Abstract
Recombinant murine retroviruses are widely used as delivery vectors for gene therapy. However, once integrated into a chromosome, these vectors often suffer from profound position effects, with vector silencing observed in vitro and in vivo. To overcome this problem, we investigated whether the HS4 chromatin insulator from the chicken β-globin locus control region could protect a retrovirus vector from position effects. When used to flank a reporter vector, this element significantly increased the fraction of transduced cells that expressed the provirus in cultures and in mice transplanted with transduced marrow. These results demonstrate that a chromatin insulator can improve the expression performance of a widely used class of gene therapy vectors by protecting these vectors from chromosomal position effects.
Most gene therapy strategies involving hematopoietic stem cells require both a high level of gene transfer and persistent transgene expression in specific target lineages. Recent advances in nonhuman primate models demonstrate that gene transfer rates of approximately 10% in reconstituting hematopoietic stem cells can be routinely achieved with virus vectors based on murine leukemia virus and related oncoretroviruses (1–4). However, achieving persistent, uniform gene expression from murine leukemia virus-based vectors has been problematic. Much research has focused on defining the elements of the virus long terminal repeat (LTR) that are responsible for provirus silencing in vivo (5–7), and identifying the most appropriate promoters and enhancers(8, 9). Expression of integrated provirus is also affected by chromatin structure. Because the bulk of the mammalian genome is packaged into transcriptionally silent heterochromatin (10), and murine leukemia virus-based vectors insert at random sites in the genome, a large portion of murine leukemia virus insertions result in gene silencing. This can lead to highly variable expression among clones, with complete silencing of provirus expression in a significant fraction of clones either immediately after insertion or following cell expansion. The progeny of a single clone containing a unique integration event can also be affected by the surrounding chromatin to varying degrees (10), a phenomenon known as position effect variegation. Position-dependent silencing and position effect variegation are particularly troublesome for retrovirus vectors containing the human β- or γ-globin genes (8, 9, 11, 12).
The mammalian genome is organized into discrete chromosomal domains, in part through the use of sequences termed chromatin insulators (13). These elements, first described in Drosophila and more recently in several vertebrate species, help define the boundary between differentially regulated loci and serve to shield promoters from the influence of neighboring regulatory elements (14, 15). Insulators function in a polar manner (e.g., they must be located between the cis effectors and promoter) and do not have stimulatory or inhibitory transcriptional effects on their own, distinguishing them from classical enhancers and silencers. The first and best characterized vertebrate chromatin insulator is located within the chicken β-globin locus control region. This element, which contains a DNase-I hypersensitive site (cHS4), appears to constitute the 5′ boundary of the chicken β-globin locus (15). A 1.2-kb fragment containing the cHS4 element displays classic insulator activities, including the ability to block the interaction of globin gene promoters and enhancers in cell lines (16), and the ability to protect expression cassettes in Drosophila (16), transformed cell lines (17), and transgenic mammals (18, 19) from position effects. Much of this activity is contained in a 250-bp fragment. Within this stretch is a 49-bp cHS4 core (20) that interacts with the zinc finger DNA binding protein CTCF implicated in enhancer-blocking assays (21).
We investigated the ability of the cHS4 chromatin insulator to block position effects on expression of oncoretrovirus vectors. For this purpose, we flanked a reporter vector with the cHS4 element and analyzed vector expression in cell lines, primary marrow progenitor cultures, and murine bone marrow transplantation assays. We found that the insulator had no effect on vector titer and stability and, in one orientation, was able to protect both reporter genes from negative position effects to varying degrees. These results demonstrate the ability of this element to insulate an oncoretrovirus vector in vitro and in vivo. Thus, such elements may prove valuable for achieving efficient, persistent expression from such vectors in clinical gene therapy applications.
Materials and Methods
Retrovirus Vectors.
The retrovirus vector constructs (diagrammed in Fig. 1) were generated by using the murine stem cell virus-based vector MGPN2 (22), which expresses the enhanced green fluorescence protein (GFP) gene from the viral 5′ LTR promoter (LTR→GFP cassette) and the neomycin phosphotransferase (Neo) gene from a phosphoglycerate kinase (Pgk) promoter (Pgk→Neo cassette). The cHS4 element was isolated as a 1,203-bp XbaI fragment from the plasmid pJC5-4 (16) and was inserted in either the 5′-3′ or 3′-5′ orientation in the NheI site of the 3′ LTR to create vectors INS4(+) and INS4(−), respectively. From this location, the insulator fragment is copied into the 5′ LTR during provirus integration to generate a flanking “double-copy” configuration (23). This configuration was confirmed by Southern blot analysis of pools of transduced cells using HindIII, which cuts once in the insulator fragment (data not shown). Retrovirus vector producer lines were generated essentially as described (24) using the amphotropic packaging line PA317 (25) and the ecotropic packaging line GP + E86 (26). Virus titers were determined by serial dilution and transfer of G418 resistance to naive NIH 3T3 cells as described (27). Clones with the highest titers were further analyzed by Southern blot analysis for intact provirus (methods described below), and for the presence of replication-competent virus by a standard marker-rescue assay (24). Vector-containing supernatant was collected from semiconfluent plates after 48 h of culture at 33°C and was passed through a 0.44-μm filter. All cultures were otherwise maintained at 37°C in DMEM supplemented with 10% heat-inactivated characterized FBS, 2 mM l-glutamine, 1 mM sodium pyruvate, 0.1 mM nonessential amino acids, and antibiotics.
