggs were collected, and 90- to 270-min-old embryos were prepared for microinjection in the manner described by Morris (23). The embryos were injected with a mixture of the pH[cn][Vg-DefA] and pHSHH1.9 plasmids at a final concentration of 0.5 mg/ml and 0.3 mg/ml, respectively, in 5 mM KCl and 0.1 mM NaH2PO4 (pH 6.8). After DNA microinjection, 16- to 20-h-old embryos were exposed to heat shock at 39°C for 60 min and then placed at 27°C, 85% relative humidity, and hatched 5 or 6 days after the heat shock. Adult mosquitoes resulting from generation zero (G0)-injected embryos were crossed with virgin females or males of the khw mutant strain. The G1 progeny were then screened for complementation of the white-eye phenotype.
Southern Blot and Gene Amplification Analyses.
Southern blot and gene amplification analyses were done as described (24). Genomic DNA from individual mosquitoes was purified by the method of Bender et al. (25). After digestion with the ApaI and XbaI or BglII restriction endonucleases, the DNA fragments were separated by electrophoresis in a 1% agarose gel, transferred to nitrocellulose filters, and hybridized with the DefA DNA probe.
Total RNA from individual mosquitoes was purified by the Trizol technique (GIBCO/BRL). After drying, the RNA pellet was resuspended in diethyl pyrocarbonate (DEPC)-treated, distilled H2O. Aliquots equivalent to 1/10th of a mosquito were used for reverse transcription (RT) and amplification. RT-PCR was performed by using the Superscript one-step RT-PCR kit (GIBCO/BRL). Tubes containing RNA were incubated for 30 min at 45°C for the RT reaction. Amplification conditions included rapid heating to 94°C for 2 min followed by 15 cycles of 60°C for 40 sec, 72°C for 1 min, and 94°C for 45 sec. The RT-PCR forward reaction primer was Vg228F (5′-GAACACACAATCGGAACAGCTG-3′); the reverse primer was Df229R (5′-ATTCCGGCAGACGCACACCTT-3′). The following primers were used for the forward reaction of the genomic DNA amplification experiment: Vg73F (5′-AGATTGATTTATTTTATATGCTTCCTGA-3′), Vg74F (5′-ATCTTCAATTCACATCTGTAGTCTCAA-3′), and Df189F (5′-CCATGCAGCCCCTCACTGTCATT-3′), and the reverse primer was Df190R (5′-ACCATTTAACAAAATTATGC-3′).
Immunoblot Analysis.
Protein extracts from whole mosquitoes and mosquito tissues were prepared by homogenization in Trizol solution (GIBCO/BRL) according to the protocol of the manufacturer. Hemolymph from individual mosquitoes was collected by opening the hemocoel and freeing the hemolymph to mix with the dissection buffer (22). This mixture was immediately frozen in liquid nitrogen and stored at −80°C. Samples were loaded directly onto Tris-Tricine-SDS 10–20% acrylamide gradient gel (Bio-Rad). Anti-DefA IgG was prepared by Cho et al. (12). Immunoblots were developed by using the ECL detection system (Amersham Pharmacia).
Insect Immunization.
Mosquitos were infected by intrathoracic injections of 0.25 μl of logarithmic phase Escherichia coli, strain LE392, suspended in mosquito saline (150 mM NaCl/4 mM KCl/3 mM CaCl2/1.8 mM NaHCO3/0.6 mM MgCl2/25 mM Hepes, pH 7.0). The bacterial suspension was heated at 100°C for 10 min before injection. Defensin was purified from infected mosquitoes 24 h after bacteria induction.
Extraction and Purification of Defensin Peptide.
Peptide extraction and purification were done as described by Bulet et al. (26) with minor modifications. In brief, hemolymph from 50 mosquito females was collected, and the samples were filtered through UltraFree-MC (30,000 molecular weight filter unit; Millipore) for 15 min at 14,000 × g. The cell-free hemolymph was acidified (0.05% trifluoroacetic acid) and loaded onto Sep-Pak C18 cartridge (Waters). After washing with 5 ml of acidified water (0.05% trifluoroacetic acid), elution was performed with 40% acetonitrile in acidified water (0.05% trifluoroacetic acid), the samples were concentrated in a vacuum centrifuge (Savant), and the pellet reconstituted with MilliQ water before monitoring the antibacterial activity.