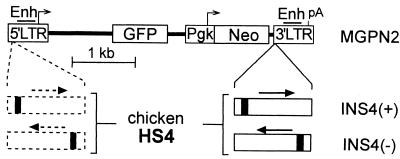
Figure 1.
Reporter vector constructs. Control reporter vector MGPN2 (22), based on the oncoretrovirus vector murine stem cell virus (6), has two reporter cassettes: GFP transcribed from the virus 5′ LTR promoter, and Neo transcribed from a Pgk promoter. A 1.2-kb fragment containing the cHS4 element (16) was inserted in either orientation into the 3′ LTR to generate vectors INS4(+) and INS4(−). In this location, the cHS4 fragment is copied into the 5′LTR to generate a flanking double-copy (23), as indicated by the dashed lines. The core DNase-I hypersensitive site for the cHS4 fragment is indicated as a solid block. Enh, enhancer; pA, polyadenylation site.
Retrovirus Vector Transductions.
NIH 3T3 cells were transduced by 24 h of culture in supernatant from amphotropic producer clones containing 8 μg/ml polybrene at a multiplicity of infection of less than 1 (to assure single-copy integration). Distinct clones were isolated under G418 selection and were expanded after 7–10 days. Mouse bone marrow progenitors were transduced as described (28). Marrow was harvested from the femora of 6- to 12-week-old B6 × D2 F1 female donors treated 2 days previously with 5-fluorouracil (150 mg/kg i.p.). Cells were preinduced at 1 × 106 cells/ml in Iscove's modified Dulbecco's media containing 10% defined FBS, l-glutamine, sodium pyruvate, non-essential amino acids, antibiotics, 5% IL-3 culture supplement (Collaborative Biomedical Products, Bedford, MA), 100 ng/ml recombinant human IL-6 (Sandoz Pharmaceutical), and 50 units/ml recombinant mouse stem cell factor (PeproTech, Rocky Hill, NJ). After 48 h of culture at 37°C in 5% CO2, the marrow cells were transferred to irradiated (15 Gy), subconfluent GP + E86 producer cells at a density of 5–10 × 106 cells per 10-cm plate in 10 ml of the above media further supplemented with 8 μg/ml polybrene. After an additional 48 h of culture, the non-adherent bone marrow cells were carefully collected on ice, were washed in cold Hanks' buffered saline solution, and either were used directly in colony assays (described below) or were transplanted at a dose of 5–10 × 105 cells per recipient into irradiated (1,050 cGy) syngeneic recipients.
Progenitor Colony Assays.
Based on an established protocol (29), marrow cells were suspended at ≈2.5 × 104 cells/ml in plating medium consisting of Iscove's modified Dulbecco's media, 30% defined FBS, 1% wt/vol BSA, l-glutamine, 10−4 M β-mercaptoethanol, antibiotics, and 0.9% methylcellulose. Myeloid progenitors (colony-forming units, granulocyte/macrophage) were induced by addition of 5% IL-3 and were scored after 7–10 days of incubation at 37°C, 5% CO2. Selection was carried out with 0.8 mg/ml active G418. Untransduced marrow was routinely included as a control to assure that G418 selection was complete. Marrow cells were used directly after bone marrow transduction or were collected from transplanted mice either by needle aspiration under anesthesia or at time of death.
Southern Blot Analysis.
Genomic DNA was isolated by standard methods (30) and was quantified by spectrophotometry. Approximately 10–15 μg was digested with KpnI, which cuts once in each of the virus LTRs, separated on 0.8% agarose gels, and blotted onto nylon filters. The blots were probed with a radiolabeled 923-bp PstI fragment for Neo, which detects a 3,566-bp provirus band for vector MGPN2 and a 4,769-bp provirus band for vectors INS4(+) and INS4(−). To control for loading, the blots were stripped and reprobed with a radiolabeled 583-bp EcoRI-HindIII fragment (coordinates 18,300–18,883, GenBank accession no. MMBGCXD) from a noncoding region of the mouse β-globin loci, which is specific for a 3,941-bp KpnI fragment. Signal intensities were quantified by PhosphorImager (Molecular Dynamics).
Flow Cytometry Analysis.
For analysis of progenitor colonies, cells were collected directly into Hank's balanced salt solution supplemented with 2% FBS and washed to remove undissolved methylcellulose. White blood cells were depleted of red blood cells by hypotonic shock. Splenocytes were prepared by mechanical disruption of the spleen and passage through a nylon filter. Bone marrow was prepared by aspiration of isolated femurs. Pelleted cells were resuspended in Hank's balanced salt solution/FBS and were kept on ice in the dark before analysis by flow cytometry on a FACScan (Beckton Dickinson) using cellquest software.
Results
Construction and Initial Characterization of Reporter Vectors.
For these studies we chose a reporter vector (MGPN2) (22) with two distinct expression cassettes: a GFP gene transcribed from the virus LTR promoter (LTR→GFP) and a Neo gene transcribed from a Pgk promoter (Pgk→Neo). As seen in Fig. 1, we inserted a 1.2-kb fragment containing the cHS4 element (16) into the 3′LTR of this vector in both orientations to generate vectors INS4(+) and INS4(−). This double-copy configuration (23) allows the cHS4 fragment to be copied into the 5′LTR during provirus integration, effectively flanking the reporter vector (a presumed prerequisite for proper function) (13). As reported in Table 1, producer clones were readily generated for each insulated vector with titers similar to that achieved with the control vector (2 × 106 vs. 3 × 106 colony-forming units/ml), demonstrating that the insulator fragments had no adverse effects on virus production. Southern blot analysis of transduced cell pools and clones further demonstrated that the insulator fragment had no effect on vector stability (data not shown; Fig. 3).
Table 1.
Reporter vector titer and expression in NIH 3T3 clones
Titer reported for best of 12 clones expressed as G418r colony-forming units/ml on naive NIH 3T3 calls.
Mean relative fluorescence determined by flow cytometric analysis of 9–12 independent transduced and G418-selected NIH 3T3 cell clones.
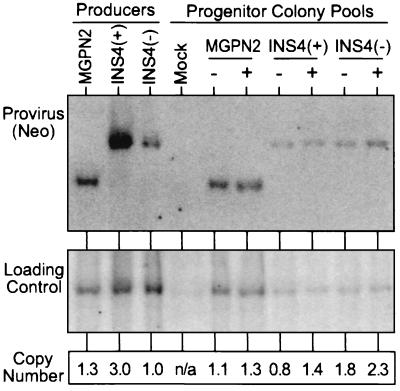
Figure 3.
Quantification of gene transfer rates by Southern blot analysis. DNA was isolated from the pools of progenitor colonies grown in the absence (−) and presence (+) of G418 selection used in the analysis presented in Fig. 2a, was digested with KpnI that cuts once in each LTR, and was analyzed by Southern blotting. The blot was first hybridized with a probe for Neo (Upper), was stripped, and was rehybridized with a loading control probe (mouse β-globin). The band intensities were quantified with a PhosphorImager and were used to calculate the relative number of provirus copies per genome in each sample (Lower). Controls included DNA from a pool of untransduced progenitor colonies (Mock), and DNA from producer clones for the three vectors.
Expression of the LTR→GFP cassette was first analyzed in transduced NIH 3T3 cell clones by flow cytometry. As reported in Table 1, the average mean relative fluorescence for the clones transduced with vector INS4(+) (90 ± 29 relative fluorescent units) was more than twice that observed for the clones transduced with the uninsulated vector MGPN2 (32 ± 9 units) or the vector insulated in the opposite orientation, vector INS4(−) (40 ± 18 units).
Insulation in Primary Myelopoietic Progenitor Colonies.
In light of the encouraging results in NIH 3T3 cells, we next turned to a mouse bone marrow transduction and myelopoietic progenitor colony assay in which the problem of vector silencing is more pronounced. In this case, expression of the Pgk→Neo cassette was analyzed by measuring the frequency of colony formation under G418 selection, and expression of the LTR→GFP cassette was analyzed by flow cytometry of colony pools. As seen for one exemplary experiment presented in Fig. 2a, the samples transduced with the insulated vector INS4(+) consistently generated more G418r colonies (76%) compared with the samples transduced with the uninsulated control vector MGPN2 (55%) or vector INS4(−) (48%). Likewise, the sample transduced with vector INS4(+) generated a higher frequency of GFP+ colony cells in the absence of G418 selection (80%) compared with the samples transduced with vector MGPN2 (34%) or vector INS4(−) (46%). Quantitative Southern blot analysis presented in Fig. 3 demonstrated that these differences in expression were not simply attributable to differences in transduction rates because there was only an estimated 0.8 provirus copies per genome in the unselected sample transduced with vector INS4(+), compared with 1.1 copies per genome for the unselected sample transduced with the control vector MGPN2 and the 1.8 copies per genome for the sample transduced with vector INS4(−).