Antibacterial Activity Assay.
Antibacterial activity was monitored by a liquid growth inhibition assay as described (26). Purified antibacterial peptide samples were dried in wells of a microtiter plate and were incubated at room temperature with 100 μl of a suspension of mid-logarithmic phase Micrococcus luteus. After overnight incubation, microbial growth was assessed by an increase in OD595.
Results and Discussion
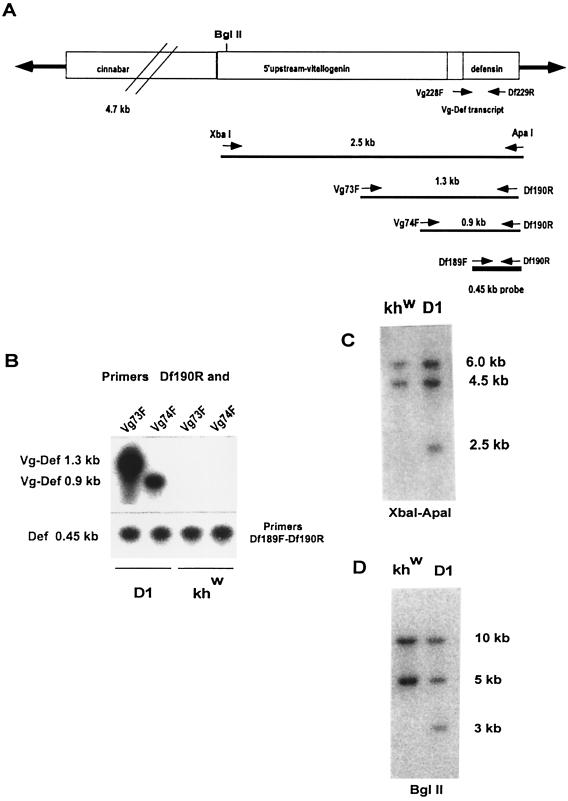
A promoter–reporter plasmid was constructed by subcloning the Vg-DefA sequence into the transformation vector pH[cn] (6), which includes the cn gene from D. melanogaster flanked by the terminal inverted repeats of the Hermes transposon (Fig. 1A). More than 3,000 preblastoderm embryos of the Ae. aegypti white-eyed, khw strain (20) were injected with the pH[cn]Vg-DefA construct and the Hermes transposase-helper plasmid (19). Transformed G1 progeny were identified by colored eyes and were used to establish transformed lines. Five independent, stable transgenic lines were recovered from 600 G0 individuals. One of these lines, D1, was used for detailed analysis and characterization of the Vg-DefA transgene expression. The characterization of other transgenic lines will be described elsewhere. In the D1 transgenic line, stability of the Vg-DefA transgene insertion was monitored from the G2 to G9 generations. For the genomic PCR amplification analyses, we used a combination of one of the direct primers with Vg-specific sequences (Vg73F or Vg74F) and a reverse primer with DefA-specific sequence (Df190R) (Fig. 1A). The use of these primers allowed us to detect the Vg-DefA transgene in genomic DNA samples without cross amplification of endogenous Vg and DefA genes (Fig. 1B). Both combinations of primers (Vg73F–Df190R) and (Vg74F–Df190R) revealed the presence of specific amplified DNA fragments of expected sizes only in the transgenic mosquitoes (Fig. 1B). Southern blot analyses of G4 and G9 progeny further confirmed the integration of the transgene. The presence of an additional band was revealed in the D1 transgenic line as compared to the nontransformed khw parental line, by using either XbaI–ApaI or BglII restriction enzymes, followed by hybridization with a DefA-specific probe (Fig. 1 C and D). Thus, both genomic PCR amplification and Southern analyses demonstrated stable integration of the Vg-DefA transgene into the Ae. aegypti genome. In addition, our genetic analysis data using test crosses between mosquitoes of the D1 transgenic line and either white-eyed khw strain or wild-type UGAL strain were consistent with the insertion and subsequent Mendelian segregation of the Vg-DefA transgene in a single chromosome (data not shown).
Figure 1.