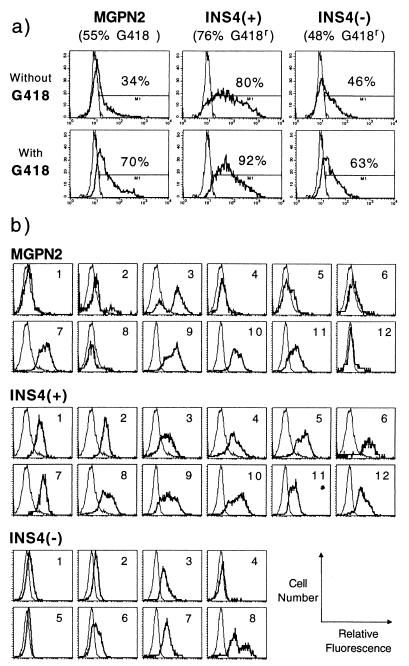
Figure 2.
Expression analysis of transduced mouse bone marrow progenitors. (a) Mouse bone marrow was transduced with the indicated vectors and was plated for myeloid progenitor colony formation in the presence and absence of the neomycin drug analog G418. The percentage of drug-resistant colonies (reported under the vector name) was calculated as follows: (number of colonies grown with G418)/(number of colonies grown without G418) × 100. Pools of colonies were subsequently analyzed for expression of GFP by flow cytometry. The percentage of GFP+ cells is reported above the indicated gates. Data are from one exemplary experiment; see Table 2 for summary of multiple experiments. (b) Flow cytometric analysis of GFP expression in individual colonies grown under G418 selection; each histogram is for a single colony. thick line; experimental samples; thin line, untransduced control. y axis, cell number; x axis, log relative fluorescence.
As summarized in Table 2, the insulated vector INS4(+) consistently expressed both the Pgk→Neo and the LTR→GFP cassettes in a statistically higher fraction of progenitor colonies (77 ± 9% and 46 ± 24%, respectively) than the uninsulated vector MGPN2 (53 ± 9% and 18 ± 14%, respectively) or the vector INS4(−) (51 ± 11% and 21 ± 21%, respectively) through repeated experiments. There was only a moderate increase in the fraction of cells expressing GFP when the colonies transduced with the insulated vector INS4(+) were first selected under G418 (from 46 ± 24% to 55 ± 26%), whereas preselection greatly increased the fraction of GFP+ cells for samples transduced with uninsulated vector MGPN2 (from 18 ± 14% to 47 ± 17%) and vector INS4(−) (from 21 ± 21% to 30 ± 28%). The fraction of cells expressing GFP in the G418-selected samples did not differ significantly. However, close inspection of the flow cytometry histograms in Fig. 2a indicates that, even with G418 selection, the majority of the cells transduced with vectors MGPN2 and INS4(−) expressed only low levels of GFP, compared with the cells transduced with the insulated vector INS4(+), which were frequently 10-fold brighter than the control cells.
Table 2.
Reporter vector expression in progenitor colonies
| Vector | Replicates | Neo expression
|
GFP expression
|
||||
|---|---|---|---|---|---|---|---|
| Unselected
|
G418-selected
|
||||||
| G418r* | P† | % GFP+‡ | P | % GFP+ | P | ||
| MGPN2 | 4 | 53 ± 9 | 18 ± 14 | 47 ± 17 | |||
| INS4(+) | 4 | 77 ± 9 | 0.03 | 46 ± 24 | 0.02 | 55 ± 26 | 0.25 |
| INS4(−) | 3 | 51 ± 11 | 0.86 | 21 ± 21 | 0.16 | 30 ± 28 | 0.08 |
Percent G418r colonies = [colony count with G418]/[colony count without G418] × 100.
Probability compared to MGPN2 control using t test.
Percent GFP+ cells determined by flow cytometric analysis of colony pools.
To examine expression of the LTR→GFP cassette on a clonal level, individual progenitor colonies grown under G418 selection were isolated and analyzed by flow cytometry. As seen in Fig. 2b, several colonies transduced with the uninsulated vector MGPN2 either failed to express significant levels of GFP (e.g., colonies 1, 4, 5, 6, 12), or exhibited a clearly variegated phenotype (e.g., colonies 2, 3, 8). Similar patterns were also observed for the colonies transduced with vector INS4(−). In contrast, all 12 colonies transduced with vector INS4(+) expressed relatively uniform, easily detectable levels of GFP.
Insulation in Transplanted Mice.