Stable integration of the Vg-Defensin transgene into the Aedes aegypti genome. (A) Schematic diagram of the transformation vector construct, pH[cn][Vg-DefA], used in this study to transform mosquitoes. The Hermes inverted terminal repeats are represented as solid arrows flanking the 4.7-kb genomic DNA fragment of the D. melanogaster cn+ marker gene and the 2.5-kb Vg-DefA fusion gene as the promoter–reporter part of the construct. The arrows and numbers below the diagram show the relative extents and the expected sizes of the fragments generated by XbaI–ApaI restriction digest and by gene amplification using different pairs of primers specific to the Vg-DefA fusion sequence. The 0.45-kb DefA DNA sequence used as a probe in these experiments is shown as a black box below the map. (B) Gene amplification analysis of genomic DNA isolated from G4 mosquito progeny of the D1 transgenic line and the control khw strain. To confirm the specificity of amplification for the Vg-DefA transgene, the gene amplification products were analyzed by Southern blot hybridization using the DefA DNA as a probe. By using the Vg-DefA primer combinations, Vg73F-Df190R and Vg74F-Df190R, the transgenic mosquito DNA showed specific amplification of fragments of the expected sizes, 1.3-kb and 0.9-kb, respectively. No amplification was observed in the genomic DNA from the control strain (Upper). As a positive control for loading and for the integrity of isolated genomic DNA, DefA-specific primer pairs, Df189F-Df190R, were used, and these produced a band of the expected size, 0.45-kb, in all samples (Lower). (C) Southern blot analysis of genomic DNA prepared from G4 progeny of transformed mosquitoes of the D1 line and the host strain (khw), digested with XbaI–ApaI and hybridized with the DefA DNA probe. A diagnostic hybridization signal of the expected size, 2.5 kb, is associated with the insertion of the Vg-DefA transgene into the mosquito genome and is evident in DNA isolated from transformed mosquitoes, but not in the host strain. The two higher molecular weight signals of 6 kb and 4.5 kb, seen in DNA digests of both the transgenic and the host strains, represented two copies of endogenous DefA genes. (D) Southern blot analysis of genomic DNA isolated from G9 progeny of the D1 transgenic line and the host khw strain, digested with BglII and hybridized with DefA DNA probe. The unique hybridization signal of 3.0-kb associated with Vg-DefA insertion is seen only in the D1 transgenic line.
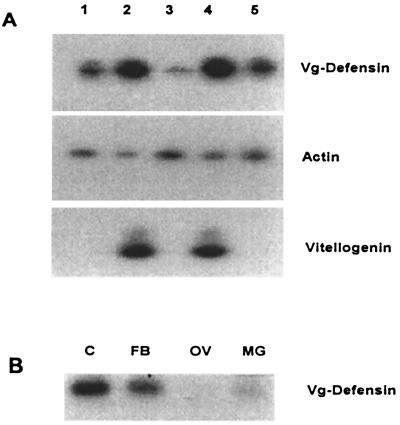
Expression of the Vg-DefA transgene was investigated by using RT-PCR technique. To provide a unique marker for the transgenic Vg-DefA mRNA that would allow its specific detection by RT-PCR, a 70-bp leader sequence from the Vg transcript was cloned upstream of the 5′-end of the DefA-coding sequence. A direct primer (Vg228F) designed from this Vg leader sequence and a reverse primer (Df229R) based on the 3′-end coding region of DefA were used in the RT-PCR analyses of the Vg-DefA transgene expression (Fig. 1A). RT-PCR demonstrated that the Vg-DefA transgene transcript was present at very low levels in previtellogenic transgenic female mosquitoes (Fig. 2A, lane 3). However, transcription of the transgene was activated to a high level after a blood meal, and its maximal expression was observed at 24 h post blood meal (hPBM) corresponding to the peak expression of the Vg gene (Fig. 2A, lane 4). Unlike Vg mRNA, the Vg-DefA mRNA was still detectable 3 days after a single blood meal, when expression of the endogenous Vg gene is no longer evident (Fig. 2A, lane 5). Low but detectable levels of the Vg-DefA mRNA were observed in transgenic mosquitoes even at 22 days after a single blood meal (data not shown). This unusually long persistence of the Vg-DefA transgene transcript may be caused by a combination of a low level of its constitutive expression as well as a high stability of its mRNA, a property characteristic for defensin mRNA (15).