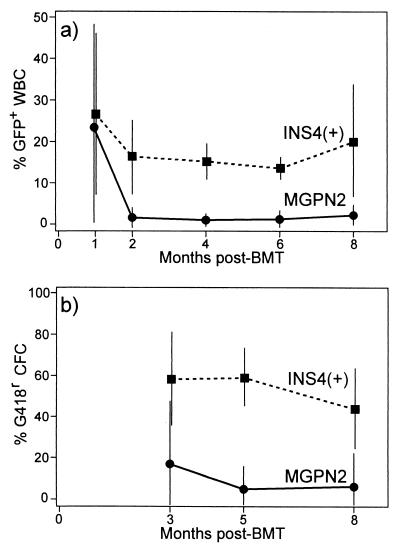
As the most critical test of insulator function, we compared expression of the uninsulated vector MGPN2 and the insulated vector INS4(+) in a long-term mouse bone marrow transduction and transplantation model. Expression of the LTR→GFP cassette was assessed by flow cytometric analysis of peripheral nucleated white blood cells (WBC). As seen in Fig. 4a, only a small fraction of cells expressed GFP long-term after transplantation of marrow transduced with the parental vector MGPN2, averaging only 2 ± 3% at 8 months (Table 3). In contrast, approximately 15–20% of WBC expressed GFP long-term after transplantation with marrow transduced with the insulated vector INS4(+), averaging 19 ± 15% at 8 months. As summarized in Table 3, the fraction of GFP+ cells were similar in peripheral blood, spleen, and bone marrow.
Figure 4.
Vector expression in mice transplanted with transduced marrow. Marrow was transduced with either the uninsulated vector MGPN2 or the insulated vector INS4(+) and was used to transplant syngeneic recipients following 1025 cGy whole body irradiation. (a) At the indicated months posttransplant, blood samples were collected and the percentage of white blood cells (WBC) expressing GFP was determined by flow cytometry as described in Fig. 2a. (b) Likewise, marrow aspirates were collected at the indicated months posttransplant by survival surgery and the percentage of progenitors [colony-forming cells (CFC)] expressing Neo was determined by colony formation under G418 selection as described in Fig. 2a. Data for vector MGPN2 include eight mice from three separate experiments, and data for vector INS4(+) include nine mice from three separate experiments. Only those mice with detectable provirus at 8 months posttransplant were included in the analysis.
Table 3.
Reporter vector expression in long-term reconstituted mice
| Vector, number | Copy number* | Hematopoietic tissues, % GFP+†
|
Marrow progenitor colonies
|
||||
|---|---|---|---|---|---|---|---|
| WBC | Spleen | Marrow | % G418r‡ | % GFP+ | Variegation§ | ||
| MGPN2, n = 8 | 0.4 ± 0.2 | 2 ± 3 | 1 ± 1 | 1 ± 1 | 6 ± 16 | 3 ± 2 | Not determined |
| INS4 (+), n = 9 | 1.0 ± 0.3 | 19 ± 15 | 12 ± 7 | 16 ± 11 | 45 ± 20 | 18 ± 9 | 9 of 23 |
Provirus copy number estimated by Southern blot analysis of at time of death.
Percent GFP+ cells determined by flow cytometry.
Percent G418r colonies = (colony count + G418)/[colony count − G418] × 100.
Defined by lack of GFP expression in most or all of cells in individual colonies analyzed after G418 selection.
The differences in the frequency of expression was even more pronounced for the Pgk→Neo cassette. In this case, marrow aspirates were obtained by serial aspirations posttransplant and were analyzed for myeloid progenitor colony formation under G418 selection. As seen in Fig. 4b, only a small fraction of the progenitors were able to form colonies under G418 selection after transplantation with marrow transduction with the uninsulated vector MGPN2, with an average of only 6 ± 16% at 8 months (Table 3). In contrast, typically more than half of the progenitors from the mice transplanted with marrow transduced with the insulated vector INS4(+) were able to form colonies under G418 selection, with an average of 45 ± 20% at the time of death. At 8 months posttransplant, the recipients were killed and the level of provirus was determined by Southern blot analysis of splenocytes as in Fig. 3. As summarized in Table 3, there was an average of 0.4 ± 0.2 vector copies per genome for the mice transduced with MGPN2, and 1.0 ± 0.3 vector copies per genome in the mice transduced with vector INS4(+). These rates of gene transfer are consistent with PCR analysis of progenitor colonies grown from these mice in the absence of G418 selection (data not shown). Thus, the differences in the rates of vector expression were not attributable solely to differences in transduction rates but, rather, represent a difference in the rate with which the reporter cassettes were expressed.