Figure 2.
RT-PCR analyses of the Vg-DefA transgene expression. (A) Blood meal activation of the Vg-DefA transgene in mRNA samples isolated from transgenic males (1), blood-fed females (2), previtellogenic (3) and vitellogenic females 24 (4), and 72 hPBM (5). The same RNA samples also were analyzed using actin-specific primers as a control for RNA integrity and loading (Middle), and vitellogenin-specific primers, as a control showing the expression of the endogenous Vg gene after blood meal activation (Bottom). (B) Tissue-specific expression of the Vg-DefA transgene in vitellogenic females. Specific amplification was observed only in RNA samples isolated from the fat body (FB) and carcass (C), containing the fat body of the thorax and head. No amplification was found in the ovary (OV) or midgut (MG).
In addition to a strong activation of the Vg-DefA transgene by a blood meal, its expression also was similar to that of the Vg gene with respect to its tissue specificity. The Vg-DefA mRNA was detected only in the fat body and not in the ovary or midgut (Fig. 2B). However, unlike the Vg gene, the Vg-DefA transgene was expressed at a low level in transgenic male mosquitoes (Fig. 2A, lane 1). The tissue specificity of the Vg-DefA transgene expression in mosquito males was not assessed. It is possible that regulatory elements required for a complete repression of the Vg gene expression in males were not present in the 2.1-kb 5′-upstream region of the Vg gene used for the construction of the Vg-DefA transgene.
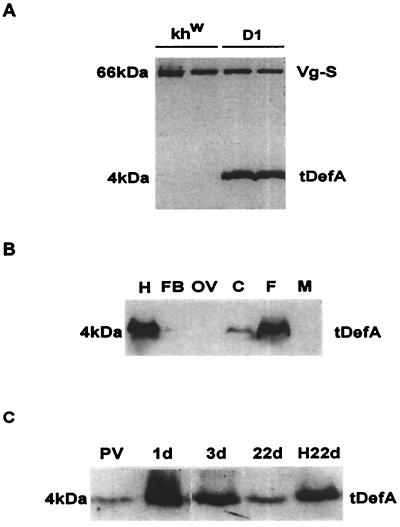
Immunoblot analyses with anti-DefA antibodies (12) showed that in response to a blood meal the fat body of the transgenic mosquito produced a mature peptide with the expected molecular weight of 4 kDa (Fig. 3A). Transgenic defensin (tDefA) was observed in blood-fed transgenic mosquitoes but not in blood-fed mosquitoes from the khw host strain (Fig. 3A). Likewise, defensin was not induced by blood meal in wild-type mosquitoes of Rockefeller strain (12). Thus, in the transgenic mosquitoes, the Vg-DefA mRNA was properly translated and the prepro-defensin was correctly processed into a mature defensin protein in the blood-activated, rather than infection-activated fat body. In turn, this suggested that the machinery required for translational and posttranslational immune factor processing was present in the previtellogenic fat bodies not challenged by infection. In agreement with the RT-PCR data on Vg-Def transcription, tDefA was produced exclusively in the fat body of transgenic female mosquitoes and accumulated in the hemolymph at a high level (Fig. 3B). The low level of tDefA in the fat body compared to the high level of this protein in the hemolymph suggested a high rate of its secretion by the fat body (Fig. 3B, lanes H and FB). This is similar to the fat body production of several hemolymph proteins such as yolk protein precursors and lipophorin, which have been shown to be rapidly secreted and not stored in this tissue (8, 28). Interestingly, although male mosquitoes contained low levels of the Vg-DefA mRNA, the tDefA protein in males was below any detectable level (Fig. 3B, lane M).
Figure 3.
Immunoblot analyses of tDefA peptide expression in transgenic mosquitoes after blood meal activation. (A) Protein extracts from individual 24 hPBM females of the host khw strain and the D1 transgenic line were tested by using polyclonal antibody to Ae. aegypti DefA and mAb to Ae. aegypti vitellogenin small subunit (27). Expression of the 4-kDa DefA peptide was observed in transgenic mosquitoes but not in the khw host strain. The expression of 66-kDa Vg-small subunit (Vg-S), used as a positive control for blood meal activation, was present at the same level in both the transgenic line and the host strain. (B) Immunoblot analysis of tissue- and sex-specific expression of tDefA peptide in transgenic mosquitoes. Protein extracts from hemolymph (H), fat bodies (FB), ovary (OV), carcass (C), 24 hPBM whole mosquito female, (F), and mosquito male (M) were analyzed. (C) Time course of tDefA protein accumulation in transgenic mosquitoes during the vitellogenic cycle after a single blood feeding. Previtellogenic female (PV), vitellogenic females 1 (1d), 3 (3d), and 22 (22d) days PBM and the hemolymph collected from 22-day-old female mosquitoes (H22d) were tested using DefA-specific antibodies. One mosquito-equivalent was loaded in each lane, except for the hemolymph sample from 22-day-old females, in which a four-mosquito equivalent was used.
The time course of the tDefA protein levels in the transgenic female mosquitoes showed that it was present at a low level in previtellogenic females and exhibited a dramatic increase in response to blood feeding, reaching its maximum at 24 hPBM (Fig. 3C). This is consistent with the kinetics of the Vg protein (8). A peculiar feature of the Vg gene is that in contrast to vitellogenic carboxypeptidase gene (VCP), another yolk protein gene activated by a blood meal, Vg is expressed at a very low level in the fat body of the previtellogenic female (29) and traces of Vg protein are detectable in the previtellogenic females (A.S.R., unpublished observation). However, in contrast to Vg, tDefA was still present at high amounts in female mosquitoes 3 days after a blood meal (Fig. 3C, lane 3d). Importantly, a sufficiently high concentration of tDefA accumulated in the hemolymph of postvitellogenic transgenic mosquitoes so that it was still present 22 days after a single blood meal (Fig. 3C). Older mosquitoes have not been evaluated. The effect of a second blood meal appeared to be additive in terms of the concentration of tDefA in the hemolymph of transgenic female mosquitoes (data not shown).
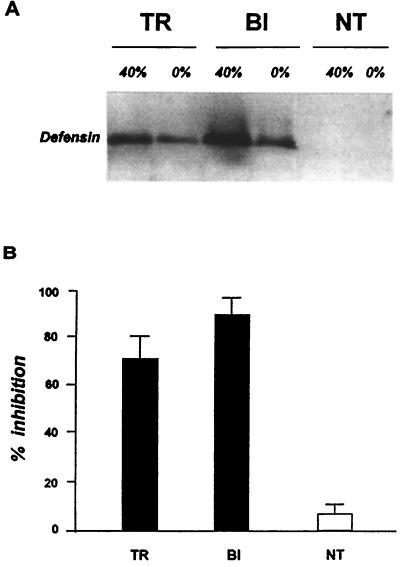
Next, we tested whether the tDefA protein produced by transgenic mosquitoes in response to a blood meal was biologically active. For these experiments, defensins were extracted and purified from transgenic 24 hPBM females, bacteria-challenged females of the nontransgenic khw strain as a positive control, and nontransgenic khw 24 hPBM female mosquitoes as a negative control (Fig. 4A). The antibacterial activity of tDefA and control defensin proteins was assayed by the liquid growth inhibition assay against Micrococcus luteus (26). The purified tDefA protein produced by transgenic mosquitoes had a high level of antibacterial activity comparable to that of defensin isolated from bacteria-challenged control mosquitoes (Fig. 4B). In contrast, the defensin fraction separated in the same manner from the hemolymph of blood-fed nontransgenic khw females showed only a background level of antibacterial activity. Thus, in the blood-activated rather than the infection-activated fat body of transgenic mosquitoes, the Vg-DefA mRNA was not only correctly translated and the prepro-defensin properly processed, but the mature tDefA acquired its biological activity with respect to its natural antimicrobial characteristics. We conclude that the mosquito fat body had all of the components of the protein processing machinery required to produce a biologically active defense protein without an immune challenge.
Figure 4.