GFP expression was also analyzed in the myeloid progenitor colonies derived from the marrow collected at the time of death. As summarized in Table 3, the frequency of GFP expression in colonies grown in the absence of selection was strikingly similar to that observed in WBC [3 ± 2% vs. 2 ± 3% for vector MGPN2 and 18 ± 19% vs. 19 ± 15% for vector Ins4(+)]. However, in the case of vector INS4(+), the LTR→GFP cassette was only expressed about 40% of the time compared with the Pgk→Neo cassette [18 ± 9% vs. 45 ± 20% respectively (Table 3)]. In contrast to the studies with primary transduced progenitors (Fig. 2b), flow cytometric analysis of individual, G418-selected colonies derived from the long-term reconstituted mice also revealed a variegated phenotype in 9 of 23 clones containing vector INS4(+).
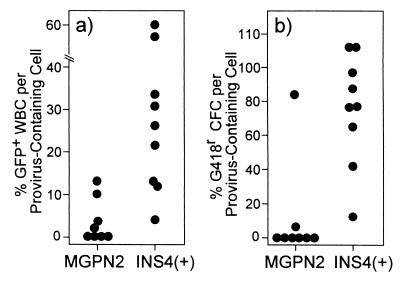
As seen in Fig. 5, the differences in expression between the uninsulated and insulated vectors were particularly apparent when the fraction of vector-expressing cells at 8 months posttransplant was normalized to the fraction of cells containing provirus. In the mice transplanted with marrow transduced with the uninsulated vector MGPN2, only 4 ± 5% of the provirus-containing WBC expressed GFP and only 11 ± 30% of the provirus-containing marrow progenitors expressed Neo. This is in striking contrast to the mice transplanted with marrow transduced with the insulated vector INS4(+). In this case, 29 ± 21% of the provirus-containing WBC expressed GFP whereas 73 ± 34% of the provirus-containing marrow progenitors expressed Neo. These levels of expression were maintained even after secondary transplantation (data not shown).
Figure 5.
Normalized vector expression in long-term reconstituted mice. Southern blot analysis similar to that shown in Fig. 3 was performed on spleens collected at 8 months posttransplant, and the copies of provirus per genome were determined (for average copy numbers, see Table 3). The fraction of hematopoietic cells containing at least one copy per cell was then calculated by using the Poisson distribution (assuming the provirus was distributed randomly), and was used to normalize the expression data at the 8-month time point from Fig. 4. (a) Scatter plot showing the fraction of WBC expressing GFP divided by the calculated fraction of cells containing provirus for individual mice. (b) Scatter plot showing the fraction of progenitors [colony-forming cells (CFC)] that were able to form colonies under G418 selection divided by the calculated fraction of cells containing provirus for individual mice.
Discussion
For gene therapy to be successful, the transfer vector must reach the target tissue and be expressed at therapeutic levels. For vectors that integrate into the host genome, the effects of surrounding chromatin may severely limit vector expression. Several studies suggest that the cHS4 element exhibits properties that could be useful for protecting therapeutic vectors from such negative position effects (16–21), including a relatively small size and ability to function in a wide range of species. Our initial studies of vector expression in NIH 3T3 cells demonstrated that the cHS4 element did not affect vector titer and stability, which is a critical prerequisite for its use with retrovirus vectors. We found that, in transduced mouse myeloid progenitors (Fig. 2; Table 2), the 1.2-kb cHS4 fragment in the (+) orientation increased the fraction of myeloid progenitors expressing Neo by nearly 50%. The fraction of colony cells grown in the absence of G418 selection also showed a 2.5-fold increase in GFP expression relative to the uninsulated vector control. The simplest interpretation of these data is that the flanking insulator reduced the incidence of clonal silencing for both the LTR→GFP and Pgk→Neo cassettes. Flow cytometric analysis of individual transduced progenitor colonies revealed that the flanking insulator also reduced the incidence of position-effect variegation at the clonal level. These results are similar to those recently reported by Rivella et al., in which flanking a retrovirus reporter vector with the cHS4 fragment increased the probability that integrated provirus was expressed (31). They also found that this increased incidence of expression was associated with a dramatic decrease in the level of de novo methylation of the 5′ long terminal repeat, providing a possible mechanism for the insulating activity in such vectors.
The most striking evidence that the insulating activity of the cHS4 fragment functions in retrovirus vectors came from the in vivo studies. In the case of the uninsulated vector MGPN2, there was a precipitous drop in the fraction of WBC expressing the LTR→GFP during the first 2 months posttransplant, and a very low fraction for marrow progenitors expressing the Pgk→Neo at the earliest time point tested (Figs. 4 and 5; Table 3). These expression patterns are reminiscent of those reported in studies investigating the silencing of conventional Molony-based retrovirus vectors (5), as well as other studies in mice and large mammals (1–4, 32, 33). Although higher levels of expression have been reported with murine stem cell virus-based vectors similar to those used here, these studies frequently involved the use of in vivo selection or vectors with more robust GFP expression cassettes (22, 34, 35). In the studies reported here, we purposely sought to use a reporter vector with low-level expression to readily detect position effects. It is possible that this attribute may actually render this vector more highly sensitive to position effects than those used in the above-cited studies. As seen in Fig. 4 and Table 3, flanking this vector greatly improved the fraction of WBC expressing the LTR→GFP cassette and the fraction of progenitors expressing the Pgk→Neo cassette at all later time points. Further, flanking with this element prevented the precipitous drop in LTR→GFP expression in the first 2 months posttransplant.