Analysis of antibacterial activity of tDefA peptide isolated from transgenic mosquitoes. (A) Immunoblot analysis of peptide fractions used to assay the antibacterial activity. Protein samples obtained from transgenic 24 hPBM females of D1 line (TR), bacterially induced nontransgenic females of the khw host strain (BI), and nontransgenic 24 hPBM females of the khw host strain (NT) were isolated by using acid extraction and chromatography purification on Sep-Pak C18 cartridges and analyzed by immunoblotting. The analyzed samples were prepared from the 0% acetonitrile fraction in acidified water (0%) as a loading control, and the 40% acetonitrile fraction (40%), enriched for DefA peptide. (B) Aliquots from 40% fractions containing the major portion of defensin, isolated from transgenic (TR) and bacteria-induced (BI) females, were analyzed by the liquid growth inhibition assay using the Gram-positive bacteria, Micrococcus luteus. An equal aliquot of the 40% acetonitrile fraction isolated from nontransgenic 24 hPBM females of the host strain (NT) was used as a negative control. Data represent the mean of three independent experiments.
In summary, a 2.1-kb Vg upstream promoter fragment was sufficient for activation of a transgene by a blood meal-triggered regulatory cascade in a tissue-specific manner. It also was adequate to drive a high level of transgene expression. Active defensin peptide was produced by the transgene and persisted for as long as 22 days after a blood meal. This stability of the transgene product may make it an effective means of controlling pathogen infection throughout most of the adult life of a mosquito. The question of whether the transgenic Ae. aegypti, carrying the functional Vg-DefA transgene, is refractory to Plasmodium gallinaceum is important and is currently being tested. However, the significance of our present work is in demonstrating the possibility of engineering a transgenic mosquito in which turning on the gene(s) encoding antipathogen factor(s) is “wired” to the blood meal activation cascade. Thus, it constitutes a crucial step toward the use of molecular transgenesis in the development of pathogen-refractory vectors.
Acknowledgments
We thank Dr. M. H. Mulks for kindly providing us with M. luteus bacteria strain, M. J. Mienaltowski and Geoff Attardo for their critical reading of the manuscript, and Alan Hays for excellent technical assistance. This work was supported by grants from the National Institutes of Health, AI24716 and AI45123 (to A.S.R.) and AI29746 and AI44238 (to A.A.J.).
Abbreviations
- Vg
vitellogenin
- DefA
defensin A
- Vg-DefA
vitellogenin-defensin A
- hPBM
hours post blood meal
- tDefA
transgenic defensin A
- cn
cinnabar
- khw
kynurenine hydroxylase-white
- RT-PCR
reverse transcription–PCR
- G
generation
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.160258197.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.160258197
References
- 1.Butler D. Nature (London) 1997;386:535–536. doi: 10.1038/386535a0. [DOI] [PubMed] [Google Scholar]
- 2.Bruno J M, Feachem R, Godal T, Nchinda T, Ogilvie B, Mons B, Mshana R, Radda G, Samba E, Schwartz M, et al. Nature (London) 1997;386:541. doi: 10.1038/386541a0. [DOI] [PubMed] [Google Scholar]
- 3.Beier J C. Annu Rev Entomol. 1998;43:519–543. doi: 10.1146/annurev.ento.43.1.519. [DOI] [PubMed] [Google Scholar]
- 4.Collins F H, James A A. Sci Med. 1996;3:52–61. [Google Scholar]
- 5.O'Brochta D A, Atkinson P W. Insect Biochem Mol Biol. 1996;26:739–753. doi: 10.1016/s0965-1748(96)00022-7. [DOI] [PubMed] [Google Scholar]