It is not clear why the Pgk→Neo cassette appeared to be better insulated than the LTR→GFP cassette. It is possible that the assay for Neo (G418 resistance) is more sensitive than the assay for GFP (flow cytometry), although both assays are thought to be highly sensitive. Alternatively, such disparate results may reflect inherent differences in the Pgk and LTR promoters. It is also not clear why the cHS4 fragment only appears to function in one orientation. This may simply reflect an orientation dependency of the HS4 insulating activity, as recently described using a repressor blocking assay system (36). However, as indicated in Fig. 1, the core DNase-I hypersensitive site is not positioned in the middle of this fragment, but instead is located at the far 5′ end. Thus, it is possible that the differences observed in the insulation of vectors INS4(+) and INS4(−) may reflect the relative distance of the cHS4 core to the reporter vector promoters. Current models suggest that insulators may function in part through the formation of chromatin loop structures mediated by sequence-specific binding proteins (13). Given the presumed geometric and steric constraints under such models, it should not be surprising that promoter function would diminish near such insulator elements.
Taken together, these results demonstrate that the cHS4 insulator can be used to improve the expression performance of retrovirus vectors in vitro and in vivo. Because the element used here was derived from chicken, it may be important to identify and functionally characterize novel insulator elements from humans and other higher order mammals to fully capitalize on such elements for clinical gene therapy.
Acknowledgments
We thank G. Felsenfeld and A. Bell for providing the cHS4 chromatin insulator fragment and for helpful discussions, L. Cheng for the MGPN2 vector, and S. Fiering for the control mouse β-globin control probe. We also thank M. Knibbe for help with vector construction, L. Jin and C. A. Blau for help with the animal studies, and B. Wakimoto for helpful review of the manuscript. This work was supported by Grant HL 53750 from the National Institutes of Health.
Abbreviations
- LTR
long terminal repeat
- Neo
neomycin phosphotransferase
- Pgk
phosphoglycerate kinase
- GFP
green fluorescent protein
- cHS4
chicken hypersensitive site 4
- WBC
white blood cell
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.160159597.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.160159597
References
- 1.Dunbar C E, Seidel N E, Doren S, Sellers S, Cline A P, Metzger M E, Agricola B A, Donahue R E, Bodine D M. Proc Natl Acad Sci USA. 1996;93:11871–11876. doi: 10.1073/pnas.93.21.11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kiem H P, Heyward S, Winkler A, Potter J, Allen J M, Miller A D, Andrews R G. Blood. 1997;90:4638–4645. [PubMed] [Google Scholar]
- 3.Bodine D M, Dunbar C E, Girard L J, Seidel N E, Cline A P, Donahue R E, Orlic D. Ann NY Acad Sci. 1998;850:139–150. doi: 10.1111/j.1749-6632.1998.tb10471.x. [DOI] [PubMed] [Google Scholar]
- 4.Kiem H P, Andrews R G, Morris J, Peterson L, Heyward S, Allen J M, Rasko J E, Potter J, Miller A D. Blood. 1998;92:1878–1886. [PubMed] [Google Scholar]
- 5.Challita P M, Kohn D B. Proc Natl Acad Sci USA. 1994;91:2567–2571. doi: 10.1073/pnas.91.7.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hawley R G, Lieu F H, Fong A Z, Hawley T S. Gene Ther. 1994;1:136–138. [PubMed] [Google Scholar]
- 7.Halene S, Wang L, Cooper R M, Bockstoce D C, Robbins P B, Kohn D B. Blood. 1999;94:3349–3357. [PMC free article] [PubMed] [Google Scholar]
- 8.Rivella S, Sadelain M. Semin Hematol. 1998;35:112–125. [PubMed] [Google Scholar]
- 9.Emery D W, Stamatoyannopoulos G. Ann NY Acad Sci. 1999;872:94–107. doi: 10.1111/j.1749-6632.1999.tb08456.x. [DOI] [PubMed] [Google Scholar]
- 10.Karpen G H. Curr Opin Genet Dev. 1994;4:281–291. doi: 10.1016/s0959-437x(05)80055-3. [DOI] [PubMed] [Google Scholar]
- 11.Raftopoulos H, Ward M, Leboulch P, Bank A. Blood. 1997;90:3414–3422. [PubMed] [Google Scholar]
- 12.Emery D W, Morrish F, Li Q, Stamatoyannopoulos G. Hum Gene Ther. 1999;10:877–888. doi: 10.1089/10430349950018283. [DOI] [PubMed] [Google Scholar]
- 13.Bell A C, Felsenfeld G. Curr Opin Genet Dev. 1999;9:191–198. doi: 10.1016/S0959-437X(99)80029-X. [DOI] [PubMed] [Google Scholar]
- 14.Udvardy A, Maine E, Schedl P. J Mol Biol. 1985;185:341–358. doi: 10.1016/0022-2836(85)90408-5. [DOI] [PubMed] [Google Scholar]
- 15.Prioleau M N, Nony P, Simpson M, Felsenfeld G. EMBO J. 1999;18:4035–4048. doi: 10.1093/emboj/18.14.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chung J H, Whiteley M, Felsenfeld G. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- 17.Pikaart M J, Recillas-Targa F, Felsenfeld G. Genes Dev. 1998;12:2852–2862. doi: 10.1101/gad.12.18.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, DeMayo F J, Tsai S Y, O'Malley B W. Nat Biotechnol. 1997;15:239–243. doi: 10.1038/nbt0397-239. [DOI] [PubMed] [Google Scholar]
- 19.Taboit-Dameron F, Malassagne B, Viglietta C, Puissant C, Leroux-Coyau M, Chereau C, Attal J, Weill B, Houdebine L M. Transgenic Res. 1999;8:223–235. doi: 10.1023/a:1008919925303. [DOI] [PubMed] [Google Scholar]
- 20.Chung J H, Bell A C, Felsenfeld G. Proc Natl Acad Sci USA. 1997;94:575–580. doi: 10.1073/pnas.94.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bell A C, West A G, Felsenfeld G. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 22.Cheng L, Du C, Murray D, Tong X, Zhang Y A, Chen B P, Hawley R G. Gene Ther. 1997;4:1013–1022. doi: 10.1038/sj.gt.3300507. [DOI] [PubMed] [Google Scholar]
- 23.Hantzopoulos P A, Sullenger B A, Ungers G, Gilboa E. Proc Natl Acad Sci USA. 1989;86:3519–3523. doi: 10.1073/pnas.86.10.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller A D, Rosman G J. BioTechniques. 1989;7:980–890. [PMC free article] [PubMed] [Google Scholar]
- 25.Miller A D, Buttimore C. Mol Cell Biol. 1986;6:2895–2902. doi: 10.1128/mcb.6.8.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Markowitz D, Goff S, Bank A. J Virol. 1988;62:1120–1124. doi: 10.1128/jvi.62.4.1120-1124.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bodine D M, McDonagh K T, Bramdt S J, Ney P A, Agricola B, Byrne E, Nienhuis A W. Proc Natl Acad Sci USA. 1990;87:3738–3742. doi: 10.1073/pnas.87.10.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bodine D M, Karlsson S, Nienhuis A W. Proc Natl Acad Sci USA. 1989;86:8897–8901. doi: 10.1073/pnas.86.22.8897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eaves C J, Eaves A C. Blood. 1978;52:1196–1210. [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 31.Rivella S, Callegari J A, May C, Tan C W, Sadelain M. J Virol. 2000;74:4679–4687. doi: 10.1128/jvi.74.10.4679-4687.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bagley J, Aboody-Guterman K, Breakefield X, Iacomini J. Transplantation. 1998;65:1233–1240. doi: 10.1097/00007890-199805150-00015. [DOI] [PubMed] [Google Scholar]
- 33.Emery D W, Sablinski T, Shimada H, Germana S, Gianello P, Foley A, Shulman S, Arn S, Fishman J, Lorf T, et al. Transplantation. 1997;64:1414–1423. doi: 10.1097/00007890-199711270-00007. [DOI] [PubMed] [Google Scholar]
- 34.Allay J A, Persons D P, Galipeau J, Riberdy J M, Ashmun R A, Blakley R L, Sorrentino B P. Nat Med. 1998;4:1136–1143. doi: 10.1038/2632. [DOI] [PubMed] [Google Scholar]
- 35.Persons D A, Allay J A, Allay E R, Ashmun R A, Orlic D, Jane S M, Cunningham J M, Nienhuis A W. Blood. 1999;93:488–499. [PubMed] [Google Scholar]
- 36.van der Vlag J, den Blaauwen J L, Sewalt R G, van Driel R, Otte A P. J Biol Chem. 2000;275:697–704. doi: 10.1074/jbc.275.1.697. [DOI] [PubMed] [Google Scholar]