- 6.Jasinskiene N, Coates C J, Benedict M Q, Cornel A J, Rafferty C S, James A A, Collins F H. Proc Natl Acad Sci USA. 1998;95:3743–3747. doi: 10.1073/pnas.95.7.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coates C J, Jasinskiene N, Miyashiro L, James A A. Proc Natl Acad Sci USA. 1998;95:3748–3751. doi: 10.1073/pnas.95.7.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raikhel A S. Adv Dis Vector Res. 1992;9:1–39. [Google Scholar]
- 9.Romans P, Tu Z, Ke Z, Hagedorn H H. Insect Biochem Mol Biol. 1995;25:936–958. doi: 10.1016/0965-1748(95)00037-v. [DOI] [PubMed] [Google Scholar]
- 10.Hoffmann J A, Kafatos F C, Janeway C A, Ezekowitz R A B. Science. 1999;284:1313–1318. doi: 10.1126/science.284.5418.1313. [DOI] [PubMed] [Google Scholar]
- 11.Lowenberger C, Bulet P, Charlet M, Hetru C, Hodgeman B, Christensen B M, Hoffmann J A. Insect Biochem Mol Biol. 1995;25:867–873. doi: 10.1016/0965-1748(95)00043-u. [DOI] [PubMed] [Google Scholar]
- 12.Cho W L, Fu Y C, Chen C C, Ho C M. Insect Biochem Mol Biol. 1996;26:395–402. doi: 10.1016/0965-1748(95)00108-5. [DOI] [PubMed] [Google Scholar]
- 13.Richman A M, Bulet P, Hetru C, Barillas-Mury C, Hoffmann J A, Kafatos F C. Insect Mol Biol. 1996;5:203–210. doi: 10.1111/j.1365-2583.1996.tb00055.x. [DOI] [PubMed] [Google Scholar]
- 14.Cho W-L, Fu T-F, Chiou J-Y, Chen C-C. Insect Biochem Mol Biol. 1997;27:351–358. doi: 10.1016/s0965-1748(97)00017-9. [DOI] [PubMed] [Google Scholar]
- 15.Lowenberger C, Smart C, Bulet P, Ferdig M, Severson D, Hoffmann J, Christensen B. Insect Mol Biol. 1999;8:107–118. doi: 10.1046/j.1365-2583.1999.810107.x. [DOI] [PubMed] [Google Scholar]
- 16.Shahabuddin M, Field I, Bulet P, Hoffmann J A, Miller L H. Exp Parasitol. 1998;89:103–112. doi: 10.1006/expr.1998.4212. [DOI] [PubMed] [Google Scholar]
- 17.Lowenberger C A, Kamal S, Chiles J, Paskewitz S, Bulet P, Hoffmann J A, Christensen B M. Exp Parasitol. 1999;91:59–69. doi: 10.1006/expr.1999.4350. [DOI] [PubMed] [Google Scholar]
- 18.Richman A M, Dimopoulos G, Seeley D, Kafatos F C. EMBO J. 1997;16:6114–6119. doi: 10.1093/emboj/16.20.6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarkar A, Yardley K, Atkinson P W, James A A, O'Brochta D. Insect Biochem Mol Biol. 1997;27:359–363. doi: 10.1016/s0965-1748(97)00018-0. [DOI] [PubMed] [Google Scholar]
- 20.Bhalla S. Mosquito News. 1968;28:380–385. [Google Scholar]
- 21.Cornel A J, Benedict M Q, Salazar-Rafferty C, Howells A J, Collins F H. Insect Biochem Mol Biol. 1997;27:993–997. doi: 10.1016/s0965-1748(97)00084-2. [DOI] [PubMed] [Google Scholar]
- 22.Hays A R, Raikhel A S. Roux's Arch Dev Biol. 1990;199:114–121. doi: 10.1007/BF02029559. [DOI] [PubMed] [Google Scholar]
- 23.Morris A C. In: Molecular Biology of Insect Disease Vectors. Crampton J M, Beard C B, Louis C, editors. London: Chapman & Hall; 1997. pp. 423–429. [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 25.Bender W, Spierer P, Hogness D. J Mol Biol. 1983;168:17–33. doi: 10.1016/s0022-2836(83)80320-9. [DOI] [PubMed] [Google Scholar]
- 26.Bulet P, Dimarq J L, Hetru C, Lagueux M, Charlet M, Hegy G, van Dorsolaer A, Hoffmann J A. J Biol Chem. 1993;268:14893–14897. [PubMed] [Google Scholar]
- 27.Raikhel A S, Pratt L H, Lea A O. J Insect Physiol. 1986;32:879–890. [Google Scholar]
- 28.Sun, J., Hiraoka, T., Dittmer, N. T., Cho, K.-H. & Raikhel, A. S. (2000) Insect Biochem. Mol. Biol., in press. [DOI] [PubMed]
- 29.Deitsch K W, Chen J-S, Raikhel A S. Insect Biochem Mol Biol. 1995;25:449–454. doi: 10.1016/0965-1748(94)00082-a. [DOI] [PubMed] [Google Scholar]